Introduction
Human cervical cancer is one of the most common and
lethal malignancies in women worldwide (1,2).
Despite the fact that cervical cancer incidence has remarkably
reduced with the application of Papanicolaou smear screening,
cervical cancer remains imperil women's health, with an estimated
275,000 deaths annually (3,4).
Regarding the etiology of cervical cancer, there's evidence that
cervical cancer is associated with persistent oncogenic human
papillomavirus (HPV) infections inducing atypical hyperplasia and
abnormal maturation leading to cervical intraepithelial neoplasia
(CIN) (5). In reality, the
development of cervical cancer is a lengthy course from CIN1 to
CIN3. Although the cure rate for cervical cancer is up to 80–90% in
the early stage, the prognosis of patients with advanced stage or
recurrence is still poor (6).
Therefore, it is necessary to uncover the molecular mechanism and
to identify effective prognostic markers in cervical cancer.
Long non-coding RNAs (lncRNAs), a cluster of RNAs
longer than 200 nucleotides, are important member of the ncRNA
family, yielding little or no protein (7). It is proved that lncRNA play
important roles in physiological and pathological processes,
including genome imprinting, gene transcription, cellular
differentiation and immune response (8,9).
Emerging evidence has been shown that lncRNA modulate gene activity
in response to external oncogenic stimuli and DNA damage,
indicating the potential involvement of lncRNAs in the pathogenesis
of human diseases, especially in cancer (10). Recently, accumulating evidences
show that aberrantly expressed lncRNAs could be associated with
cervical cancer progression (11,12).
Understanding of molecular mechanisms of lncRNAs in cervical cancer
process may be greatly valuable for cervical cancer therapy. Recent
studies indicate that aberrant expression of lncRNA nuclear
paraspeckle assembly transcript 1 (NEAT1) has been documented in
different types of solid tumors (including lung cancer (13), oesophageal cancer (14), colorectal cancer (15) and hepatocellular carcinoma
(16). However, whether NEAT1 is
involved in cervical cancer remains to be elucidated.
In the current study, we aimed to explore the
expression and clinical significance of NEAT1 in cervical cancer.
Firstly, we demonstrated that overexpression of NEAT1 was found in
cervical cancer, and its level was associated with prognosis. Next,
we downregulated the NEAT1 in HeLa/Caski cells to assess the effect
of NEAT1 on cervical cancer cell growth, cell cycle, apoptosis,
migration and invasion. Then, the molecular mechanisms underlying
NEAT1 in cervical cancer were investigated. Our findings suggest
that NEAT1 serves as an important role in cervical cancer
development.
Materials and methods
Patients and clinical samples
Human cervical cancer specimens and adjacent normal
tissues were obtained from 68 patients admitted to Department of
Obstetrics and Gynecology, Xidian Group Hospital (Xi'an, China)
between August 2013 and June 2016. All included patients did not
accept any local or systemic treatment. The samples were determined
by pathologic examination and immediately placed in liquid nitrogen
followed by storage at −80°C after surgical resection. This study
was approved by the Ethics Review Board of Department of Obstetrics
and Gynecology, Xidian Group Hospital. Written informed consent was
gathered from all participants.
RNA extraction and cDNA synthesis
Total RNA from clinical samples and cells were
extracted using TRIzol reagent (Takara Bio, Inc., Otsu, Japan)
according to the manufacturer's instructions. Concentration and
purification (1.8<OD260/280<2.0) of RNA were detected using a
NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Then, the agarose gel electrophoresis was used
to determine RNA integrity. cDNA was generated from 1 µg RNA
template using Reverse Transcription kit (Invitrogen; Thermo Fisher
Scientific, Inc.).
Quantitative polymerase chain
reactions (qPCR) expression assay
qPCR was performed in a volume of 30 µl using
SYBR-Green PCR kit protocol in the ABI StepOnePlus System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). PCR cycling process
was set as follows: initial denaturation at 98°C for 5 min; 35
cycles of denaturation at 95°C for 15 sec, annealing at 55°C for 1
min, and elongation at 72°C for 1 min; and a final elongation at
72°C for 10 min. U6 snRNA was used for normalization and
2−ΔΔCq method was used to determine the relative
expression of NEAT1.
Cell culture and transfection
Cervical cancer cell lines SiHa, Caski, HeLa were
purchased from Institute of Biochemistry and Cell Biology at the
Chinese Academy of Sciences (Shanghai, China). SiHa cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco,
Shanghai, China) containing 10% fetal bovine serum (FBS; Gibco).
Caski and HeLa cells were grown in RPMI-1640 medium (Gibco)
supplemented with 10% FBS. All cells were incubated under 5%
CO2, saturated humidity at 37°C for 3 days. MicroRNA
(miRNA/miR) mimic/inhibitor and si-lncRNA were designed and
synthesized by GenePharma (Shanghai, China). For cell transfection,
cells were seeded into 24-well plates and were transfected with
miRNA or siRNA (20 nM) with Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for
transfection according to the manufacturer's instructions when cell
density reached 60–70% confluence. Cells were harvested and
subjected to subsequent analyses after 48 h transfection.
Plasmid construction and luciferase
reporter assay
Potential wild type and mutant binding sequence of
NEAT1 was synthesized by PCR-amplification and cloned into the
pmirGLO vector (Promega Corporation, Madison, WI, USA). Cells were
placed into 24-well plates and were cotransfected with wild
type/mutant pmirGLO-NEAT1 and miRNA using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were
collected and lysed with passive lysis buffer for luciferase
detection by Dual-Luciferase Reporter Assay System (Promega
Corporation) 48 h after transfection.
Cell proliferation assay
Cell viability was measured using Cell Counting
Kit-8 (CCK-8; Dojindo, Tokyo, Japan) following the manufacturer's
instructions. Briefly, 1×103 cells/well cells were
seeded onto 96-well plates, and the cell viability was measured
every 24 h. The absorbance was detected at 450 nm by using an ELx
800 Microplate Reader (Bio-Tek Instruments Inc., Winooski, VT,
USA).
Flow cytometry analysis
Cell cycle and apoptosis were measured by flow
cytometry FACSDiva (BD Biosciences, San Jose, CA, USA). For cell
cycle, cells were gathered and fixed with 70% ethanol at −20°C
overnight. Then, cells were stained propidium iodide by the
CycleTEST PLUS DNA Reagent Kit (BD Biosciences) according to the
manufacturer's introduction. Cell apoptosis was detected using
double staining with propidium iodide and Annexin V-FITC from
fluorescein Apoptosis kit (BD Biosciences) according to the
manufacturer's introduction. Data analysis was performed using
FlowJo software (Tree Star, Inc., Ashland, OR, USA).
Colony-forming assay and matrigel
invasion assay
The transfected cells were seeded into 6-well plates
at density of 1×103 cells/well. After cultured for 10
days, cells were fixed with 10% paraformaldehyde and were stained
with 0.1% crystal violet solution (Beijing Solarbio Science &
Technology Co., Ltd., Beijing, China) followed by counting the
number of colonies. For invasion assay, matrigel invasion chamber
(pore size: 8 µm, 24-well; BD Biosciences, Bedford, MA, USA) was
used. Briefly, a total of 1×105 transfected cells were
seeded in the upper wells of chambers with DMEM containing 0.1%
FBS. Normal medium (10% FBS) was added to the lower chamber. 24 h
later, the invasive cells were stained with 2% crystal violet
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and counted in 5
high-power fields under the microscopic fields.
Wound healing assay
Transfected cervical cancer cells were grown in
12-well plates when cell density reached 90% confluence. A sterile
200 µl pipette tip was used to scratch the cell monolayer. The
Cells were washed with phosphate-buffered saline to remove cell
debris followed by cultured in medium with 2% FBS. The wound
distance was measured at 0, 24 h using a microscope.
Statistical analysis
Statistical analyses were conducted using SPSS
version 17.0 software (SPSS, Inc., Chicago, IL, USA). All the data
were expressed as mean ± standard deviation and derived from at
least three independent experiments. Pearson's χ2 test
was used to assess the associations between NEAT1 expression and
the clinicopathological characteristics. The correlation between
the expression of NEAT1 and miR-101 level was determined by
Spearman's correlation analysis. Student's t-test and one-way ANOVA
were performed to compare the differences between two groups.
Tukey's test was used when it was necessary following a one-way
ANOVA. P<0.05 was considered to indicate a statistically
significant difference.
Results
Association between NEAT1 expression
and clinicopathologic factors in cervical cancer
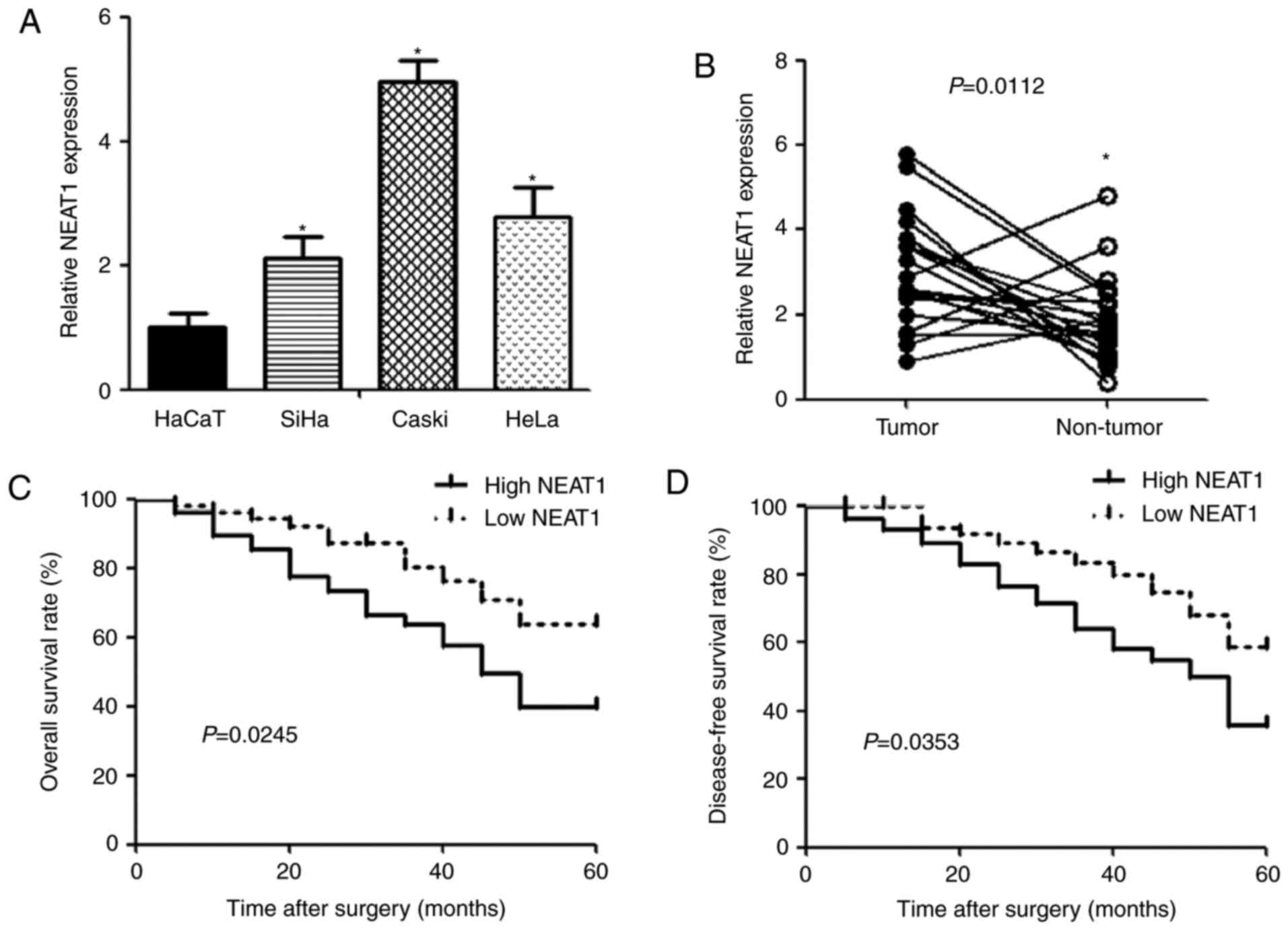
qPCR was performed to detect the expression of NEAT1
in human cervical cancer cell lines (SiHa, Caski and HeLa) and
human keratinocyte cell line HaCaT. The expression of NEAT1 was
observed to be significantly upregulated in cervical cancer cells
compared with HaCaT cells (Fig.
1A). Similarly, NEAT1 expression in cervical cancer tissues was
higher than that in noncancerous tissues (Fig. 1B). To evaluate the correlation
between NEAT1 and clinicopathologic characteristics in cervical
cancer, patients were divided into low (n=34) and high (n=34)
groups according to NEAT1 expression. We found that NEAT1 level was
positively associated with unfavorable factor, including size of
carcinoma (P=0.0014), FIGO stage (P=0.0036), depth of cervical
invasion (P=0.0034) and lymphatic metastasis (P=0.0023) (Table I). In addition, cervical cancer
patients with high NEAT1 expression had significantly shorter
overall (P=0.0245) and disease-free (P=0.0353) survival time than
those with low NEAT1 expression (Fig.
1C and D).
 | Table I.Correlations between NEAT1 expression
in cervical cancer and clinical characteristics. |
Table I.
Correlations between NEAT1 expression
in cervical cancer and clinical characteristics.
|
|
| Relative NEAT1
expression |
|
|---|
|
|
|
|
|
|---|
| Group | No. | Low | High | P-value |
|---|
| Age |
|
|
|
|
|
>50 | 36 | 20 | 16 | 0.3311 |
|
≦50 | 32 | 14 | 18 |
|
| Size of carcinoma
(cm) |
|
|
|
|
|
>4 | 29 | 8 | 21 | 0.0014 |
|
≦4 | 39 | 26 | 13 |
|
| Differentiation |
|
|
|
|
| Well and
moderate | 37 | 22 | 15 | 0.0883 |
| Poor | 31 | 12 | 19 |
|
| Histology |
|
|
|
|
|
Squamous | 50 | 27 | 23 | 0.2716 |
|
Adenocarcinoma | 18 | 7 | 11 |
|
| FIGO stage |
|
|
|
|
|
Ib-IIa | 36 | 24 | 12 | 0.0036 |
|
IIb-IIIa | 32 | 10 | 22 |
|
| Depth of cervical
invasion |
|
|
|
|
|
>2/3 | 30 | 9 | 21 | 0.0034 |
|
≦2/3 | 38 | 25 | 13 |
|
| Lymphatic
metastasis |
|
|
|
|
| No | 44 | 28 | 16 | 0.0023 |
|
Yes | 24 | 6 | 18 |
|
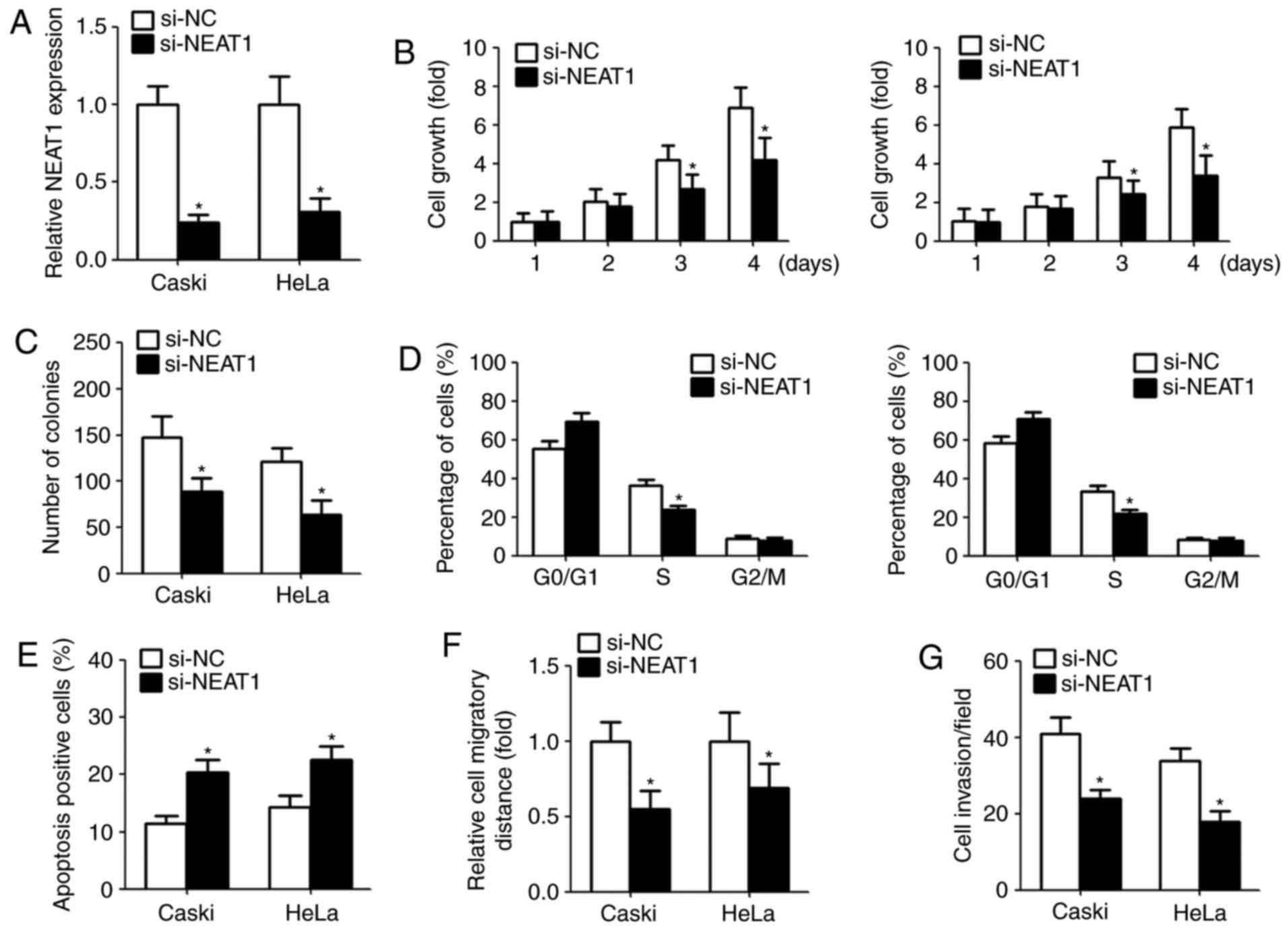
Downregulation of NEAT1 decreases
cervical cancer cells proliferation and induces cell apoptosis
Given that the aberrant overexpression of NEAT1 in
cervical cancer, we investigate the function of NEAT1 on cell
proliferation, apoptosis, colony-forming ability, migration and
invasion by transfection with NEAT1 siRNA. The suppression
efficiency in cells transfected with NEAT1 siRNAs is shown in
Fig. 2A. CCK-8 assay indicated
that NEAT1 knockdown significantly repressed the cell growth and
colony-forming capacity (Fig. 2B and
C). Moreover, we assessed the cell cycle and cell apoptosis
after transfection. Flow cytometry assay showed that downregulation
of NEAT1 in cervical cancer cells obviously decreased the
percentage of S phase and induced apoptosis compared with the
control group (Fig. 2D and E).
NEAT1 knockdown inhibits cervical
cancer migration and invasion
To explore the effect of NEAT1 on migration and
invasion, we performed wound healing and Matrigel invasion assays,
respectively. Wound healing assay indicated that downregulation of
NEAT1 obviously suppressed the width of wound closure (Fig. 2F). Similarly, following the
knockdown of NEAT1 expression, the number of invasive cells was
reduced compared with the control (Fig. 2G).
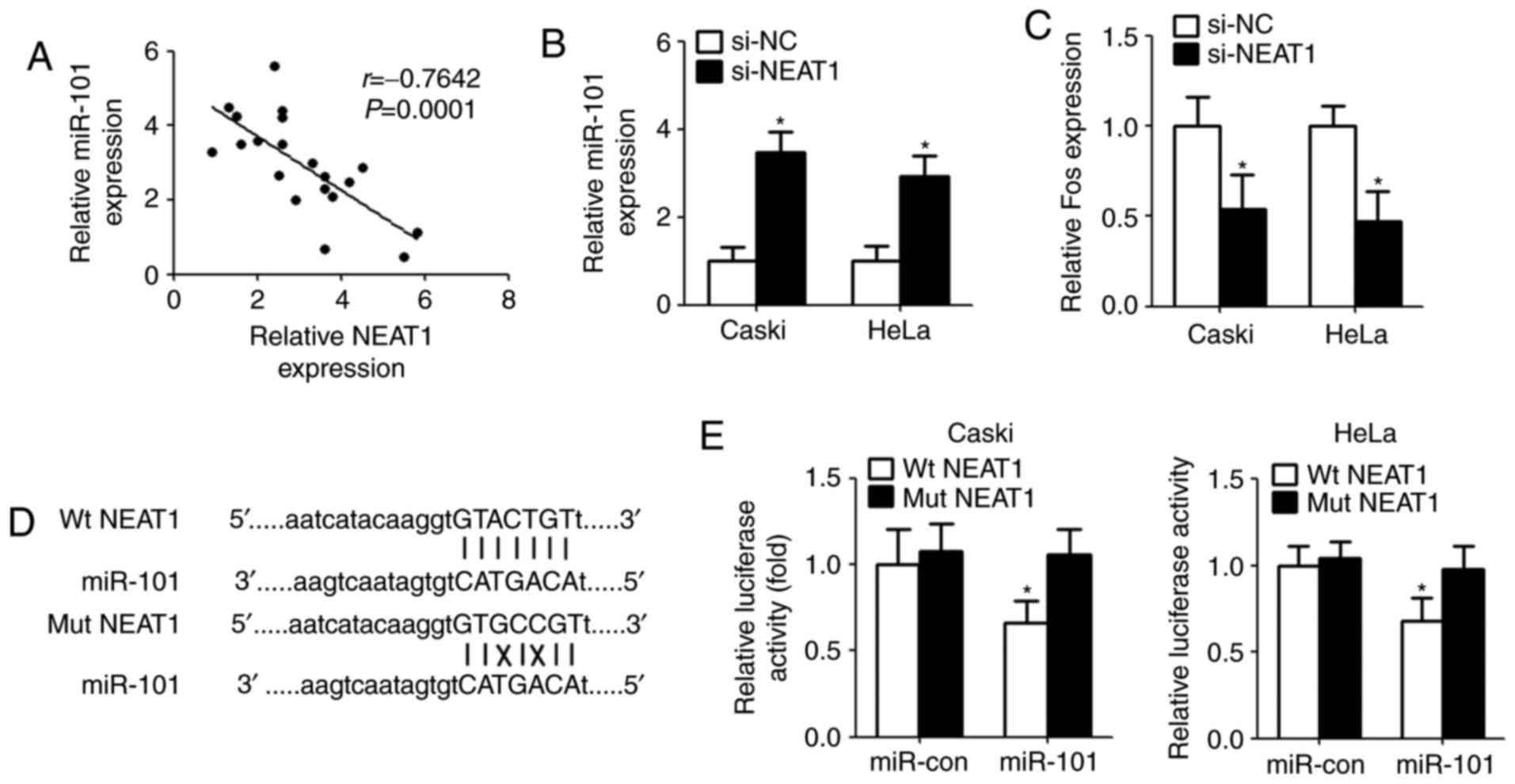
NEAT1 functions as a sponge for
miR-101 in cervical cancer cells
It was elucidated that lncRNAs serve as ‘molecular
sink’ to regulate the biological functions of miRNAs (17). In order to identify the potential
miRNA interacting with NEAT1, bioinformatics method (starBase v2.0
algorithm, http://starbase.sysu.edu.cn/index.php) was used to
predict the miRNA. The results showed that miR-101 could directly
bind with NEAT1. Then, qPCR was conducted after bioinformatics
analyses. The results demonstrated that there is a significant
negative correlation between NEAT1 and miR-101 in clinical
specimens (Fig. 3A). Next, we
measured the levels of miR-101 expression in Caski and HeLa cells
after transfection with NEAT1 siRNA. Compared with controls,
miR-101 expression was remarkably increased in cells transfected
with NEAT1 siRNA (Fig. 3B). As
expected, downregulation of NEAT1 inhibited Fos level that is a
target gene of miR-101 (Fig. 3C).
For further confirmation, we cloned the wild type/mutant fragments
including the paired bases into a pmiR-GLO vector (Fig. 3D). Luciferase reporter assay
indicated that overexpression of miR-101 significantly reduced the
luciferase activity of wild type pmirGLO-NEAT1, but not of mutant
pmirGLO-NEAT1 (Fig. 3E).
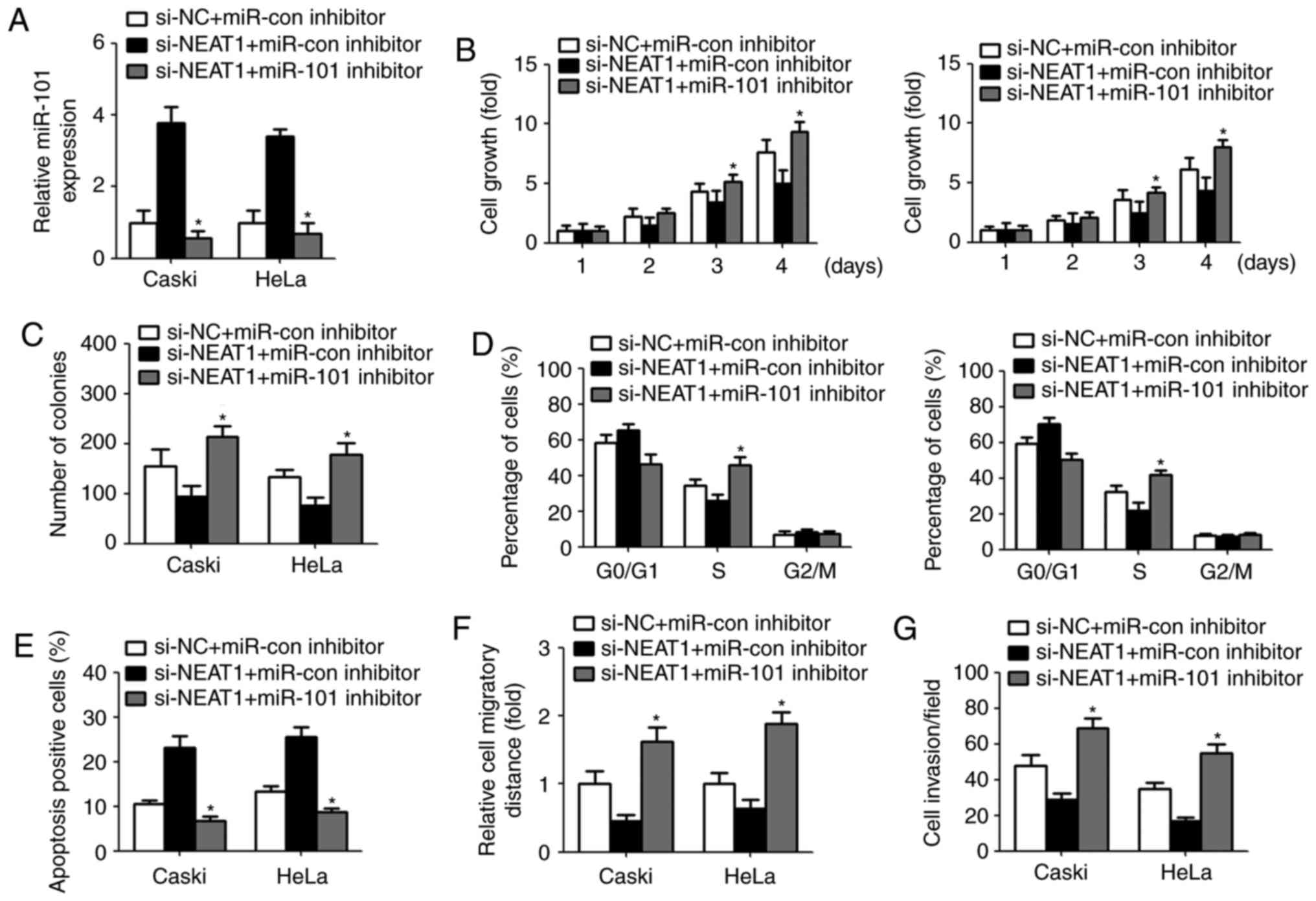
The inhibitory effect of NEAT1 on
cervical cancer cells was reversed by miR-101 inhibitor
To investigate whether NEAT1 exert its biological
effect by modulating miR-101, we repressed the miR-101 expression
by transfection with miR-101 inhibitor in cervical cancer cells.
qPCR analysis displayed a significant decreased miR-101 level after
transfection with miR-101 inhibitor (Fig. 4A). CCK-8 assay revealed that
downregulation of miR-101 restored NEAT1 siRNA-mediated inhibition
of cell growth and colony-forming ability (Fig. 4B and C). Similarly, transfection
with miR-101 inhibitor recovered the percentage of S phase and
decreased cell apoptosis (Fig. 4D and
E). Furthermore, NEAT1 siRNA-induced suppression of cell
migration and invasion were abrogated due to miR-101 inhibitor
transfection (Fig. 4F and G).
Discussion
Increasing evidences have implicated that aberrant
expression of lncRNA is closely related to the occurrence and
development of malignant tumors (10). Therefore, a further understanding
of precise regulatory mechanism of lncRNA in cervical cancer
progression is conductive to development of new diagnostic, more
targeted, and effective, therapeutic treatments. NEAT1, an
essential structural component of a nuclear domain called
paraspeckles, is located on chromosome 11q13.1 and has been
implicated in the development of corpus luteum and mammary gland as
well as myeloid differentiation (18,19).
Cumulatively, recent studies indicate that NEAT1 was aberrantly
expressed in a number of solid tumors, including non-small lung
cancer, colorectal cancer, ovarian cancer, prostate cancer and
hepatocellular cancer (13,15,16,20,21).
However, the molecular mechanisms of NEAT1 underlying the
progression and development of malignancy remain poorly
understood.
Our present study attempted to investigate the
association between NEAT1 and the clinical characteristics in human
cervical cancer. Previous studies reveal that NEAT1 is most
abundant lncRNA in cervical tissue, and minor allele ‘C’ of
rs512715 in NEAT1 was associated with an increased risk of cervical
cancer (22). In this study, our
result demonstrated that NEAT1 level was upregulated in cervical
cancer cells and tissues. Patients in the high NEAT1 group had
significantly shorter overall/disease-free survival than those in
the low NEAT1 group. It was confirmed to be an independent
prognostic marker, suggesting that NEAT1 acts as a promoter in the
development of cervical cancer.
In addition, our results demonstrated that NEAT1
knockdown inhibited cervical cancer cell growth via decreasing S
phase and increasing cell apoptosis. We also found that
downregulation of NEAT1 inhibited the migratory and invasive
ability of cervical cancer cells. These results are in accordance
with above clinical data, indicating that NEAT1 is a novel booster
in the progression of cervical cancer. NEAT1 have been widely
investigated and plays an oncogenic role in various solid tumor. It
is recognized that the mechanism by which NEAT1 mediates its
function is complex and involves multiple factors, including
sponging of tumor-suppressive microRNAs (23–25).
miR-101 usually acts as a suppressor that inhibits the
tumorigenicity of various cancer cell (26–29).
Recent studies indicated that miR-101 is obviously downregulated in
cervical cancer, miR-101 is a kind of miRNAs which act as a
potential tumor suppressor in cervical cancer (30). Furthermore, miR-101 inhibits the
G1-to-S phase transition of cervical cancer cells by targeting the
proto-oncogene Fos (31).
Bioinformatics prediction indicated that NEAT1 may
act as a miR-101 sponge. Our results demonstrated a significant
negative correlation between NEAT1 expression and miR-101 level in
clinical samples. In vitro, downregulation of NEAT1 elevated
miR-101 level leading to a reduction of Fos expression. Luciferase
reporter assay confirmed that NEAT1 could directly bind with
miR-101 to control its expression in cervical cancer cells. Recent
study suggests that NEAT1 promotes breast cancer cell growth and
DNA synthesis through targeting miR-101. Our results lend credence
to the previous study suggesting that NEAT1 interacted with miR-101
by directly targeting. Moreover, transfection with miR-101
inhibitor abolished NEAT1 siRNA-mediated effect on cell
proliferation, apoptosis, migration and invasion. These results
imply that NEAT1, acting as a miR-101 sponge, accelerate cervical
cancer progression via suppressing miR-101.
In conclusion, current study found that NEAT1 acts a
sponge of miR-101 in cervical cancer, and NEAT1 level is
upregulated in cervical cancer, where it correlates with
unfavorable clinicopathologic factors and poor prognosis. NEAT1
plays a crucial role in cervical cancer cell proliferation,
apoptosis, migration and invasion targeting miR-101. Taken
together, NEAT1 is an important molecular biomarker for predicting
prognosis and a potential target for cervical cancer therapy.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen J, Fu Z, Ji C, Gu P, Xu P, Yu N, Kan
Y, Wu X, Shen R and Shen Y: Systematic gene microarray analysis of
the lncRNA expression profiles in human uterine cervix carcinoma.
Biomed Pharmacother. 72:83–90. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sakuragi N: Refining insight into cervical
cancer progression. Lancet Oncol. 15:371–372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schiffman M, Wentzensen N, Wacholder S,
Kinney W, Gage JC and Castle PE: Human papillomavirus testing in
the prevention of cervical cancer. J Natl Cancer Inst. 103:368–383.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baak JP, Kruse AJ, Robboy SJ, Janssen EA,
van Diermen B and Skaland I: Dynamic behavioural interpretation of
cervical intraepithelial neoplasia with molecular biomarkers. J
Clin Pathol. 59:1017–1028. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee M, Kim HJ, Kim SW, Park SA, Chun KH,
Cho NH, Song YS and Kim YT: The long non-coding RNA HOTAIR
increases tumour growth and invasion in cervical cancer by
targeting the Notch pathway. Oncotarget. 7:44558–44571. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mattick JS: The genetic signatures of
noncoding RNAs. PLoS Genet. 5:e10004592009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calore F, Lovat F and Garofalo M:
Non-coding RNAs and cancer. Int J Mol Sci. 14:17085–17110. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Liu SC, Luo XH, Tao GX, Guan M,
Yuan H and Hu DK: Exosomal long noncoding RNAs are differentially
expressed in the cervicovaginal lavage samples of cervical cancer
patients. J Clin Lab Anal. 30:1116–1121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gibb EA, Becker-Santos DD, Enfield KS,
Guillaud M, Niekerk Dv, Matisic JP, Macaulay CE and Lam WL:
Aberrant expression of long noncoding RNAs in cervical
intraepithelial neoplasia. Int J Gynecol Cancer. 22:1557–1563.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jen J, Tang YA, Lu YH, Lin CC, Lai WW and
Wang YC: Oct4 transcriptionally regulates the expression of long
non-coding RNAs NEAT1 and MALAT1 to promote lung cancer
progression. Mol Cancer. 16:1042017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X, Kong J, Ma Z, Gao S and Feng X: Up
regulation of the long non-coding RNA NEAT1 promotes esophageal
squamous cell carcinoma cell progression and correlates with poor
prognosis. Am J Cancer Res. 5:2808–2815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng W, Wang Z and Fan H: LncRNA NEAT1
impacts cell proliferation and apoptosis of colorectal cancer via
regulation of akt signaling. Pathol Oncol Res. 23:651–656. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mang Y, Li L, Ran J, Zhang S, Liu J, Li L,
Chen Y, Liu J, Gao Y and Ren G: Long noncoding RNA NEAT1 promotes
cell proliferation and invasion by regulating hnRNP A2 expression
in hepatocellular carcinoma cells. Onco Targets Ther. 10:1003–1016.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ergun S and Oztuzcu S: Oncocers:
ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways.
Tumour Biol. 36:3129–3136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sasaki YT, Ideue T, Sano M, Mituyama T and
Hirose T: MENepsilon/beta noncoding RNAs are essential for
structural integrity of nuclear paraspeckles. Proc Natl Acad Sci
USA. 106:pp. 2525–2530. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen ZJ, Zhang Z, Xie BB and Zhang HY:
Clinical significance of up-regulated lncRNA NEAT1 in prognosis of
ovarian cancer. Eur Rev Med Pharmacol Sci. 20:3373–3377.
2016.PubMed/NCBI
|
|
21
|
Chakravarty D, Sboner A, Nair SS,
Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K,
Kossai M, et al: The oestrogen receptor alpha-regulated lncRNA
NEAT1 is a critical modulator of prostate cancer. Nat Commun.
5:53832014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fang J, Li Y, Zhang J, Yan M, Li J, Bao S
and Jin T: Correlation between polymorphisms in microRNA-regulated
genes and cervical cancer susceptibility in a Xinjiang Uygur
population. Oncotarget. 8:31758–31764. 2017.PubMed/NCBI
|
|
23
|
Qian K, Liu G, Tang Z, Hu Y, Fang Y, Chen
Z and Xu X: The long non-coding RNA NEAT1 interacted with miR-101
modulates breast cancer growth by targeting EZH2. Arch Biochem
Biophys. 615:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li JH, Zhang SQ, Qiu XG, Zhang SJ, Zheng
SH and Zhang DH: Long non-coding RNA NEAT1 promotes malignant
progression of thyroid carcinoma by regulating miRNA-214. Int J
Oncol. 50:708–716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang B, Liu C, Wu Q, Zhang J, Min Q,
Sheng T, Wang X and Zou Y: Long non-coding RNA NEAT1 facilitates
pancreatic cancer progression through negative modulation of
miR-506-3p. Biochem Biophys Res Commun. 482:828–834. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang R, Zhang C, Liu G, Gu R and Wu H:
MicroRNA-101 inhibits proliferation, migration and invasion in
osteosarcoma cells by targeting ROCK1. Am J Cancer Res. 7:88–97.
2017.PubMed/NCBI
|
|
27
|
Wang L, Yao J, Sun H, He K, Tong D, Song T
and Huang C: MicroRNA-101 suppresses progression of lung cancer
through the PTEN/AKT signaling pathway by targeting DNA
methyltransferase 3A. Oncol Lett. 13:329–338. 2017.PubMed/NCBI
|
|
28
|
Liu J, Pang Y, Wang H, Li Y, Sun X, Xu F,
Ren H and Liu D: miR-101 inhibits the proliferation and migration
of breast cancer cells via downregulating the expression of DNA
methyltransferase 3a. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi.
32:299–303. 2016.(In Chinese). PubMed/NCBI
|
|
29
|
Zheng HB, Zheng XG and Liu BP: miRNA-101
inhibits ovarian cancer cells proliferation and invasion by
down-regulating expression of SOCS-2. Int J Clin Exp Med.
8:20263–20270. 2015.PubMed/NCBI
|
|
30
|
Huang F, Lin C, Shi YH and Kuerban G:
MicroRNA-101 inhibits cell proliferation, invasion and promotes
apoptosis by regulating cyclooxygenase-2 in Hela cervical carcinoma
cells. Asian Pac J Cancer Prev. 14:5915–5920. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang X, Liu Y, Zeng L, Yu C, Hu Z, Zhou Q
and Yang Z: miR-101 inhibits the G1-to-S phase transition of
cervical cancer cells by targeting Fos. Int J Gynecol Cancer.
24:1165–1172. 2014. View Article : Google Scholar : PubMed/NCBI
|