Introduction
Ossification of the posterior longitudinal ligament
(OPLL) involves pathological heterotopic ossification of the
paravertebral ligament. Chronic compression of the spinal cord and
nerve root leads to spinal cord compression symptoms and
radiculopathy in patients with OPLL. The severity of the symptoms
is associated with the size and segmentation of the ossifying
ligament, and 70% of OPLL occurs in the cervical spine, whereas 15%
occurs in the thoracic spine (1).
Notably, as early as 1972, researchers reported that thoracic OPLL
(T-OPLL) can cause thoracic spinal stenosis (2). Additionally, OPLL is primarily found
in individuals in northeast Asia; the prevalence of cervical OPLL
in Japanese individuals is 1.9–4.3% (3), which is much higher than the
prevalence in Caucasians (0.01–1.7%) (4). Moreover, the prevalence is higher in
men than in women (2.7:1), and the mean age of onset is more than
40 years (5). A recent study
published the prevalence of T-OPLL in Japanese individuals is 1.6%
(6).
Treatment approaches for T-OPLL are limited, and
surgery is currently the only effective treatment. However, owing
to the unique features of blood supply and anatomical structures
associated with this disease, surgery for T-OPLL is complicated,
and the risk is extremely high. It is also difficult to avoid the
occurrence of postoperative paralysis and surgical complications.
Therefore, many studies have focused on the elucidation of the
pathogenesis of T-OPLL, which is believed to involve interactions
between genetic and environmental factors. Recent advances in
genetic laboratory technology and sequencing of the human genome
have enabled more specific studies to determine gene mutations
causing OPLL or predisposing individuals to developing this
condition. In genetic research on the occurrence and development of
OPLL, the elucidation of the intrinsic mechanism may help us to
further understand the disease, to establish assessment tools for
determining T-OPLL severity and disease probability indexes, to
achieve early disease detection and diagnosis, and to explore other
auxiliary treatments. Thus, such studies are critical for improving
therapeutic strategies for patients with this disease.
The thoracic spine experiences less activity than
the cervical spine; thus, this region of the spine is subjected to
less local biological stress than the cervical spine, and factors
that mainly affect local mechanical stress and spine degeneration
in cervical OPLL have little effect on T-OPLL. Previous studies
have suggested that multiple polymorphisms in osteogenesis-related
genes are associated with the development and progression of
cervical OPLL. Additionally, extensive linkage and association
studies have identified many genes linked to OPLL susceptibility.
In previous reports, more than 16 susceptibility genes/loci have
been shown to be linked to OPLL susceptibility (4), including collagen, type VI, α1
(COL6A1) (7–9), collagen, type XII, α2 (COL11A2)
(10), bone morphogenetic protein
2 (BMP2) (11), transforming
growth factor (TGF)-β1 (12),
interleukin (IL)-1β (13), IL-15
receptor α (IL-15RA) (14,15), and runt-related transcription
factor 2 (RUNX2) (16). However,
these findings have not been sufficiently reproducible, and no
genetic studies have assessed the causes of T-OPLL.
Thus, in this study, we used whole-genome sequencing
(WGS) with high-density single nucleotide polymorphism (SNP) data
combined with a predictive deleterious effects algorithm to
identify genes or loci associated with T-OPLL in the Han Chinese
population.
Materials and methods
Disease criteria and patients
The study protocol was approved by the ethical
committee for human subjects of the Peking University Third
Hospital. Informed consent was provided by all participating
individuals. A total of 30 unrelated northern Chinese Han T-OPLL
patients with myelopathy and/or neurological dysfunction [14 men,
(mean age, 51.71±6.38 years); 16 women (mean age, 52.63±5.97
years)] and 5 unrelated healthy controls (mean age, 52.20±1.30
years, use for analysis of susceptibility gene expression levels in
peripheral blood) were enrolled in this study from February 2010 to
July 2016. Diagnosis of T-OPLL was performed by specialists based
on clinical symptoms and radiologic examinations (including CT and
magnetic resonance imaging) of the thoracic spine. The appearance
of OPLL observed in radiographs was classified into four subtypes:
i) Segmental; ii) continuous; iii) mixed; and iv) local (17). Neurological status was evaluated by
the Japanese Orthopedic Association (JOA) score for thoracic
myelopathy (a total of 11 points). Individuals who had lumbar
spondylolisthesis, ankylosing spondylitis, diffuse idiopathic
skeletal hyperostosis, and disc herniation of the thoracic spines
were excluded in this study and did not take any drugs known to
affect bone or calcium metabolism.
WGS
Genomic DNA was extracted from peripheral blood
leukocytes using a standard method. To discover genetic variations,
we performed WGS on 30 unrelated northern Chinese Han patients.
Quality genomic DNA from the 30 samples was fragmented using a
Covaris ultrasonicator. By adjusting shearing parameters, DNA
fragments from each sample were concentrated in 500-bp peaks. These
fragments were purified, end blunted, ‘A’ tailed, and adaptor
ligated. DNA templates with adapters were then selectively enriched
using polymerase chain reaction (PCR) to obtain a sufficient amount
for the DNA library. The concentration of DNA for the libraries was
quantified using a bioanalyser (Agilent Technologies, Inc., Santa
Clara, CA, USA) and qPCR. Each qualified DNA library was sequenced
on an Illumina HiSeq Xten platform using paired-end reads according
to the manufacturer's instructions. Sequencing-derived raw image
files were processed by Illumina base calling Software with default
parameters, and sequence data for each individual were generated as
paired-end reads, defined as ‘raw data’.
Bioinformatics analysis
Bioinformatics analysis began with sequencing data
(raw data from the Illumina machine). First, clean data were
produced by data filtering of raw data. All clean data for each
sample were mapped to the human reference genome (GRCh37/HG19).
Burrows-Wheeler Aligner (18,19)
software was used to do the alignment. To ensure accurate variant
calling, we followed recommended best practices for variant
analysis with the Genome Analysis Toolkit. Local realignment around
InDels and base quality score recalibration were performed using
GATK (20,21), with duplicate reads removed by
Picard tools. The sequencing depth and coverage for each individual
were calculated based on the alignments. In addition, a strict data
analysis QC system throughout the entire pipeline was built to
guarantee high-quality sequencing data.
To decrease noise in the sequencing data, data
filtering was carried out, including: i) removal of reads
containing the sequencing adapter; ii) removal of reads with a
low-quality base ratio (base quality less than or equal to 5) that
was more than 50%; and iii) removal of reads with an unknown base
(‘N’ base) ratio that was more than 10%. Statistical analysis of
data and downstream bioinformatics analyses were performed on the
filtered, high-quality data, referred to as the ‘clean data’.
Variant frequency was compared with the 1000G SNP database
(http://www.1000genomes.org/). Potential
deleterious effects of identified sequence variants were assessed
by various algorithms, such as SIFT (http://sift.jcvi.org/) (22), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) (23), MutationTaster (http://www.mutationtaster.org/) (24), and GERP++ (http://mendel.stanford.edu/SidowLab/downloads/gerp/)
(25).
Confirmation of variants
Sanger DNA sequencing (ABI 3730XL; Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was
used to confirm the accuracy of variants identified by WGS. The PCR
fragments were submitted for Sanger sequencing at the Beijing
Genomics Institute. Details of the two studied SNPs and the primer
sequences are listed in Table I.
PCR was performed with 20 ng genomic DNA per 15 µl reaction
mixture, containing 0.2 µM of each primer, 200 µM of
deoxyribonucleotides, 50 mM KCl, 10 mM Tris HCl (pH 8.3), 1.5 mM
MgCl2 and 0.5 units of Taq DNA polymerase in a DNA
Gradient PCR machine (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The thermocycling conditions were as follows: initial
denaturation at 95°C for 10 min; followed by 35 cycles of 95°C for
30 sec, annealing at an assay-specific temperature (48 to 65°C) for
45 sec, and elongation at 72°C for 45 sec; and a final terminal
elongation step at 72°C for 5 min. The PCR products were analyzed
by direct sequencing using a BigDye Terminator v3.1 Cycle
Sequencing kit (Thermo Fisher Scientific, Inc.) with POP-7™ Polymer
in a 3730XL DNA Analyzer with Sequencing Analysis Software version
5.2 (Thermo Fisher Scientific, Inc.).
 | Table I.Details of the two SNPs in
COL6A1 and IL17RC and their associated primers. |
Table I.
Details of the two SNPs in
COL6A1 and IL17RC and their associated primers.
| Gene | SNP ID | Nucleotide
substitution (M/m) | Primer
sequence |
|---|
| COL6A1 | rs201153092 | G/A | Forward
5′-TGAAAGGGTGAGTGTCCAA-3′ |
|
|
|
| Reverse
5′-GTGCCCAGTCCACTAAAGAG-3′ |
| IL17RC | rs199772854 | C/A | Forward
5′-CCCAACTGCCAGACTTCCT-3′ |
|
|
|
| Reverse
5′-GCCACAGCCTGCGTAAAA-3′ |
Protein conservation analysis
CLUSTAL W (http://www.genome.jp/tools/clustalw/) was used to
compare homologous protein sequences among multiple species to
analyse the consistency of amino acid sequences, particularly
mutation sites, in seven representative species.
Plasma COL6A1 and IL17RC ELISAs
Plasma collection and storage were carried out using
standard methods. Plasma COL6A1 (cat. no. HG21134) and IL17RC (cat.
no. HG1762) levels were quantified using commercially available
ELISA kits (Trust Specialty Zeal, San Francisco, CA, USA). All
samples were assayed according to the manufacturer's instructions
and were run in duplicate. The optical density of each well was
determined using a microplate reader at 450 nm. No interference and
no cross reactivity were expected based on the manufacturer's
instructions.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was purified from blood using the SK1321
RNA Blood Mini kit (Sangon Biotech Co., Ltd., Shanghai, China). An
on-column DNase digest (Sangon Biotech Co., Ltd.) was performed
before the clean-up step to eliminate residual genomic DNA. cDNA
was synthesized from total RNA (2 µg) using a RevertAid Premium
Reverse Transcriptase kit from Thermo Fisher Scientific, Inc.
Relative quantitative RT-PCR was applied to quantify the mRNAs
levels of COL6A1, IL17RC and GAPDH using SYBR-Green Real-Time PCR
master mix on the LightCycler480 Real-Time System from Roche
(Basel, Switzerland). All experiments were performed in triplicate
and normalized to GAPDH.
Statistical analysis
All statistical analyses were performed using SPSS
v17.0 software (SPSS, Inc., Chicago, IL, USA). Descriptive data for
continuous variables are presented as the mean ± standard
deviation. Student's t-tests were used to determine the age and JOA
score differences between patients with or without OPLL gene
mutations. The OPLL subtype differences between patients with or
without OPLL gene mutations were determined using one-way analysis
of variance with a post hoc Fisher's test. P<0.05 was considered
to indicate a statistically significant difference.
Results
WGS analysis
After standard SNP quality control, we performed WGS
on 30 DNA samples with an average of 119,583.62 Mb raw bases from
the Illumina Hiseq Xen sequencer. After removing low-quality reads,
we obtained an average of 793,363,211 clean reads (119,004.48 Mb).
The clean reads of each sample had high Q20 and Q30, indicating
high sequencing quality. The average GC content was 41.01%
(Table II). Strict data quality
control (QC) was performed during the entire analysis pipeline to
obtain accurate data (Table
III).
 | Table II.Summary of whole-genome sequencing
data. |
Table II.
Summary of whole-genome sequencing
data.
| Sample | Raw reads | Raw bases (Mb) | Clean reads | Clean bases
(Mb) | Clean data rate
(%) | Clean read Q20
(%) | Clean read Q30
(%) | GC content (%) |
|---|
| P1 |
806578038 | 120986.71 |
802018468 | 120302.77 | 99.43 | 98.5 | 96.46 |
40.85 |
| P2 |
803614916 | 120542.24 |
800270858 | 120040.63 | 99.58 | 98.57 | 96.61 | 41.02 |
| P3 | 1166765398 | 175014.81 | 1162305438 | 174345.82 | 99.62 | 98.58 | 96.64 | 41 |
| P4 |
742579698 | 111386.95 |
738204906 | 110730.74 | 99.41 | 98.51 | 96.49 | 40.83 |
| P5 |
737867244 | 110680.09 |
734411142 | 110161.67 | 99.53 | 98.52 | 96.61 | 41.23 |
| P6 |
695402032 | 104310.3 |
691857040 | 103778.56 | 99.49 | 98.5 | 96.47 | 40.88 |
| P7 |
724494036 | 108674.11 |
721418588 | 108212.79 | 99.58 | 98.49 | 96.41 | 40.94 |
| P8 |
776034852 | 116405.23 |
773219638 | 115982.95 | 99.64 | 98.48 | 96.5 | 41.28 |
| P9 |
759932006 | 113989.8 |
755805520 | 113370.83 | 99.46 | 98 | 96.46 | 40.92 |
| P10 |
781101726 | 117165.26 |
777211156 | 116581.67 | 99.5 | 98.53 | 96.53 | 40.93 |
| P11 |
828771876 | 124315.78 |
824405368 | 123660.81 | 99.47 | 98.57 | 96.66 | 41.3 |
| P12 |
845882256 | 126882.34 |
842286794 | 126343.02 | 99.57 | 98.52 | 96.58 | 41.25 |
| P13 |
865879474 | 129881.92 |
860936622 | 129140.49 | 99.43 | 98.59 | 96.72 | 41.37 |
| P14 |
737673162 | 110650.97 |
734221322 | 110133.2 | 99.53 | 98.53 | 96.55 | 40.88 |
| P15 |
740541398 | 111081.21 |
735208756 | 110281.31 | 99.28 | 98.52 | 96.45 | 40.92 |
| P16 |
792275714 | 118841.36 |
788504808 | 118275.72 | 99.52 | 98.49 | 96.4 | 40.94 |
| P17 |
752134540 | 112820.18 |
749032976 | 112354.95 | 99.59 | 98.47 | 96.35 | 40.96 |
| P18 |
728158076 | 109223.71 |
724409280 | 108661.39 | 99.49 | 98.49 | 96.44 | 40.92 |
| P19 |
792928182 | 118939.23 |
789057438 | 118358.62 | 99.51 | 98.5 | 96.42 | 40.95 |
| P20 |
777804644 | 116670.7 |
774841888 | 116226.28 | 99.62 | 98.53 | 96.51 | 41 |
| P21 |
833487852 | 125023.18 |
829696094 | 124454.41 | 99.55 | 98.47 | 96.34 | 40.98 |
| P22 |
759919330 | 113987.9 |
756111904 | 113416.79 | 99.5 | 98.58 | 96.62 | 40.9 |
| P23 |
728707168 | 109306.08 |
725408206 | 108811.23 | 99.55 | 98.57 | 96.6 | 40.86 |
| P24 |
742897576 | 111434.64 |
739389748 | 110908.46 | 99.53 | 98.58 | 96.62 | 40.9 |
| P25 | 1094810746 | 164221.61 | 1090602492 | 163590.37 | 99.62 | 97.39 | 95.8 | 41.17 |
| P26 |
772278386 | 115841.76 |
769004376 | 115350.66 | 99.58 | 98.59 | 96.65 | 40.88 |
| P27 |
752896392 | 112934.46 |
748911760 | 112336.76 | 99.47 | 98.56 | 96.62 | 41.18 |
| P28 |
756830924 | 113524.64 |
752633012 | 112894.95 | 99.45 | 98.61 | 96.78 | 41.16 |
| P29 |
840021700 | 126003.26 |
835148570 | 125272.29 | 99.42 | 98.62 | 96.8 | 41.13 |
| P30 |
778453666 | 116768.05 |
774362180 | 116154.33 | 99.47 | 98.55 | 96.59 | 40.82 |
| Average |
797224100 | 119583.62 |
793363211 | 119004.48 | 99.51 | 98.48 | 96.52 | 41.01 |
 | Table III.Data quality control. Strict data
quality control was performed across the whole analysis pipeline
for the clean data, the mapping data and the variant calling Y
pass. |
Table III.
Data quality control. Strict data
quality control was performed across the whole analysis pipeline
for the clean data, the mapping data and the variant calling Y
pass.
| Samples | Clean read1 Q20
(%) | Clean read1 Q30
(%) | GC content (%) | Mapping rate
(%) | Duplicate rate
(%) | Mismatch rate
(%) | Average sequencing
depth (X) | Coverage (%) | Coverage at least
4X (%) |
|---|
| P1 | Y(98.50) | Y(96.46) | Y(40.85) | Y(99.54) | Y(11.84) | Y(0.63) | Y(35.10) | Y(99.09) | Y(98.69) |
| P2 | Y(98.57) | Y(96.61) | Y(41.02) | Y(99.63) | Y(10.85) | Y(0.56) | Y(35.63) | Y(99.06) | Y(98.68) |
| P3 | Y(98.58) | Y(96.64) | Y(41.00) | Y(99.72) | Y(9.32) | Y(0.59) | Y(52.43) | Y(99.82) | Y(99.63) |
| P4 | Y(98.51) | Y(96.49) | Y(40.83) | Y(99.55) | Y(11.78) | Y(0.63) | Y(32.39) | Y(99.08) | Y(98.66) |
| P5 | Y(98.52) | Y(96.61) | Y(41.23) | Y(99.53) | Y(10.67) | Y(0.56) | Y(32.64) | Y(99.79) | Y(99.41) |
| P6 | Y(98.50) | Y(96.47) | Y(40.88) | Y(99.61) | Y(10.75) | Y(0.60) | Y(30.78) | Y(99.07) | Y(98.60) |
| P7 | Y(98.49) | Y(96.41) | Y(40.94) | Y(99.66) | Y(11.05) | Y(0.58) | Y(32.06) | Y(99.04) | Y(98.61) |
| P8 | Y(98.48) | Y(96.50) | Y(41.28) | Y(99.54) | Y(10.08) | Y(0.56) | Y(34.62) | Y(99.14) | Y(98.71) |
| P9 | Y(98.00) | Y(96.46) | Y(40.92) | Y(99.60) | Y(11.48) | Y(0.61) | Y(33.34) | Y(99.07) | Y(98.67) |
| P10 | Y(98.53) | Y(96.53) | Y(40.93) | Y(99.63) | Y(12.61) | Y(0.58) | Y(33.90) | Y(99.17) | Y(98.69) |
| P11 | Y(98.57) | Y(96.66) | Y(41.30) | Y(99.55) | Y(12.50) | Y(0.54) | Y(35.89) | Y(99.80) | Y(99.48) |
| P12 | Y(98.52) | Y(96.58) | Y(41.25) | Y(99.55) | Y(10.47) | Y(0.55) | Y(37.59) | Y(99.82) | Y(99.52) |
| P13 | Y(98.59) | Y(96.72) | Y(41.37) | Y(99.57) | Y(13.70) | Y(0.53) | Y(36.96) | Y(99.12) | Y(98.76) |
| P14 | Y(98.53) | Y(96.55) | Y(40.88) | Y(99.65) | Y(11.22) | Y(0.59) | Y(32.56) | Y(99.09) | Y(98.67) |
| P15 | Y(98.52) | Y(96.45) | Y(40.92) | Y(99.63) | Y(10.90) | Y(0.55) | Y(32.68) | Y(99.09) | Y(98.66) |
| P16 | Y(98.49) | Y(96.40) | Y(40.94) | Y(99.58) | Y(11.39) | Y(0.57) | Y(34.71) | Y(99.78) | Y(99.43) |
| P17 | Y(98.47) | Y(96.35) | Y(40.96) | Y(99.57) | Y(10.13) | Y(0.56) | Y(33.54) | Y(99.76) | Y(99.40) |
| P18 | Y(98.49) | Y(96.44) | Y(40.92) | Y(99.54) | Y(10.86) | Y(0.64) | Y(32.12) | Y(99.09) | Y(98.66) |
| P19 | Y(98.50) | Y(96.42) | Y(40.95) | Y(99.60) | Y(10.99) | Y(0.55) | Y(35.05) | Y(99.12) | Y(98.73) |
| P20 | Y(98.53) | Y(96.51) | Y(41.00) | Y(99.47) | Y(9.63) | Y(0.57) | Y(34.80) | Y(99.80) | Y(99.46) |
| P21 | Y(98.47) | Y(96.34) | Y(40.98) | Y(99.46) | Y(10.91) | Y(0.57) | Y(36.67) | Y(99.78) | Y(99.47) |
| P22 | Y(98.58) | Y(96.62) | Y(40.90) | Y(99.52) | Y(10.27) | Y(0.54) | Y(33.78) | Y(99.11) | Y(98.69) |
| P23 | Y(98.57) | Y(96.60) | Y(40.86) | Y(99.54) | Y(9.64) | Y(0.54) | Y(32.65) | Y(99.07) | Y(98.63) |
| P24 | Y(98.58) | Y(96.62) | Y(40.90) | Y(99.53) | Y(10.30) | Y(0.54) | Y(33.10) | Y(99.05) | Y(98.63) |
| P25 | Y(97.39) | Y(95.80) | Y(41.17) | Y(99.78) | Y(8.65) | Y(0.58) | Y(49.71) | Y(99.11) | Y(98.84) |
| P26 | Y(98.59) | Y(96.65) | Y(40.88) | Y(99.52) | Y(11.30) | Y(0.54) | Y(33.95) | Y(99.79) | Y(99.43) |
| P27 | Y(98.56) | Y(96.62) | Y(41.18) | Y(99.59) | Y(13.03) | Y(0.53) | Y(32.52) | Y(99.10) | Y(98.67) |
| P28 | Y(98.61) | Y(96.78) | Y(41.16) | Y(99.56) | Y(13.03) | Y(0.54) | Y(32.56) | Y(99.77) | Y(99.42) |
| P29 | Y(98.62) | Y(96.80) | Y(41.13) | Y(99.80) | Y(13.33) | Y(0.51) | Y(36.23) | Y(99.12) | Y(98.75) |
| P30 | Y(98.55) | Y(96.59) | Y(40.82) | Y(99.72) | Y(11.83) | Y(0.60) | Y(34.09) | Y(99.80) | Y(99.44) |
Variant identification
To prioritize potential pathogenic variants, we
focused on the identification of rare [multiple allele frequency
(MAF) ≤0.005, based on the BGI database] and damaging variants
predicted by at least two algorithms (e.g., SIFT, Polyphen-2,
MutationTaster, and GERP++). Two deleterious variants were
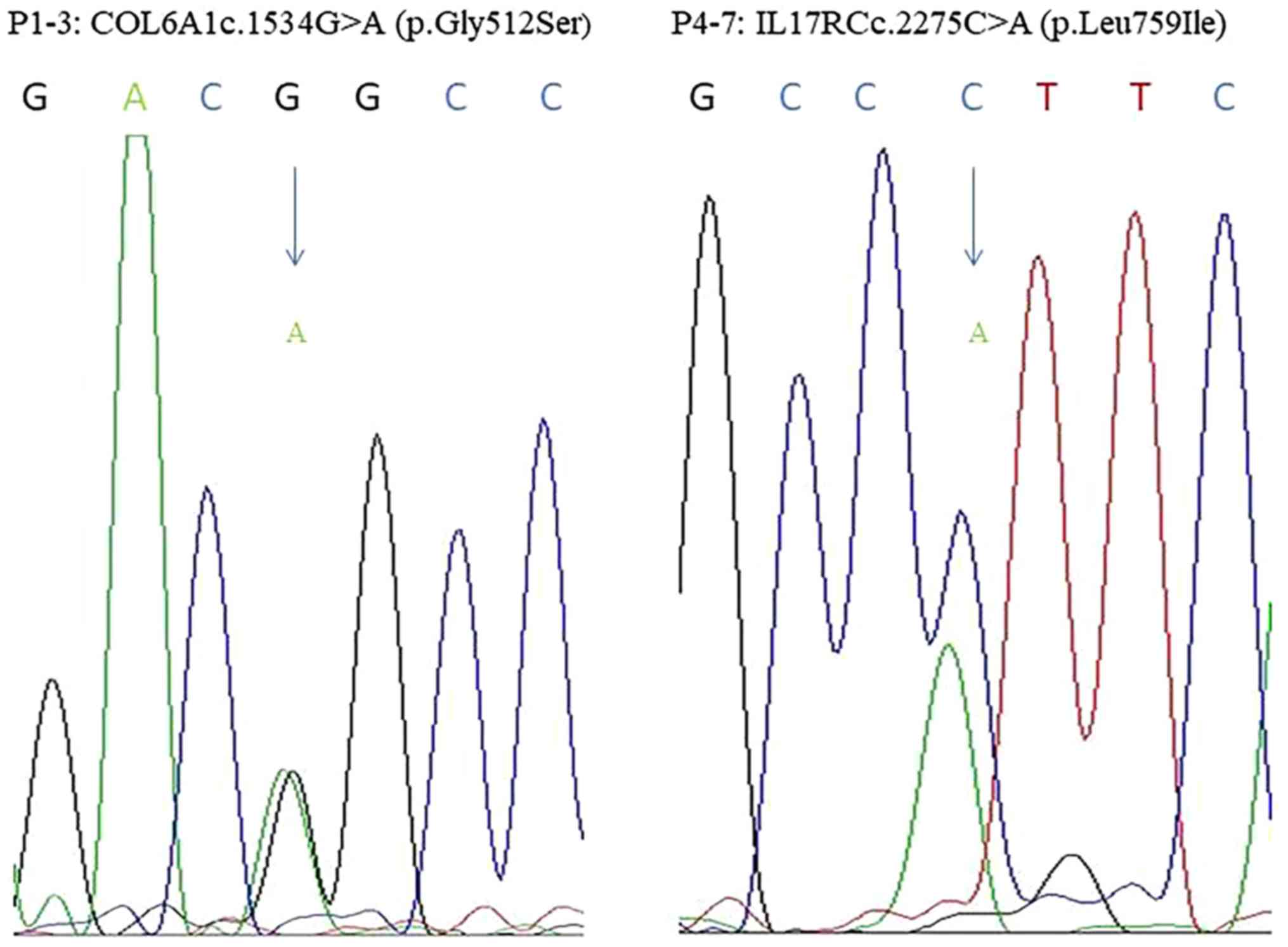
identified in seven unrelated patients (Table IV), and these findings were
further confirmed by directional Sanger sequencing (Fig. 1). Indicating that the test results
for WGS are accurate.
 | Table IV.Clinical features of patients with or
without the OPLL gene. |
Table IV.
Clinical features of patients with or
without the OPLL gene.
| ID | Sex/age | Gene | SNP ID | Chromosome | Nucleotide
change | Protein change | 1000G (EAS) | SIFT | PP2 | Mutation
taster | GERR++ | OPLL
morphology | OPLL |
|---|
| P1 | F/66 | COL6A1 | rs201153092 | 21 | c.1534G>A | p.Gly512Ser | 0 | D | D | D | R | Local | T11-12 |
| P2 | F/60 | COL6A1 | rs201153092 | 21 | c.1534G>A | p.Gly512Ser | 0 | D | D | D | R | Continuous | T1-7 |
| P3 | M/44 | COL6A1 | rs201153092 | 21 | c.1534G>A | p.Gly512Ser | 0 | D | D | D | R | Mixed | T3-7, T8-9 |
| P4 | F/49 | IL17RC | rs199772854 | 3 | c.2275C>A | p.Leu759Ile | 0 | D | D | N | R | Segmental | T2-12 |
| P5 | M/52 | IL17RC | rs199772854 | 3 | c.2275C>A | p.Leu759Ile | 0 | D | D | N | R | Continuous | T3-8 |
| P6 | F/51 | IL17RC | rs199772854 | 3 | c.2275C>A | p.Leu759Ile | 0 | D | D | N | R | Continuous | T2-7 |
| P7 | M/59 | IL17RC | rs199772854 | 3 | c.2275C>A | p.Leu759Ile | 0 | D | D | N | R | Continuous | T4-8 |
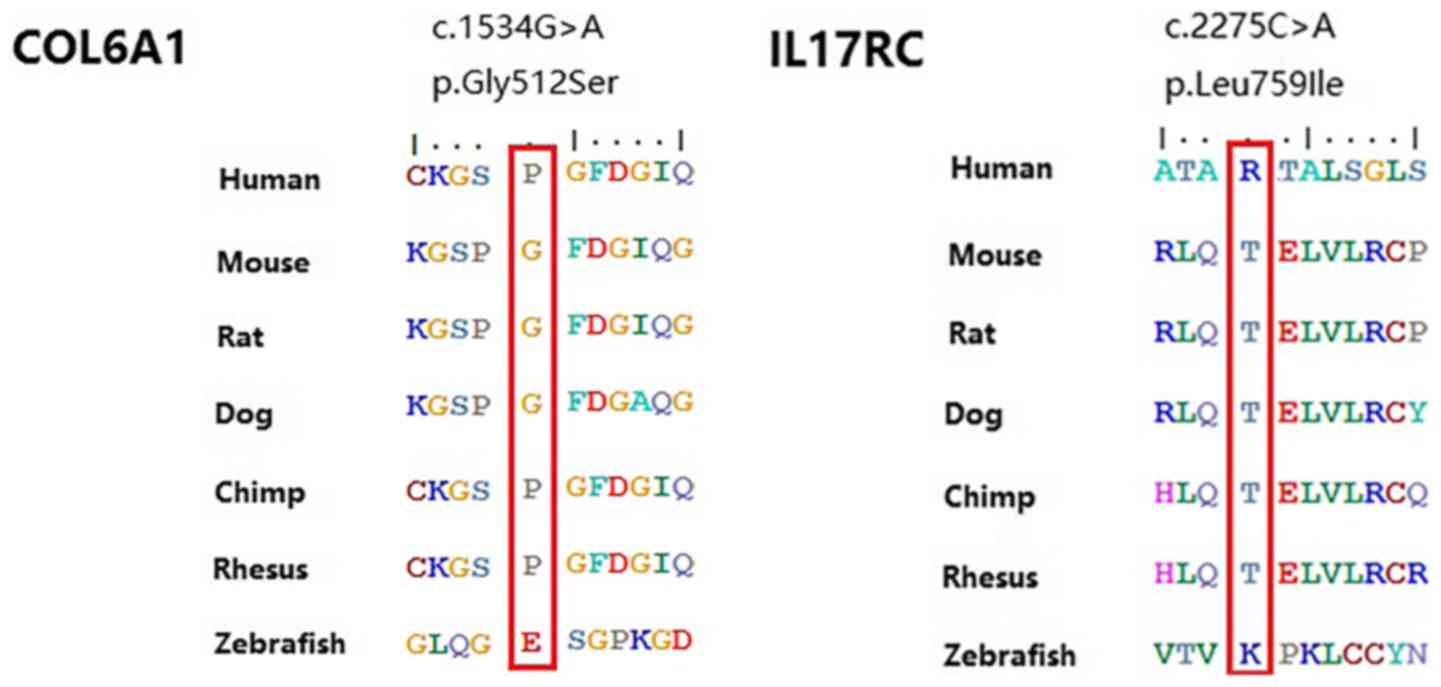
c.1534G>A (p.Gly512Ser) was located in the exon
regions of 42.9% of patients with OPLL. Moreover, this variant was
found to be evolutionarily conserved in three primates.
c.2275C>A (p.Leu759Ile) was located in intron regions of 57.1%
of patients with OPLL. However, this variant was not found in six
other vertebrate species (Fig. 2).
These two heterozygous mutations were found to be relatively
conserved, suggesting highly genetic heterogeneities and complex
pathogenesis.
Analysis of COL6A1 and IL17RC levels
in patients' blood samples
To elucidate the functional roles of loci and genes
associated with T-OPLL, we measured the expression levels of genes
in three patients carrying p.Gly512Ser mutations in COL6A1
and four patients carrying p.Leu759Ile mutations in IL17RC
compared with 5 healthy controls by enzyme-linked immunosorbent
assay (ELISA). We found that plasma COL6A1 concentrations
(20.90±0.64 µg/l) were significantly higher (P<0.05),
approximately 4-fold higher than those in healthy controls
(4.79±1.18 µg/l). Moreover, plasma IL17RC concentrations (6.13±0.36
µg/l) were also significantly higher (P<0.05), approximately
2-fold higher than those in healthy controls (2.89±0.30 µg/l). We
performed RT-qPCR analysis using peripheral blood cells and found
that COL6A1 mRNA levels in three patients carrying the p.Gly512Ser
mutation were approximately 6-fold higher than those in healthy
controls, the difference was statistically significant (P<0.05).
The IL17RC mRNA levels in four patients carrying the p.Leu759Ile
mutation were approximately 4-fold higher than those in the healthy
controls, the difference was also statistically significant
(P<0.05). Compared with healthy controls group, we observed that
these two mutations significantly increase their respective gene
expressed, suggesting that these two potential pathogenic loci have
potential effect on their respective gene expressed cells.
Genotype-phenotype analysis
Phenotype-genotype correlations were analysed among
the seven patients with missense mutations and 23 patients without
significant mutations (Table V).
No differences were found between these two groups in terms of sex
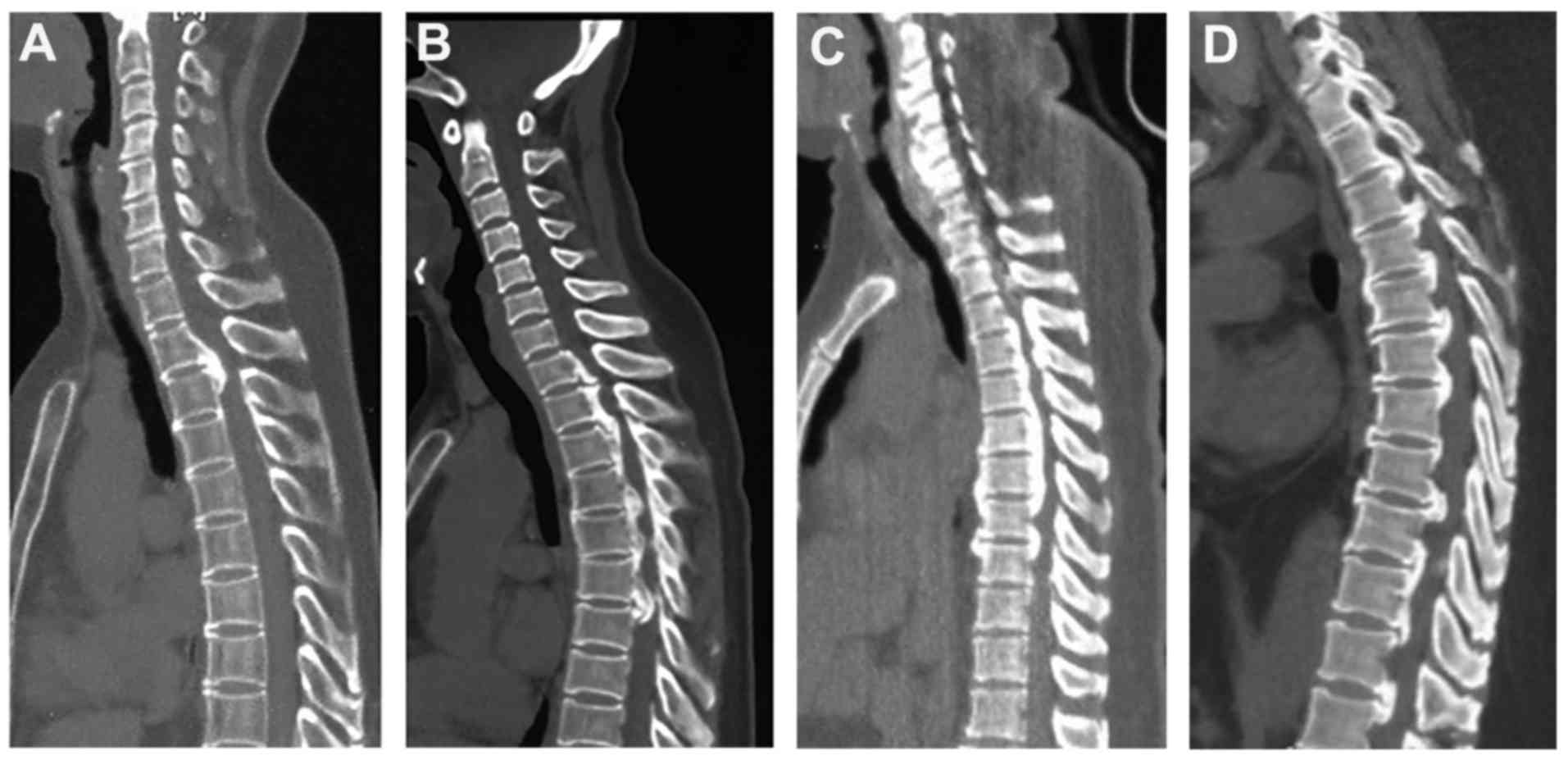
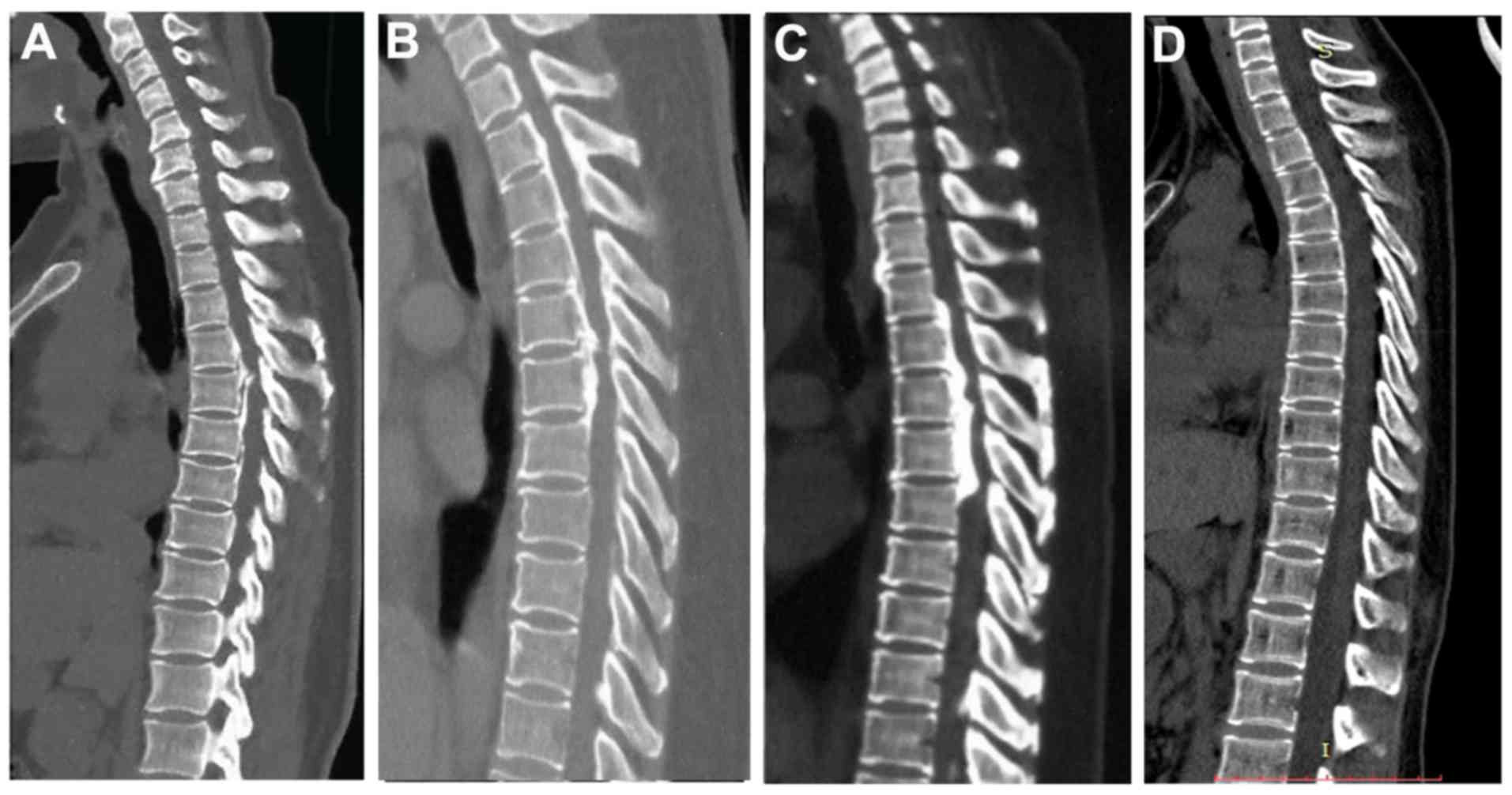
or age at diagnosis. Two-dimensional computed tomography (2D-CT)
scans of seven patients with OPLL harbouring deleterious variants
and one healthy control are shown in Figs. 3 and 4. More specifically, one patient belonged
to the Local subtype (Fig. 3A),
one patient belonged to the Mixed subtype (Fig. 3B), one patient showed the Segmental
subtype (Fig. 3C), and four
patients showed the Continuous subtype (Figs. 3D and 4A-C), Fig.
4D showed the healthy control. However, the JOA score for
thoracic myelopathy was significantly lower in patients with rare
missense mutations compared with patients without mutations
(P=0.001). Additionally, radiological analysis of OPLL morphology
(17) revealed that the frequency
of the continuous subtype was significantly higher in patients with
rare missense mutations than in patients without mutations
(P=0.033). Moreover, four mutation-positive patients (57.1%) showed
the continuous subtype, one patient (14.3%) showed the local
subtype, one patient (14.3%) showed the segmental subtype, and one
patient (14.3%) showed the mixed subtype. By contrast, three (13%)
mutation-negative patients showed the continuous subtype, six
patients (26.1%) showed the local subtype, 10 patients (43.4%)
showed the segmental subtype, and four patients (17.4%) without
mutations showed the mixed subtype.
 | Table V.Clinical features of patients with or
without OPLL gene mutations. |
Table V.
Clinical features of patients with or
without OPLL gene mutations.
| Indices | Positive mutation
(n=7) | Negative mutation
(n=23) | P-value |
|---|
| Age (years) | 54.43±7.56 | 51.52±5.57 | 0.275 |
| Male/female | 3/4 | 10/13 | 0.660 |
| OPLL subtype
(%) |
|
|
|
|
Continuous | 4 (57.1) | 3 (13) | 0.033 |
|
Local | 1 (14.3) | 6
(26.1) | 0.468 |
|
Segmental | 1 (14.3) | 10 (43.4) | 0.171 |
|
Mixed | 1 (14.3) | 4
(17.4) | 0.671 |
| JOA Score | 3.29±0.95 | 4.26±0.45 | 0.001 |
Discussion
COL6A1, which encodes the α1 chain of type VI
collagen, is located on chromosome 21q22.3 and spans approximately
23.3 kb. Additionally, as a major structural component of
microfibrils, COL6A1 plays a role in maintaining the integrity of
various tissues and has been shown to be associated with OPLL in
Japanese and Chinese populations (8,9).
Previously, COL6A1 mutations associated with OPLL were found
in intronic regions. However, in the present study, we report for
the first time a mutation, c.1534G>A(p.Gly512Ser)/COL6A1, in the
exonic region that is associated with T-OPLL. This mutation may be
involved in transcriptional regulation, and analysis using the
SIFT, Polyphen-2, and MutationTaster algorithms suggested that this
mutation may have an adverse impact on the structure and function
of the encoded protein. Additionally, COL6A1 may serve as a
scaffold for osteoblastic or pre-osteoblastic cells or chondrocytes
that subsequently undergo membranous or endochondral ossification
(26). Therefore, the molecular
variants of extracellular proteins may be involved in the ectopic
bone formation that is observed in patients with OPLL (27).
The IL17RC gene encodes a single-pass type I
transmembrane protein located on chromosome region 3p25.3 to
3p24.1, spanning approximately 16,550 bp (28). OPLL results in increased bone
formation in ligament tissue, and there is some evidence showing a
correlation between OPLL and increased systemic bone mineral
density. IL17RC accelerates osteoblast differentiation; a
recent study indicated that dysfunction of both IL17RC and the
IL-17 cytokine/IL-17R signalling axis is indispensable for
osteoblastogenesis (29).
Moreover, the TGF-β signalling pathway is known to be associated
with the formation of bone mass and matrix (30). The IL17RC gene may also be
involved in bone metabolism through canonical TGF-β signalling
(31). However, this variant was
not found in six other vertebrate species, suggesting high genetic
heterogeneities. In this study, we identified
c.2275C>A(p.Leu759Ile)/IL17RC, which had not previously been
reported to be associated with OPLL, using the standard potential
pathogenic variant method, thereby linking IL17RC to OPLL
and suggesting that this mutation, as well as the mutation in
COL6A1, may be involved in bone development.
At present, several lines of evidence suggest that
OPLL seems to occur and develop as a result of systemic and local
factors in combination with a genetic abnormality (32–34).
The progression of the disease also affects the gene expression in
peripheral blood cells, several genes are highly expressed in
peripheral blood of patients with OPLL (7,32).
In addition, peripheral blood is easily accessible and routinely
used for diagnostic laboratory analysis and thus is a good resource
for additional tests that might define extent of T-OPLL. Therefore,
what we observed an increased level of COL6A1 and IL17RC with two
heterozygous mutations in the patient's peripheral blood revealed
its potential role in the pathogenicity of T-OPLL.
There was a limitation in the present study. The
sample size is small, larger scale studies is necessary. However,
the T-OPLL is primarily found in individuals in northeast Asia, the
prevalence of T-OPLL in Japanese individuals is only 0.8–1.6%
(6,35), the prevalence of this disease is
very rare. To the best of our knowledge, our 30 sample size is the
largest cases of single center. To accomplish this issue,
international collaboration is the good way to go.
In summary, in this study, we identified mutations
in Han Chinese patients with T-OPLL for the first time using WGS.
From our analysis, we found two new potential pathogenic loci for
OPLL: c.1534G>A(p.Gly512Ser) in the COL6A1 gene, which
has previously been reported to be associated with OPLL, and
c.2275C>A(p.Leu759Ile) in the IL17RC gene, which had not
previously been reported to be associated with OPLL. The results of
the current study provide insights into the molecular aetiology of
OPLL. Further genetic and functional studies, including studies
with more participants of other ethnicities, are needed to confirm
these positive findings.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81472041). Thanks are due to
Medical Research Center of Peking University Third Hospital for
providing technical guidance.
References
|
1
|
Kim KH, Kuh SU, Park JY, Lee SJ, Park HS,
Chin DK, Kim KS and Cho YE: Association between BMP-2 and COL6A1
gene polymorphisms with susceptibility to ossification of the
posterior longitudinal ligament of the cervical spine in Korean
patients and family members. Genet Mol Res. 13:2240–2247. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nagashima C: Cervical myelopathy due to
ossification of the posterior longitudinal ligament. J Neurosurg.
37:653–660. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yonemori K, Imamura T, Ishidou Y, Okano T,
Matsunaga S, Yoshida H, Kato M, Sampath TK, Miyazono K, ten Dijke P
and Sakou T: Bone morphogenetic protein receptors and activin
receptors are highly expressed in ossified ligament tissues of
patients with ossification of the posterior longitudinal ligament.
Am J Pathol. 150:1335–1347. 1997.PubMed/NCBI
|
|
4
|
Ikegawa S: Genetics of ossification of the
posterior longitudinal ligament of the spine: A mini review. J Bone
Meta. 21:127–132. 2014. View Article : Google Scholar
|
|
5
|
Tsuji T, Chiba K, Hosogane N, Fujita N,
Hikata T, Iwanami A, Watanabe K, Ishii K, Toyama Y, Nakamura M and
Matsumoto M: Epidemiological survey of ossification of the
posterior longitudinal ligament by using clinical investigation
registration forms. J Orthop Sci. 21:291–294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fujimori T, Watabe T, Iwamoto Y, Hamada S,
Iwasaki M and Oda T: Prevalence, concomitance and distribution of
ossification of the spinal ligaments: Results of Whole Spine CT
Scans in 1500 Japanese Patients. Spine (Phila Pa 1976).
41:1668–1676. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen X, Guo J, Cai T, Zhang F, Pan S,
Zhang L, Wang S, Zhou F, Diao Y, Zhao Y, et al: Targeted
next-generation sequencing reveals multiple deleterious variants in
OPLL-associated genes. Sci Rep. 6:269622016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kong Q, Ma X, Li F, Guo Z, Qi Q, Li W,
Yuan H, Wang Z and Chen Z: COL6A1 polymorphisms associated with
ossification of the ligamentum flavum and ossification of the
posterior longitudinal ligament. Spine (Phila Pa 1976).
32:2834–2838. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tanaka T, Ikari K, Furushima K, Okada A,
Tanaka H, Furukawa K, Yoshida K, Ikeda T, Ikegawa S, Hunt SC, et
al: Genomewide linkage and linkage disequilibrium analyses identify
COL6A1, on chromosome 21, as the locus for ossification of the
posterior longitudinal ligament of the spine. Am J Hum Genet.
73:812–822. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koga H, Sakou T, Taketomi E, Hayashi K,
Numasawa T, Harata S, Yone K, Matsunaga S, Otterud B, Inoue I and
Leppert M: Genetic mapping of ossification of the posterior
longitudinal ligament of the spine. Am J Hum Genet. 62:1460–1467.
1998. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang H, Liu D, Yang Z, Tian B, Li J, Meng
X, Wang Z, Yang H and Lin X: Association of bone morphogenetic
protein-2 gene polymorphisms with susceptibility to ossification of
the posterior longitudinal ligament of the spine and its severity
in Chinese patients. Eur Spine J. 17:956–964. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kamiya M, Harada A, Mizuno M, Iwata H and
Yamada Y: Association between a polymorphism of the transforming
growth factor-beta1 gene and genetic susceptibility to ossification
of the posterior longitudinal ligament in Japanese patients. Spine
(Phila Pa 1976). 26:1264–1267. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ogata N, Koshizuka Y, Miura T, Iwasaki M,
Hosoi T, Shiraki M, Seichi A, Nakamura K and Kawaguchi H:
Association of bone metabolism regulatory factor gene polymorphisms
with susceptibility to ossification of the posterior longitudinal
ligament of the spine and its severity. Spine (Phila Pa 1976).
27:1765–1771. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim DH, Jeong YS, Chon J, Yoo SD, Kim HS,
Kang SW, Chung JH, Kim KT and Yun DH: Association between
interleukin 15 receptor, alpha (IL15RA) polymorphism and Korean
patients with ossification of the posterior longitudinal ligament.
Cytokine. 55:343–346. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo Q, Lv SZ, Wu SW, Tian X and Li ZY:
Association between single nucleotide polymorphism of IL15RA gene
with susceptibility to ossification of the posterior longitudinal
ligament of the spine. J Orthop Surg Res. 9:1032014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Y, Zhao Y, Chen Y, Shi G and Yuan W:
RUNX2 polymorphisms associated with OPLL and OLF in the Han
population. Clin Orthop Relat Res. 468:3333–3341. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsuyama N: Ossification of the posterior
longitudinal ligament of the spine. Clin Orthop Relat Res. 1–84.
1984.
|
|
18
|
Li H and Durbin R: Fast and accurate
long-read alignment with Burrows-Wheeler transform. Bioinformatics.
26:589–595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
DePristo MA, Banks E, Poplin R, Garimella
KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA,
Hanna M, et al: A framework for variation discovery and genotyping
using next-generation DNA sequencing data. Nat Genet. 43:491–498.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sim NL, Kumar P, Hu J, Henikoff S,
Schneider G and Ng PC: SIFT web server: Predicting effects of amino
acid substitutions on proteins. Nucleic Acids Res. 40:W452–W457.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schwarz JM, Cooper DN, Schuelke M and
Seelow D: MutationTaster2: Mutation prediction for the
deep-sequencing age. Nat Methods. 11:361–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Davydov EV, Goode DL, Sirota M, Cooper GM,
Sidow A and Batzoglou S: Identifying a high fraction of the human
genome to be under selective constraint using GERP++. PLoS Comput
Biol. 6:e10010252010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wiberg C, Klatt AR, Wagener R, Paulsson M,
Bateman JF, Heinegård D and Mörgelin M: Complexes of matrilin-1 and
biglycan or decorin connect collagen VI microfibrils to both
collagen II and aggrecan. J Biol Chem. 278:37698–37704. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tsukahara S, Miyazawa N, Akagawa H,
Forejtova S, Pavelka K, Tanaka T, Toh S, Tajima A, Akiyama I and
Inoue I: COL6A1, the candidate gene for ossification of the
posterior longitudinal ligament, is associated with diffuse
idiopathic skeletal hyperostosis in Japanese. Spine (Phila Pa
1976). 30:2321–2324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ho AW and Gaffen SL: IL-17RC: A partner in
IL-17 signaling and beyond. Semin Immunopathol. 32:33–42. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang H, Kim HJ, Chang EJ, Lee ZH, Hwang
SJ, Kim HM, Lee Y and Kim HH: IL-17 stimulates the proliferation
and differentiation of human mesenchymal stem cells: Implications
for bone remodeling. Cell Death Differ. 16:1332–1343. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mohammad KS, Chen CG, Balooch G, Stebbins
E, McKenna CR, Davis H, Niewolna M, Peng XH, Nguyen DH,
Ionova-Martin SS, et al: Pharmacologic inhibition of the TGF-beta
type I receptor kinase has anabolic and anti-catabolic effects on
bone. PLoS One. 4:e52752009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mukherjee S, Schaller MA, Neupane R,
Kunkel SL and Lukacs NW: Regulation of T cell activation by Notch
ligand, DLL4, promotes IL-17 production and Rorc activation. J
Immunol. 182:7381–7388. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Niu CC, Lin SS, Yuan LJ, Chen LH, Yang CY,
Chung AN, Lu ML, Tsai TT, Lai PL and Chen WJ: Correlation of blood
bone turnover biomarkers and Wnt signaling antagonists with AS,
DISH, OPLL, and OYL. BMC Musculoskelet Disord. 18:612017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ikegawa S: Genomic study of ossification
of the posterior longitudinal ligament of the spine. Proc Jpn Acad
Ser B Phys Biol Sci. 90:pp. 405–412. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nakajima M, Takahashi A, Tsuji T, Karasugi
T, Baba H, Uchida K, Kawabata S, Okawa A, Shindo S, Takeuchi K, et
al: A genome-wide association study identifies susceptibility loci
for ossification of the posterior longitudinal ligament of the
spine. Nat Genet. 46:1012–1016. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ohtsuka K, Terayama K, Yanagihara M, Wada
K, Kasuga K, Machida T and Matsushima S: A radiological population
study on the ossification of the posterior longitudinal ligament in
the spine. Arch Orthop Trauma Surg. 106:89–93. 1987. View Article : Google Scholar : PubMed/NCBI
|