Introduction
Lung cancer is the leading cause of
cancer-associated mortality in the Chinese population. In 2015, an
estimated 4292,000 new cancer cases and 2814,000 cancer deaths
occurred in China, with lung cancer being the most common cancer
and the leading cause of cancer death (1). Lung cancer is among the most common
types of cancer, followed by stomach, colorectal, liver and
esophageal cancer. In 2015, lung cancer was the most frequent type
of cancer in male, followed by stomach, liver, colorectal and
esophageal cancer; among females, breast cancer was the most
frequent type of cancer, followed by lung, colorectal, stomach and
liver cancer (1–3). Therefore, the identification of
potential biomarkers for the early diagnosis and prognosis of lung
cancer is imperative.
Collagen and calcium-binding epidermal growth factor
domain-containing protein (CCBE) 1 has been identified as a novel
lymphangiogenic signaling factor in zebrafish (4). Mutations in CCBE1 have been reported
to cause thoracic duct and dorsal lymphatic vessel malformation;
however, they had no effect on blood vasculature (5). During embryogenesis, CCBE1 and
vascular endothelial growth factor-C (VEGF-C) were demonstrated to
stimulate angiogenic sprouting and lymphangiogenic budding from the
venous endothelium (4). CCBE1 was
revealed to be necessary for lymphangiogenesis; however, it was not
identified as a component of the VEGF-C/VEGF receptor 3 (VEGFR3)
signaling pathway or the sex determining region Y-box 18
(SOX18)/prospero homeobox protein 1 (Prox1) transcriptional pathway
(6). Therefore, it was suggested
that CCBE1 may function as an independent regulator of
lymphangioblast budding and, possibly, migration (7). CCBE1 mutations in zebrafish, similar
to VEGF-C, Prox1 and SOX18 mutations, were demonstrated to cause
specific loss of embryonic lymphatic vasculature (7).
CCBE1-deficient mouse embryos were reported to lack
lymphatic structures, thus suggesting a critical role for CCBE1 in
lymphangiogenesis (6). In
addition, CCBE1 has been identified as one of the causal genes that
may be responsible for primary generalized lymph vessel dysplasia
associated with the autosomal recessive Hennekam syndrome in
humans, which is associated with lymphedema, lymphangiectasia,
mental retardation and unusual facial characteristics. It has been
suggested that CCBE1 mutations may cause a similar phenotype, thus
making CCBE1 a candidate gene implicated in human generalized
lymphatic dysplasia (8).
It has previously been reported that in zebrafish
CCBE1 may interact genetically with VEGF-C and VEGFR3 (4), whereas a VEGF-C mutation has been
implicated in inherited lymphedema (9). CCBE1 has been demonstrated to
function during the same developmental stages as VEGF-C to promote
lymphatic sprouting; however, the biochemical roles of CCBE1 in
lung cancer have yet to be elucidated (5). CCBE1 has been suggested to function
as a tumor suppressor in ovarian cancer, since low CCBE1 gene
expression has been associated with the degree of tumor
differentiation and the pathologic stage of the disease, as well as
with a low survival rate (8).
Furthermore, it has been demonstrated that CCBE1 expression levels
were downregulated in laryngocarcinoma, thus suggesting that CCBE1
may have potential as a novel biomarker for the estimation of
laryngeal cancer prognosis (10).
In addition, evaluation of the lymphatic expression of CCBE1 may
also have potential use in colorectal cancer prognosis (11). Moreover, a positive association
between the expression of CCBE1 and LYVE1 was reported in lung
cancer (12). The present study
investigated the expression of CCBE1 and lymphatic vessel
endothelial hyaluronan receptor 1 (LYVE1) in tissue samples from
patients with lung cancer, to evaluate the potential of CCBE1 as a
biomarker for the diagnosis of lung cancer.
Materials and methods
Patients
A total of 50 patients, including 40 patients with
lung cancer and 10 patients with pulmonary bullae, were enrolled in
the present study between May and July 2013 in Jinling Hospital
(Nanjing, China). Among the patients with lung cancer, 10 patients
exhibited evidence of lymph node metastasis (LNM) and 30 patients
did not. The patients did not receive any preoperative anticancer
treatment. Patients with lung cancer underwent pulmonary carcinoma
resection, whereas control patients underwent pulmonary bullae
resection. The study population consisted of 30 males and 20
females with a mean age of 53.35 years (range, 45–64 years). The
lung tissues were obtained from the lung bullae from lung cancer
patients and normal controls.
The present study was approved by the Ethics
Committee of Southern Medical University (Guangzhou, China).
Written informed consent was obtained from all human subjects prior
to enrollment in the present study.
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was extracted from tissue samples isolated
from patients with lung cancer or pulmonary bullae using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's
instructions. Total RNA (2 µg) was reverse transcribed (20 µl
reaction volume) into cDNA using a reverse transcription system, as
previously described (13). The
following primers, designed using the Primer Premier software
version 5.0 (Premier Biosoft International, Palo Alto, CA, USA) and
synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) were
employed: CCBE1 forward, 5′-CGACTAAATACCCGTGTCTGAAG-3′ and reverse,
5′-TCGGCACAAACGTCGTAATCT-3′; β-actin forward,
5′-GCTCGTCGTCGACAACGGCTC-3′ and reverse,
5′-CAAACATGATCTGGGTCACTTCTC-3′. Amplification was performed under
the following conditions: Initial incubation at 95°C for 10 sec,
followed by 40 cycles at 95°C for 5 sec and at 62°C for 45 sec, and
extension at 72°C for 3 min. The 25-µ LlPCR reaction system
included 1 µl temple, 2 µl primers, 2 µl dNTP, 0.5 µl
RT/Platinum™ Taq Mix (Invitrogen; Thermo Fisher
Scientific, Inc.) and distilled water. PCR products were detected
by 2% agarose gel electrophoresis as previously described (13).
Western blot analysis
The protein was extracted by
radioimmunoprecipitation assay buffer cell lysis (containing PMSF)
(Invitrogen; Thermo Fisher Scientific, Inc.). Protein concentration
was quantified using BCA protein assay kit (Beyotime Institute of
Biotechnology, Shanghai, China) 20 µg protein were separated by 10%
SDS-PAGE and transferred onto nitrocellulose membranes. The
membranes were blocked by 5% skimmed milk in PBST (1% Tween-20) at
room temperature for 2 h. Membranes were probed with anti-CCBE1
(1:1,000, cat. no. HPA041361; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), anti-LYVE1 (1:1,000, cat. no. ab10278; Abcam, Shanghai,
China) and anti-β-actin (1:5,000, cat. no. ab8227; Abcam) primary
antibodies at 4°C overnight. Subsequently, membranes were incubated
with horseradish peroxidase-conjugated aoat anti-rabbit
immunoglobulin G secondary antibodies (1:10,000, cat. no. TA130023;
OriGene Technologies, Inc., Beijing, China) at room temperature for
2 h. Protein bands were visualized using an enhanced
chemiluminescence detection kit. The blots were semi-quantified by
densitometry analysis using the Multi-Analyst® software
version 2.0.1 (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Immunohistochemistry
Immunohistochemical staining was performed using a
two-step method with heat-induced antigen-retrieval procedures
(14). The cancer tissues were
fixed in 4% paraformaldehyde. Following dehydration, the tissue was
sliced and blocked in 10% BSA (Sigma-Aldrich; Merck KGaA). The
following primary antibodies were used: Rabbit anti-human CCBE1
monoclonal antibody (cat. no. HPA041361, 1:50-1:200; Sigma-Aldrich,
Merck KGaA) and rabbit anti-human LYVE1 monoclonal antibody (cat.
no. ab33682, 1:100; Abcam). The slices (20 µm) were incubated with
horseradish peroxidase-conjugated secondary antibodies (1:100,
peroxidase conjugated goat anti-rabbit IgG, TA130023; OriGene
Technologies, Inc.) at room temperature for 2 h. The primary
antibody was applied in the negative control. The images were
visualized by light microscope (Olympus Corporation, Tokyo,
Japan).
Statistical analysis
Data are expressed as the mean ± standard deviation
SPSS software was used for analysis (version 17.0; SPSS, Inc.,
Chicago, IL, USA). One-way analysis of variance was used to analyze
the difference among three or more groups followed by Tukey's test.
χ2 analysis was used to assess disease recurrence in
patients with or without LNM. The association of LNM and disease
recurrence over time was confirmed by Kaplan-Meier analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
CCBE1 and LYVE1 expression in lung
cancer tissue
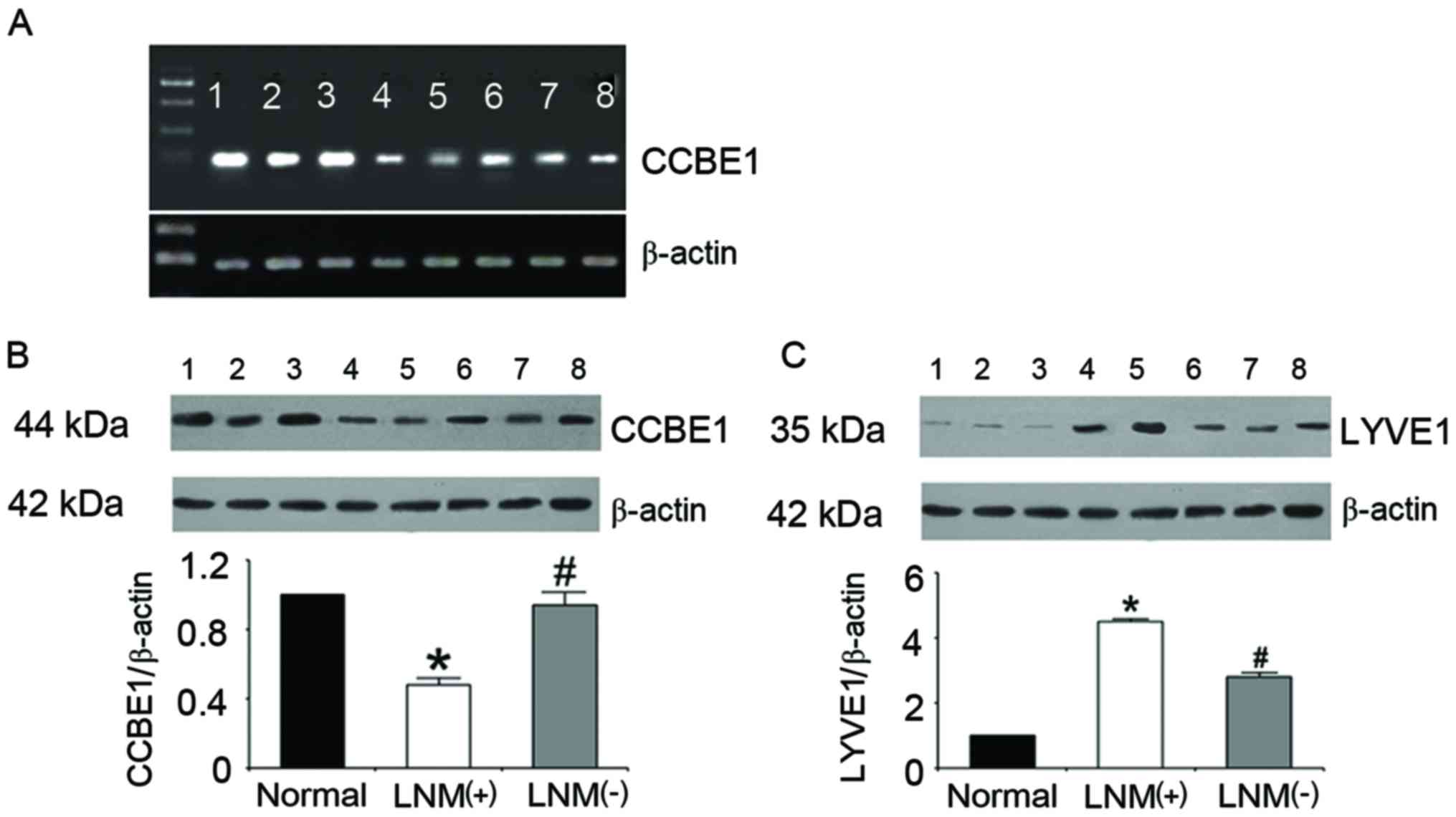
RT-PCR analysis indicated that the mRNA expression
levels of CCBE1 were reduced in lung cancer tissue compared with
normal tissue (pulmonary bullae). Furthermore, tissue samples from
patients with LNM exhibited reduced CCBE1 mRNA expression levels
compared with in samples from patients with cancer without LNM
(Fig. 1A). CCBE1 protein
expression levels exhibited a similar trend, as revealed using
western blot analysis (Fig. 1B).
Conversely, the protein expression levels of the lymphatic-specific
marker LYVE1 were revealed to be significantly upregulated in
tissue samples from patients with lung cancer compared with in
normal tissue samples. Furthermore, LYVE1 levels were significantly
increased in samples from patients with LNM compared with patients
without LNM (Fig. 1C).
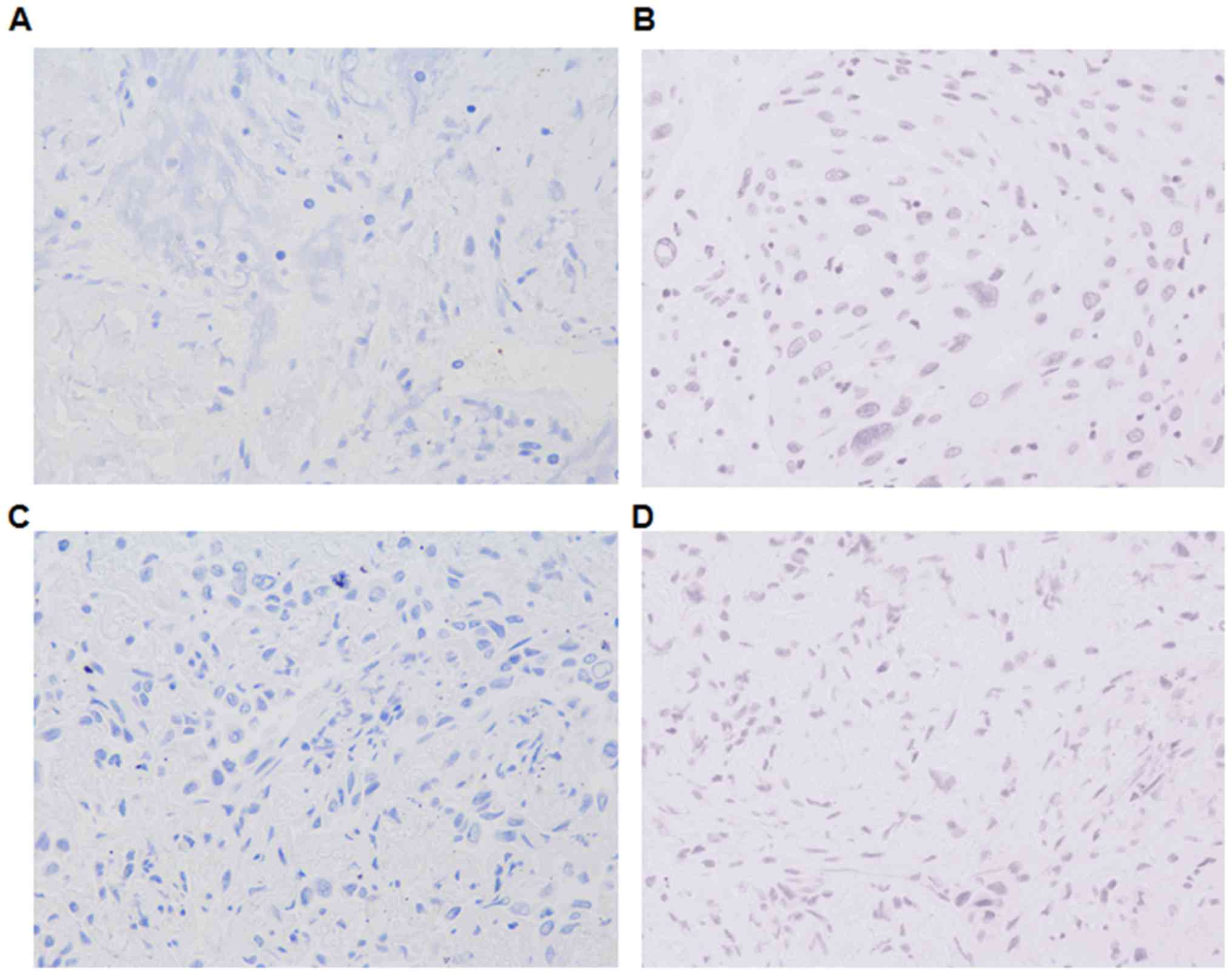
Consistent with the immunoblotting data,
immunohistochemical analysis indicated that CCBE1 expression
appeared to be downregulated in tissue samples from patients with
lung cancer compared with in normal tissue samples. In addition,
patients with LNM appeared to exhibit reduced CCBE1 expression
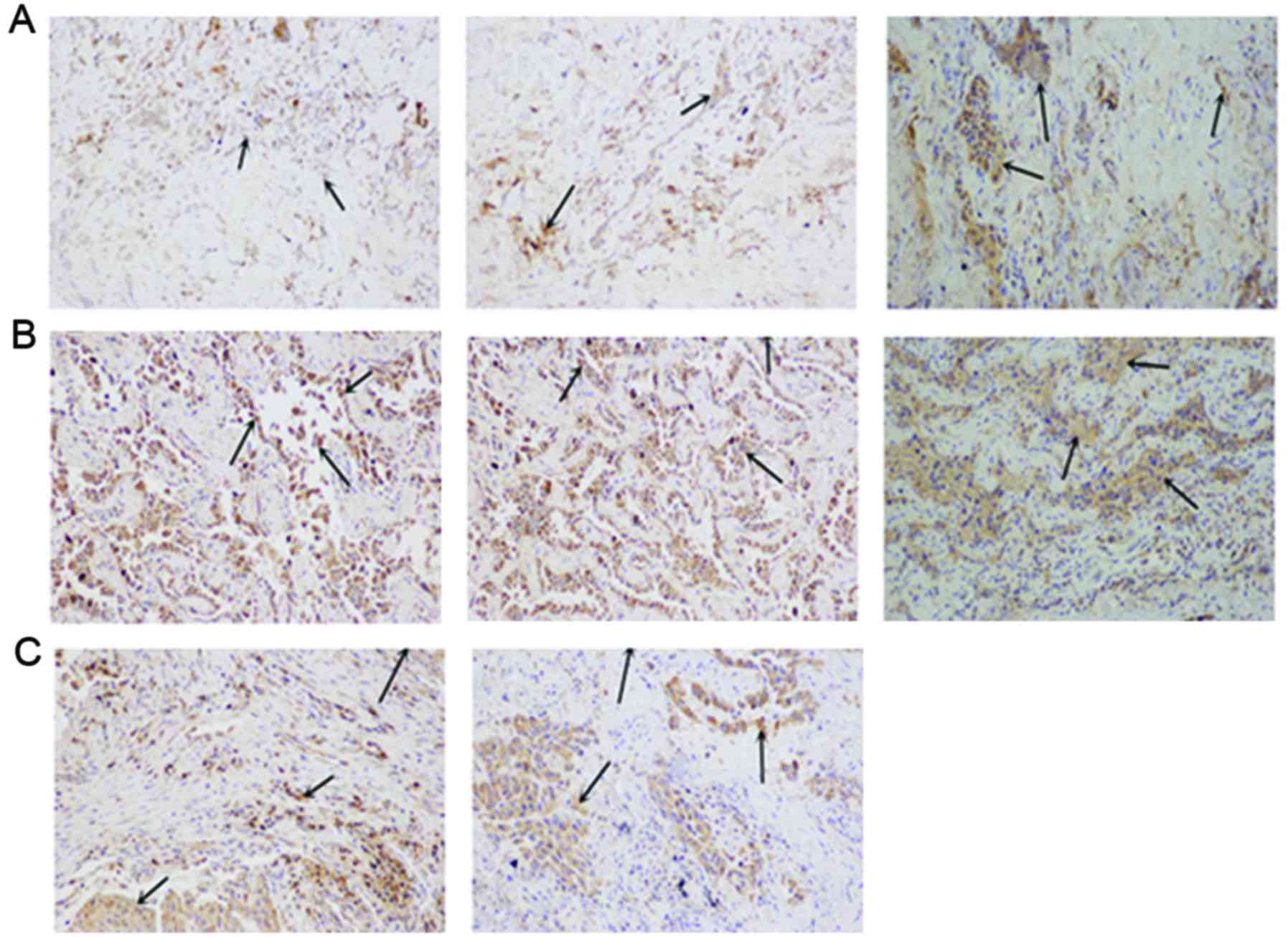
compared with patients with cancer without LNM (Fig. 2). LYVE1 appeared to be expressed at
higher levels in lung cancer samples compared with in normal tissue
samples; LYVE1 appeared to be upregulated in samples from patients
with LNM compared with in patients without LNM (Fig. 3). Thus, there was an inverse
association between expression of CCBE1 and LYVE-1 in normal and
tumor-derived tissues.
Association between CCBE1 expression
and disease prognosis
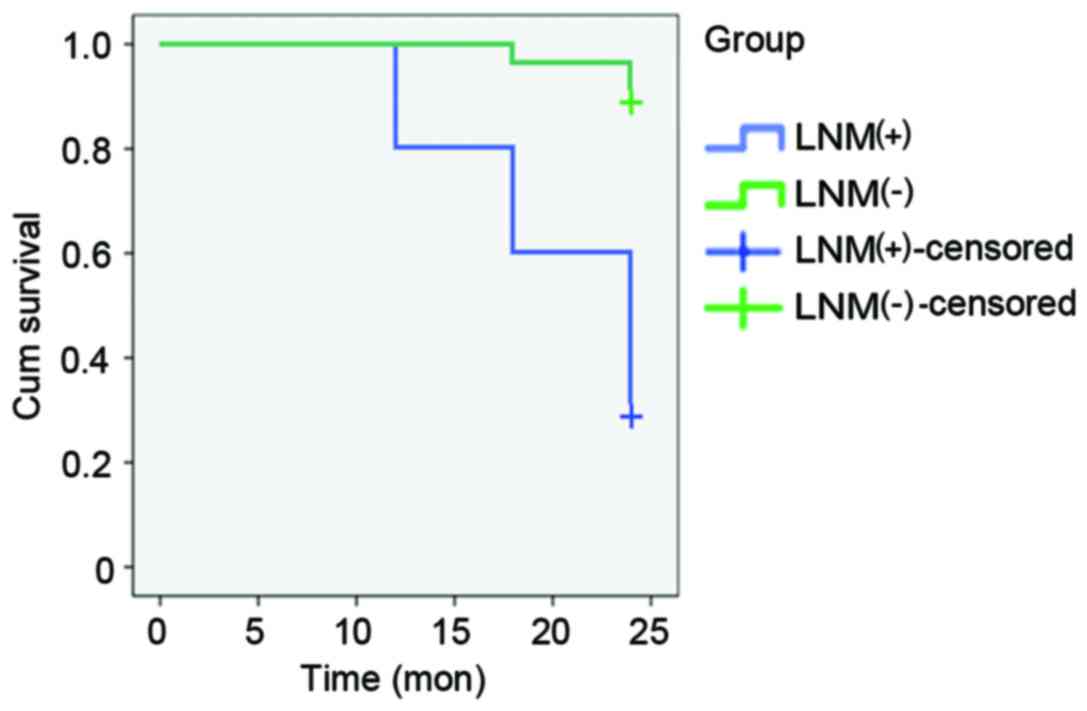
Patients with lung cancer that were enrolled in the
present study were followed up at 6-month intervals for 2 years. No
significant differences in clinical prognosis were revealed 6
months after pulmonary carcinoma resection among patients with or
without LNM; however, 12 months after surgery, two patients with
LNM exhibited recurrent lung cancer. A total of 18 months after
surgery, 2 patients with LNM exhibited disease recurrence, as well
as 1 patient without LNM. By 24 months, an additional 3 patients
with LNM exhibited disease recurrence, as well as an additional 2
patients without LNM. χ2 analysis of the proportion of
disease recurrence in patients with or without LNM revealed
significantly higher rates of recurrence among patients with LNM
(P<0.05) at 12, 18 and 24 months post-surgery
(χ2=6.32, 9.22 and 14.4, respectively; Table I). Prior to 12 months, no
significant differences were observed. The association between LNM
and disease recurrence over time was confirmed by Kaplan-Meier
analysis (Fig. 4).
 | Table I.Results of χ2 analysis of
cancer recurrence (n=40 patients). |
Table I.
Results of χ2 analysis of
cancer recurrence (n=40 patients).
| Time post-surgery
(months) | Group A LNM (+) | Group B LNM (−) | χ2
value | P-value |
|---|
| 12 |
|
| 6.32 | 0.012 |
|
Recurrence | 2 | 0 |
|
|
| No
recurrence | 8 | 30 |
|
|
| 18 |
|
| 9.22 | 0.002 |
|
Recurrence | 4 | 1 |
|
|
| No
recurrence | 6 | 29 |
|
|
| 24 |
|
| 14.40 | <0.01 |
|
Recurrence | 7 | 3 |
|
|
| No
recurrence | 3 | 27 |
|
|
Discussion
In the present study, mRNA and protein expression
levels of CCBE1 and LYVE1 were investigated in patients with lung
cancer. CCBE1 expression was revealed to be downregulated in lung
cancer tissue. Notably, tumor tissue isolated from patients with
LNM exhibited the lowest CCBE1 expression. Conversely, LYVE1
expression was revealed to be upregulated in tissue samples
isolated from patients with lung cancer; similarly, its expression
appeared to be highest in tissue samples form patients with LNM.
These results suggested that CCBE1 may have potential as a
diagnostic biomarker for lung cancer and LNM.
Previous studies have suggested that CCBE1 may
participate in extracellular matrix remodeling and cellular
migration (15). CCBE1 mutations
have been reported to cause lymphatic dysplasia,
lymphedema-cholestasis syndrome and fetal hydrops (16,17).
In ovarian cancer cell lines and primary carcinomas, CCBE1 appeared
to be downregulated, thus suggesting that it may serve a role as a
tumor suppressor (15). In the
present study, CCBE1 expression was revealed to be downregulated in
tissue samples from patients with lung cancer. Notably, samples
isolated from patients with lung cancer accompanied by LNM
exhibited the lowest CCBE1 expression levels, thus suggesting that
lung cancer progression may be associated with a decrease in CCBE1
expression.
The collagen domains in the CCBE1 structure are
implicated in the activation of VEGF-C (18), which is a critical molecule for
angiogenesis and tumor metastasis (19). LYVE1 acts as a receptor for
hyaluronan and binds to the soluble and immobilized form of the
protein. It has previously been suggested that LYVE1 may
participate in lymphatic hyaluronan transport and serve a role in
tumor metastasis (20). The
evolutionary conservation of LYVE1 suggests an important role for
this protein in lymph node metastasis (21); however, its physiological role has
yet to be elucidated. It has previously been demonstrated that
immunocytochemical staining for LYVE1 was able to recognize
lymphatic vessel endothelium and pancreatic endocrine cells
(22). In the present study, LYVE1
protein expression levels were significantly upregulated in
patients with lung cancer, particularly among patients with
LNM.
The present results suggested that CCBE1 expression
was downregulated, whereas LYVE1 expression was upregulated in lung
cancer tissue. Notably, a previous study reported a positive
correlation between the expression of CCBE1 and LYVE1 (12). However, the positive correlation
has been established in normal tissue. Therefore, it may be
hypothesized that in cancer tissue, CCBE1 downregulation can
stimulate LYVE1 to promote LNM.
The present study demonstrated the association
between LNM and disease recurrence. The present findings revealed a
direct correlation between CCBE1 expression and disease recurrence,
suggesting that reduced CCBE1 expression may be associated with an
increased risk of disease recurrence in patients with lung cancer.
Therefore, the expression of CCBE1 in tumor tissue from lung cancer
patients may have potential as a biomarker for the prognosis of
lung cancer and for evaluating the risk of LNM.
In conclusion, the present results demonstrated that
CCBE1 expression was downregulated in lung cancer, particularly in
the presence of LNM. Therefore, it may be hypothesized that CCBE1
has potential as a biomarker to evaluate lung cancer prognosis and
assess the risk of lymph node metastasis.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81172032).
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu
L and He J: Report of incidence and mortality in China cancer
registries, 2009. Chin J Cancer Res. 25:10–21. 2013.PubMed/NCBI
|
|
3
|
Parkin DM, Pisani P, Lopez AD and Masuyer
E: At least one in seven cases of cancer is caused by smoking.
Global estimates for 1985. Int J Cancer. 59:494–504. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Le Guen L, Karpanen T, Schulte D, Harris
NC, Koltowska K, Roukens G, Bower NI, van Impel A, Stacker SA,
Achen MG, et al: Ccbe1 regulates Vegfc-mediated induction of Vegfr3
signaling during embryonic lymphangiogenesis. Development.
141:1239–1249. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stacker SA, Williams SP, Karnezis T,
Shayan R, Fox SB and Achen MG: Lymphangiogenesis and lymphatic
vessel remodelling in cancer. Nat Rev Cancer. 14:159–172. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bos FL, Caunt M, Peterson-Maduro J,
Planas-Paz L, Kowalski J, Karpanen T, van Impel A, Tong R, Ernst
JA, Korving J, et al: CCBE1 is essential for mammalian lymphatic
vascular development and enhances the lymphangiogenic effect of
vascular endothelial growth factor-C in vivo. Circ Res.
109:486–491. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hogan BM, Bos FL, Bussmann J, Witte M, Chi
NC, Duckers HJ and Schulte-Merker S: Ccbe1 is required for
embryonic lymphangiogenesis and venous sprouting. Nat Genet.
41:396–398. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alders M, Hogan BM, Gjini E, Salehi F,
Al-Gazali L, Hennekam EA, Holmberg EE, Mannens MM, Mulder MF,
Offerhaus GJ, et al: Mutations in CCBE1 cause generalized lymph
vessel dysplasia in humans. Nat Genet. 41:1272–1274. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gordon K, Schulte D, Brice G, Simpson MA,
Roukens MG, van Impel A, Connell F, Kalidas K, Jeffery S, Mortimer
PS, et al: Mutation in vascular endothelial growth factor-C, a
ligand for vascular endothelial growth factor receptor-3, is
associated with autosomal dominant milroy-like primary lymphedema.
Circ Res. 112:956–960. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Li W, Zhang D, Kang S, Ding L and
Zhao S: The expression and clinical significance of CCBE1 in throat
cancer. J Shanxi Med Uni. 43:58–61. 2012.
|
|
11
|
Li W, Wang Y, Liu Y, Yin G and Feng K:
Prognosis of CCBE1 expression in colorectal cancer. Chin J
Gerontol. 7:3849–3850. 2014.
|
|
12
|
Jeltsch M, Jha SK, Tvorogov D, Anisimov A,
Leppänen VM, Holopainen T, Kivelä R, Ortega S, Kärpanen T and
Alitalo K: CCBE1 enhances lymphangiogenesis via A disintegrin and
metalloprotease with thrombospondin motifs-3-mediated vascular
endothelial growth factor-C activation. Circulation. 129:1962–1971.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu G, Li J, He L, Wang X and Hong X:
MPTP-induced changes in hippocampal synaptic plasticity and memory
are prevented by memantine through the BDNF-TrkB pathway. Br J
Pharmacol. 172:2354–2368. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu S, Cao J, Zhang T, Zhou Y, Wang K, Zhu
G and Zhou M: Electroacupuncture ameliorates the coronary occlusion
related tachycardia and hypotension in acute rat myocardial
ischemia model: Potential role of hippocampus. Evid Based
Complement Alternat Med. 2015:9259872015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barton CA, Gloss BS, Qu W, Statham AL,
Hacker NF, Sutherland RL, Clark SJ and O'Brien PM: Collagen and
calcium-binding EGF domains 1 is frequently inactivated in ovarian
cancer by aberrant promoter hypermethylation and modulates cell
migration and survival. Br J Cancer. 102:87–96. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Connell FC, Kalidas K, Ostergaard P, Brice
G, Murday V, Mortimer PS, Jeffrey I, Jeffery S and Mansour S: CCBE1
mutations can cause a mild, atypical form of generalized lymphatic
dysplasia but are not a common cause of non-immune hydrops fetalis.
Clin Genet. 81:191–197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shah S, Conlin LK, Gomez L, Aagenaes Ø,
Eiklid K, Knisely AS, Mennuti MT, Matthews RP, Spinner NB and Bull
LN: CCBE1 mutation in two siblings, one manifesting
lymphedema-cholestasis syndrome, and the other, fetal hydrops. PLoS
One. 8:e757702013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Roukens MG, Peterson-Maduro J, Padberg Y,
Jeltsch M, Leppänen VM, Bos FL, Alitalo K, Schulte-Merker S and
Schulte D: Functional dissection of the CCBE1 protein: A crucial
requirement for the collagen repeat domain. Circ Res.
116:1660–1669. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu HT, Xing AY, Chen X, Ma RR, Wang YW,
Shi DB, Zhang H, Li P, Chen HF, Li YH and Gao P: MicroRNA-27b,
microRNA-101 and microRNA-128 inhibit angiogenesis by
down-regulating vascular endothelial growth factor C expression in
gastric cancers. Oncotarget. 6:37458–37470. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jackson DG: The lymphatics revisited: New
perspectives from the hyaluronan receptor LYVE-1. Trends Cardiovasc
Med. 13:1–7. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ramani P, Dungwa JV and May MT: LYVE-1
upregulation and lymphatic invasion correlate with adverse
prognostic factors and lymph node metastasis in neuroblastoma.
Virchows Arch. 460:183–191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tomita T: Lymphatic vessel endothelial
hyaluronan receptor 1 immunocytochemical staining for pancreatic
islets and pancreatic endocrine tumors. Pancreas. 35:e18–e22. 2007.
View Article : Google Scholar : PubMed/NCBI
|