Introduction
Colorectal cancer is one of the most common types of
cancer worldwide and the incidence is increasing (1). Although progress has been made with
regard to treatment of colorectal cancer, patient survival remains
poor (2). Currently, the primary
drugs used for the treatment of colorectal cancer are oxaliplatin
and 5-fluorouracil (5-FU) (3). How
autophagy determines and regulates the sensitivity of colorectal
cancer cells to 5-FUremainsunclear and requires investigation.
Autophagy is a degradation process that has
important roles in cellular homeostasis (4). Despite its simplicity, evidence has
demonstrated that autophagy is a highly complex process, involving
protein degradation, organelle turnover and breakdown of
cytoplasmic components during nutrient starvation or stress
(5). Autophagy contributes to
apoptosis when it is executed excessively or inefficiently
(6). It has been suggested that
autophagy serves a fundamental role in tumor progression (7–10).
Insulin-like growth factor-1 (IGF-1) has been
reported to regulate cell survival, proliferation, differentiation
and metabolism (11–13). IGF-1 has an inhibitory role on
autophagy in various cell types, including human osteocarcinoma
cells (11). However, little is
known regarding the mechanisms underlying its inhibitory effect on
autophagy in drug-resistant human colorectal carcinoma cells
(HCT).
Therefore, the aim of the present study was to
analyze the inhibitory effect of IGF-1 on autophagy in
drug-resistant HCT, and its underlying mechanism.
Materials and methods
Cell culture
HCT-8 human HCT and HCT-8R5-FU-resistant HCT cells
were obtained from Bogoo Biomart (Shanghai, China) The cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.), 2 mM L-glutamine, 100 U/ml
penicillin and 100 ng/ml streptomycin at 37°C in a humidified
atmosphere of 95% air. The medium was replaced every 2 days.
Autophagy analysis using the
DsRed-microtubule-associated protein 1A/1B-light chain 3
(LC3)reporter
To develop an autophagy reporter, DsRed protein was
fused with the N-terminus of the human LC3 protein through
transfecting 293T cells with a lentivirus. Recombinant lentiviruses
expressing the DsRed-LC3 reporter were generated and applied to
infect target cells (14).
Apoptosis assays
For apoptosis assays, cells were seeded at
5×105 cells/ml in triplicate and starved with serum-free
medium for 24 h, and subsequently treated with 10 or 50 nM IGF-1
[Phoenix Biotech (Beijing) Co., Ltd., Beijing, China], 10 nM
MK-2206 (Selleck Chemicals, Houston, TX, USA) or 100 nM
3-methyladenine (MA; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
in combination with 10 µg/ml 5-FU (Kyowa Hakko Kirin Co., Ltd.,
Tokyo, Japan). After 24 h, cells were stained with 5 µl annexin
V-fluorescein isothiocyanate and propidium iodide (PI; Invitrogen,
Thermo Fisher Scientific, Inc.) for 15 min at room temperature and
analyzed by flow cytometry (BD FACSCalibur™; BD Biosciences,
Franklin Lakes, NJ, USA) using Flowjo software (version 10; Tree
Star, Inc., Ashland, OR, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
HCT-8R cells were treated with IGF-1, AKT inhibitor
and 3-MA. Total RNA was extracted using the RNeasy kit (Qiagen
GmbH, Hilden, Germany) and transcribed into cDNA with an RNA
Reverse Transcriptase kit (Takara Bio, Inc., Otsu, Japan). qPCR was
performed with a SYBR®Green PCR assay (Takara Bio, Inc.)
according to the manufacturer's protocol (95°C for 1 min, and 40
cycles of 95°C for 5 sec and 60°C for 35 sec, followed by a final
standard dissociation protocol), using the primers listed in
Table I. Expression of GAPDH
served as an internal control. The results were analyzed using the
comparative Cq method (2-ΔΔCq) (15).
 | Table I.Primers used for quantitative
polymerase chain reaction. |
Table I.
Primers used for quantitative
polymerase chain reaction.
|
| Sequence (5′-3′) |
|---|
|
|
|
|---|
| Gene | Forward | Reverse |
|---|
| ULK1 |
CTGGTCCTCTTGCTTCCGTC |
ACACCAGCCCAACAATTCC |
| BECN1 | TCCGGGCTCCCGAGG |
TTCCTCCTGGGTCTCTCCTG |
| Vps34 |
GCTTAAGATCTGGAATGAATGGCT |
TGCCAGGAGTTTTTGTGGGT |
| Atg5 |
GGGTCCCTCTTGGGGTACAT |
ACCACACATCTCGAAGCACA |
| Atg7 |
TGGTTACAAGCTTGGCTGCT |
TCAAGAACCTGGTGAGGCAC |
| LC3B |
AAGGCTTTCAGAGAGACCCTG |
CCGTTTACCCTGCGTTTGTG |
| Atg16I |
CCTGCAATAACAAATTGCTGGA |
GCCTGTTTGGTACGTCATGC |
| Atg4B |
CTCATCTACCTGGACCCCCA |
AGAATCTAGGGACAGGTTCAGGA |
| Atg12 |
CTGTGTAATTGCGTCCCCCT |
GAAGCTGCAACACAGACTGC |
| GAPDH |
AATGGGCAGCCGTTAGGAAA |
GCGCCCAATACGACCAAATC |
Western blot analysis
HCT-8R cells were treated with IGF-1, AKT inhibitor
and 3-MA. Cells (5×105 cells/ml) were lysed in
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Haimen, China). The cell extracts were collected and
diluted in SDS-loading buffer and denatured for 5 min at 95°C, and
protein determination was performed using a bicinchoninic acid
assay. The samples (30 µg) were separated using SDS-PAGE on a 12.5%
gel and blotted onto 0.2 µm polyvinylidene difluoride membranes
(Bio-Rad Laboratories, Inc., Hercules, USA). Following blocking
with 5% skimmed milk powder in TBS containing Tween 20, membranes
were incubated at 4°C overnightwithrabbit antibodies against LC3B
(1:400, catalog no. ab48394; Abcam, Cambridge, UK), protein kinase
B (1:400, AKT; catalog no. ab179463s; Abcam) and phosphorylated
(p)-AKT (1:400, catalog no. ab81283; Abcam). Subsequently,
membranes were incubated with a horseradish peroxidase-conjugated
anti-rabbit secondary antibody at room temperature for 45 min
(1:2,000; catalog no. sc-2357; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and proteins were visualized with an Enhanced
Chemiluminescence system (PerkinElmer, Inc., Waltham, MA, USA).
β-actin antibody (1:1,000; catalog no. sc-130656; Santa Cruz
Biotechnology, Inc.) was used as an internal control.
Statistical analysis
Data are expressed as the mean ± standard deviation.
A one-way analysis of variance and Scheffé post hoc test was
applied to investigate significant differences among multiple
groups. Statistical analysis was performed using SPSS software
version 11.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Autophagy assay using the DsRed-LC3
reporter
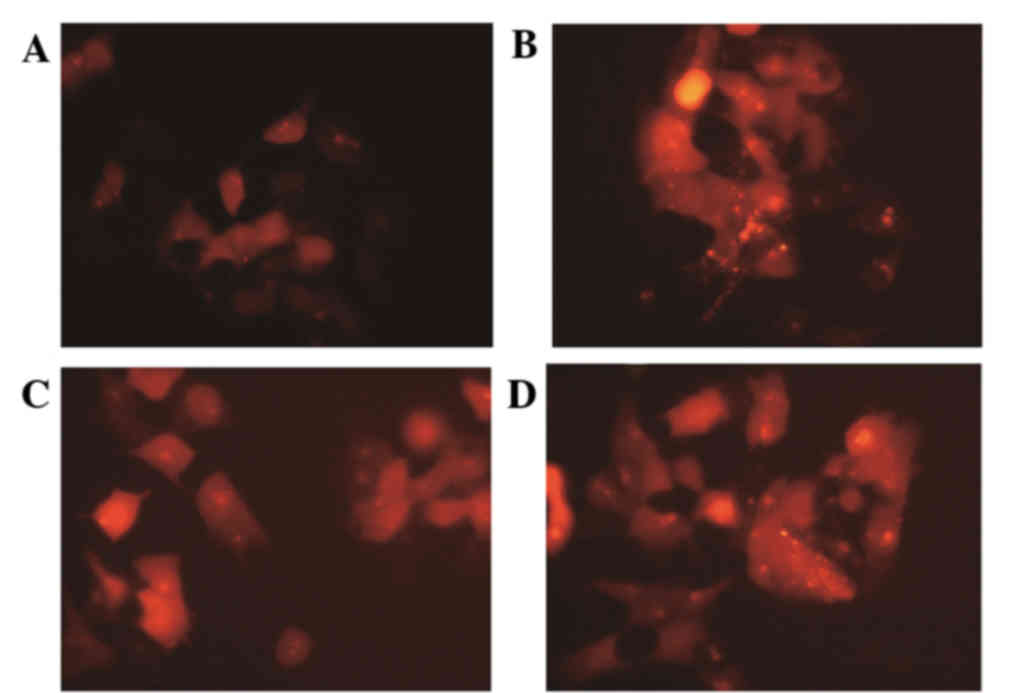
Following culture for 24 h, autophagy in the
drug-resistant cells increased. Autophagic bodies decreased
following IGF-1 treatment (Fig.
1); however, this was reversed upon the addition of an AKT
inhibitor.
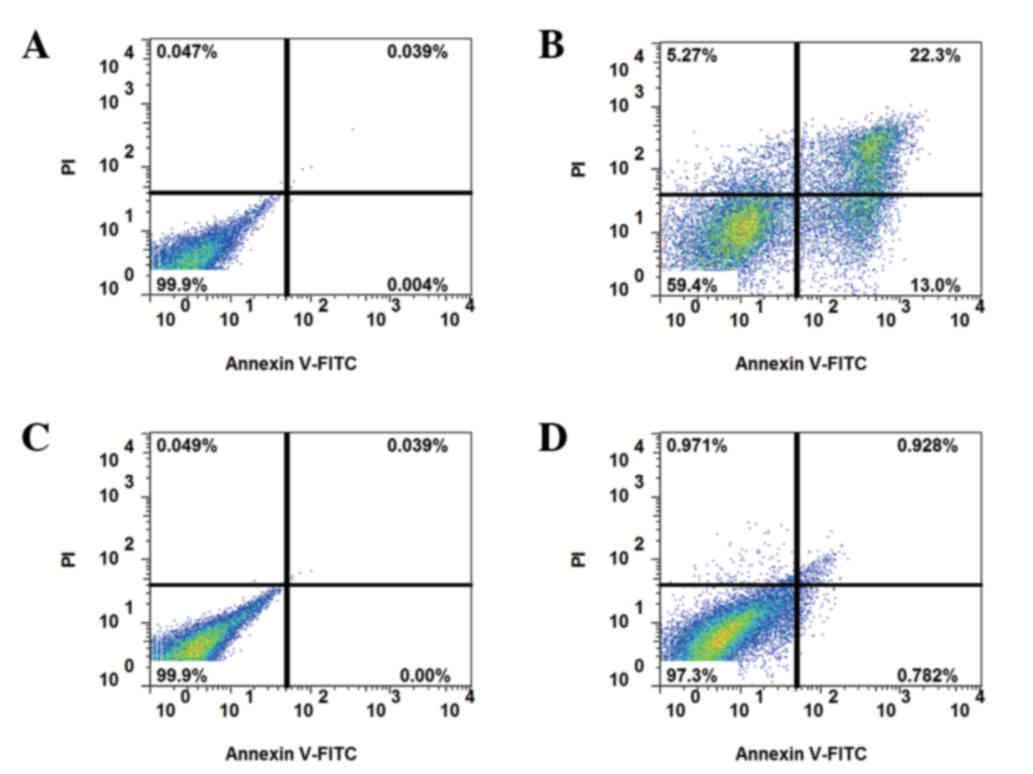
Apoptosis assays
Apoptosis was detected by annexin V-PI staining. In
non-resistant cells, apoptosis was increased by 5-FU treatment,
whereas the resistant strain exhibited reduced apoptosis at 24 h
following 5-FU treatment (Fig. 2).
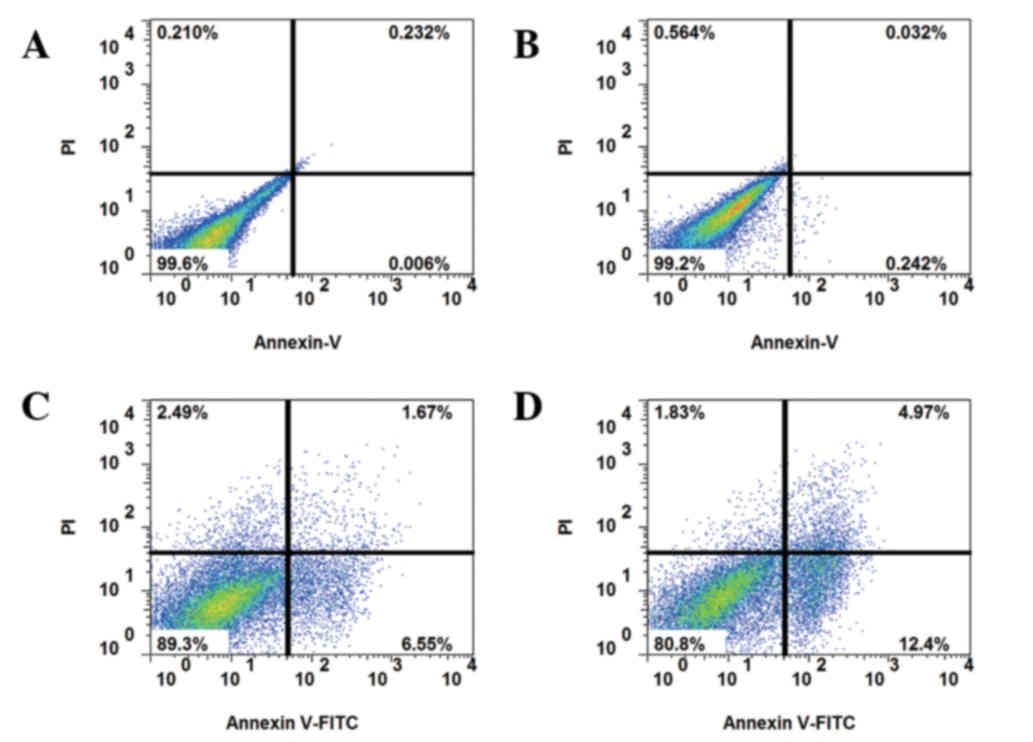
In order to reduce the inhibitory effect of serum on autophagy, the
cells were serum-starved for 24 h, and subsequently cultured with
10 or 50 nM IGF-1 and 5-FU for a further 24 h. Eventually,
apoptosis induced by 5-FU treatment was elevated by co-culture with
IGF-1 in drug-resistant HCT-8R cells (Fig. 3). With the addition of the AKT
inhibitor MK-2206 (10 nM) or autophagy agonist 3-MA (100 nM),
apoptosis was decreased (Fig.
4).
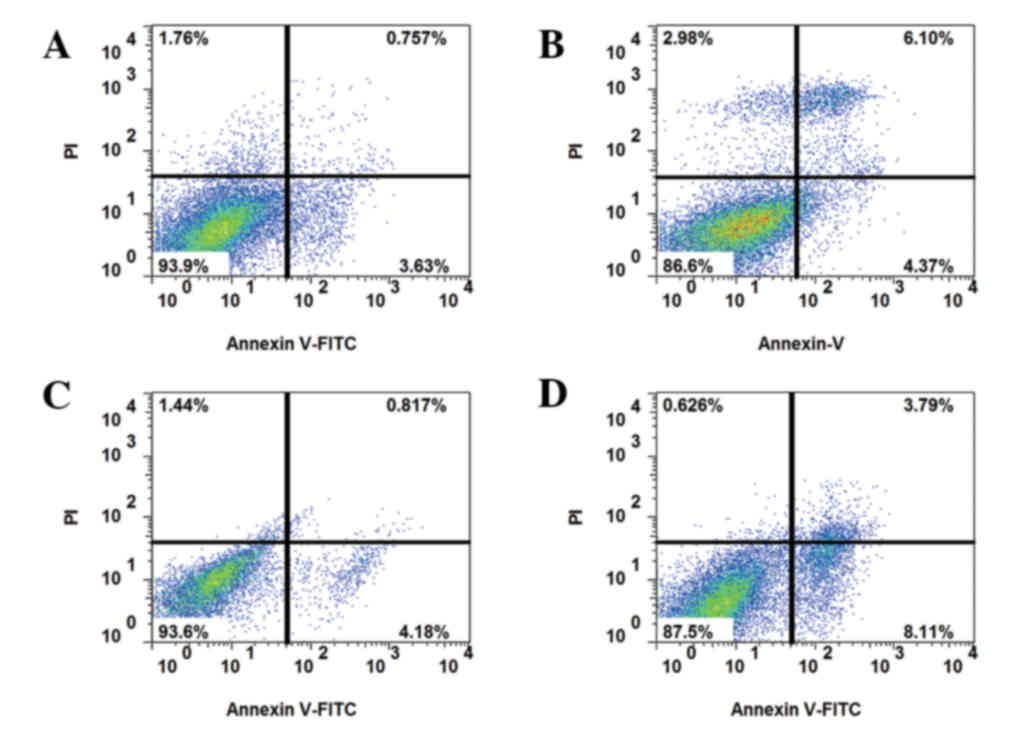 | Figure 4.Apoptosis of drug-resistant cells
following IGF-1 and AKT inhibitor treatment. Annexin V-FITC and PI
staining of (A) HCT-8R cells treated with 10 nM IGF-1, 5-FU and the
AKT inhibitor MK-2206, (B) HCT-8R cells treated with 10 nM IGF-1,
5-FU and the autophagy agonist3-MA, (C) HCT-8R cells treated with
50 nM IGF-1, 5-FU and MK-2206, and (D) HCT-8R cells treated with 50
nM IGF-1, 5-FU and 3-MA. The upper and lower right quadrants
indicate apoptotic cells. IGF-1, insulin-like growth factor 1; AKT,
protein kinase B; FITC, fluorescein isothiocyanate; PI, propidium
iodide; HCT, human colorectal carcinoma cells; R, resistant; 5-FU,
5-fluorouracil; 3-MA, 3-methyladenine. |
RT-qPCR analysis
RT-qPCR was used to assess the effects of IGF-1 on
the mRNA expression levels of autophagy-associated genes (Fig.5). In the initiation stage of
autophagy, IGF-1 treatment downregulated the mRNA expression levels
of the autophagy-associated genes unc-51 like autophagy activating
kinase 1 (ULK1), beclin-1(Becn1) and phosphatidylinositol 3-kinase
catalytic subunit type 3 (Vps34) in HCT-8cells (Fig. 5A). Treatment with IGF-1 and the AKT
inhibitor MK-2206 significantly increased the mRNA expression
levels of these genes; however, treatment with IGF-1 and 3-MA had
no significant effect. In the elongation stage, a similar pattern
was observed in the mRNA expression levels of autophagy related
(Atg)5, Atg7 and Lc3b (Fig. 5B).
In the expansion stage, IGF-1 downregulated the mRNA expression
levels of Atg4b, Atg16l and Atg12, whereas addition of MK-2206
eliminated the inhibitory effect of IGF-1 (Fig. 5C).
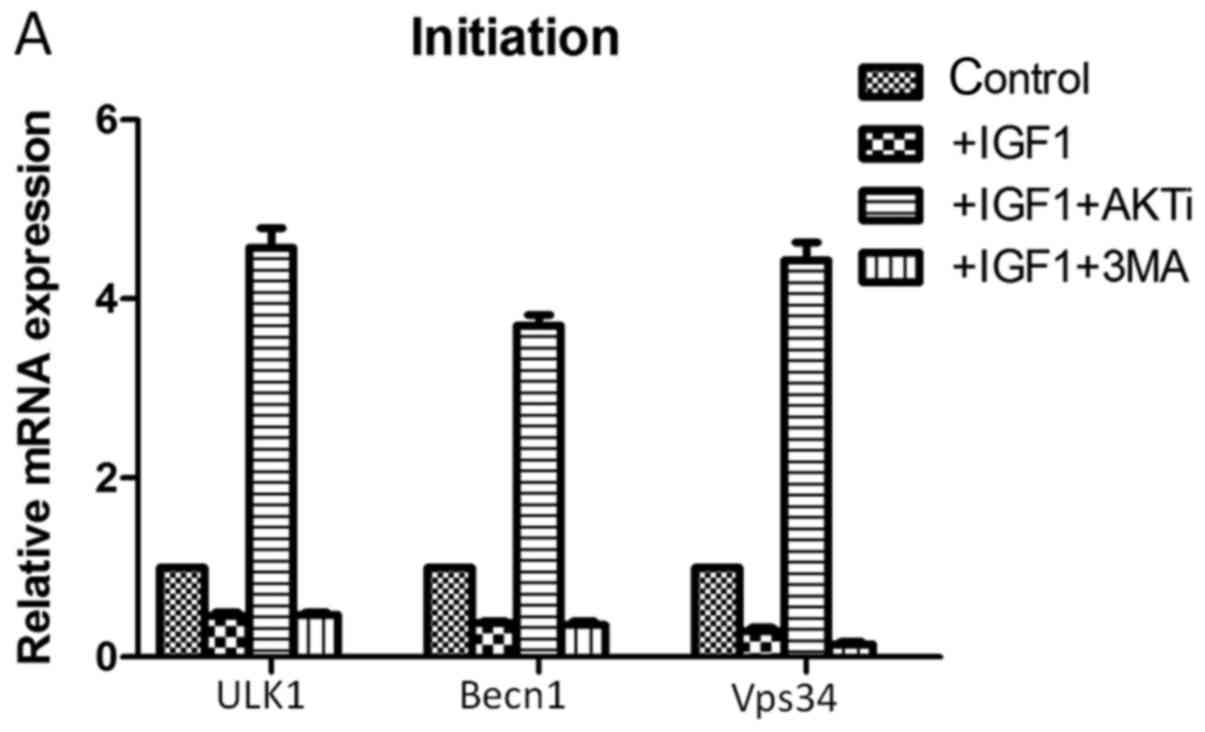 | Figure 5.Reverse transcription-quantitative
polymerase chain reaction analysis following treatment of cells
with IGF-1, AKTi and 3-MA.mRNA expression levels of genes
associated with the three stages of autophagy: (A) Initiation, (B)
elongation and (C) expansion, were assessed. IGF-1, insulin-like
growth factor 1; AKTi, protein kinase B inhibitor; 3-MA,
3-methyladenine; ULK1, unc-51 like autophagy activating kinase 1;
Becn1, beclin-1; Vps34, phosphatidylinositol 3-kinase catalytic
subunit type 3; Atg, autophagy-related; Lc3b,
microtubule-associated protein 1A/1B-light chain 3.
##P<0.05 vs. control. |
Western blot analysis
As presented in Fig.
6A, LC3 protein expression levels increased following treatment
with IGF-1, and decreased with the addition of IGF-1 and the AKT
inhibitor MK-2206. Similarly, p-AKT protein expression levels
increased with the addition of 50 nM IGF-1, and decreased with the
addition of 10 or 50 nM IGF-1 and MK-2206 (Fig. 6B).
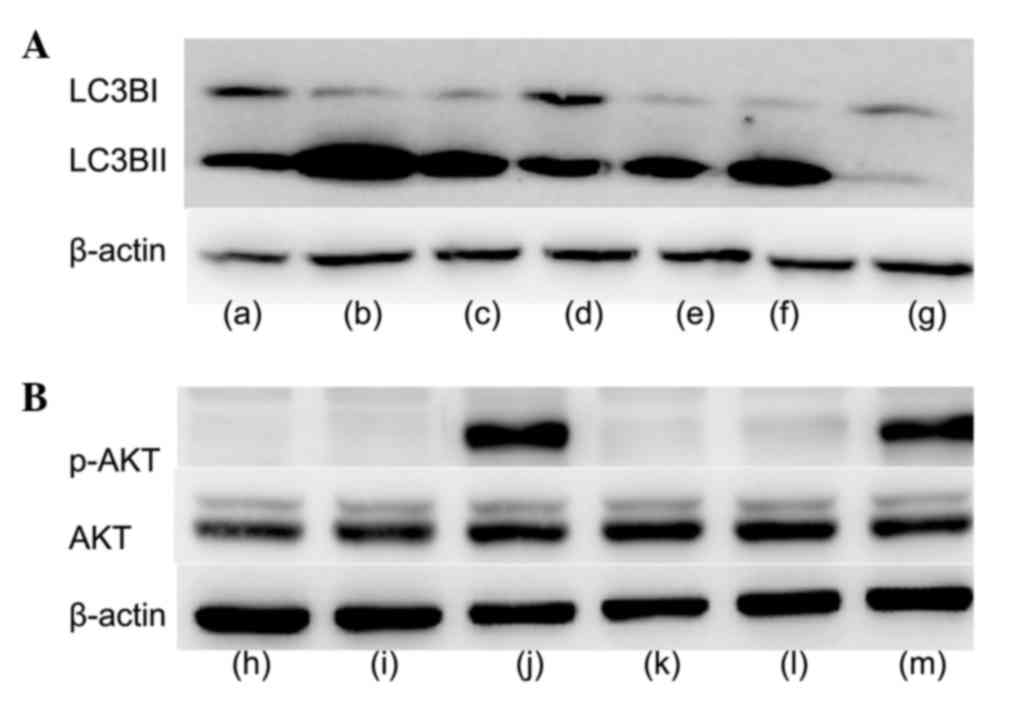 | Figure 6.Western blot analysis following
treatment of cells with IGF-1, AKTi and 3-MA. (A) Protein
expression levels of LC3B. Lane a, FBS; lane b, FBS and AKTi; lane
c, FBS and 3-MA; lane d, FBS and 50 nM IGF-1; lane e, FBS and 50 nM
IGF-1 and AKTi; lane f, FBS, 50 nM IGF-1 and 3-MA; lane g, CM. (B)
Protein expression levels of p-AKT and AKT. Lane h, FBS; lane i,
FBS and AKTi; lane j, FBS and 50 nM IGF-1; lane k, FBS, 10 nM IGF-1
and AKTi; lane l, FBS, 50 nM IGF-1 and AKTi; lane m,CM.IGF-1,
insulin-like growth factor 1;AKTi, protein kinase B inhibitor;
3-MA, 3-methyladenine; LC3B, microtubule-associated protein
1A/1B-light chain 3; FBS, fetal bovine serum; CM, complete medium;
AKT, protein kinase B; p, phosphorylated. |
Discussion
Autophagy has been reported to serve an important
role in cell death (4,6). The present study aimed to analyze the
inhibitory effect of IGF-1 on autophagy in HCT-8R drug-resistant
cells, and its potential underlying mechanisms.
In the present study, autophagic bodies decreased
following IGF-1 treatment; this effect was reversed following the
addition of an AKT inhibitor. This suggested that IGF-1 may inhibit
autophagy by activating AKT. Through apoptosis analysis, IGF-1 was
observed to increase sensitivity to apoptosis induced by 5-FU,
which decreased upon the addition of an AKT inhibitor or 3-MA.
These findings suggested that IGF-1 may inhibit autophagy via the
AKT/mammalian target of rapamycin (mTOR) signaling pathway, and
increase the sensitivity of resistant strains to 5-FU.
In a previous study, Lyu et al (16) revealed that peiminine resulted in
cell death and promoted autophagic flux in HCT-116 cells, and
suggested that peiminine enhanced autophagic flux by suppressing
the phosphorylation of mTOR via the inhibition of upstream signals.
In addition, Troncoso et al (11) reported that IGF-1 inhibits
autophagy via the 5’ adenosine monophosphate-activated protein
kinase/mTOR signaling pathway in addition to the AKT/mTOR signaling
pathway. It has additionally been reported that inhibition of the
mTOR signaling pathway induces autophagy and decreases cell
viability (17,18).
The present study revealed that IGF-1 downregulated
the mRNA expression levels of autophagy-associated genes involved
in the three autophagy stages. Atg and Becn1 are important markers
of autophagy (19–24). Previously, Jia et al
(25) investigated the effect of
IGF-1 on the expression of the autophagy-associated genes LC3 and
Becn1 in vascular smooth muscle cells (VSMCs). Transmission
electron microscopy revealed significantly reduced numbers of
vacuolated cells in IGF-1-treated VSMCs compared with the untreated
control group, andIGF-1 was demonstrated to inhibit the expression
of autophagy-associated genes via the AKT signaling pathway. In the
present study, LC3B-I and p-AKT protein expression levels increased
following IGF-1 treatment, and decreased upon the addition of IGF-1
and an AKT inhibitor. These findings indicated that IGF-1 did not
appear to promote apoptosis alone, although it increased
sensitivity to apoptosis induced by 5-FU.
In conclusion, the results of the present study
suggested that IGF-1 activated AKT and inhibited autophagy via the
AKT/mTOR signaling pathway. Following inhibition of autophagy,
drug-resistant HCT-8R cells became sensitive to 5-FU treatment, and
treatment with 5-FU in combination with IGF-1 increased
apoptosis.
References
|
1
|
Oliveira CS, Pereira H, Alves S, Castro L,
Baltazar F, Chaves SR, Preto A and Côrte-Real M: Cathepsin D
protects colorectal cancer cells from acetate-induced apoptosis
through autophagy-independent degradation of damaged mitochondria.
Cell Death Dis. 6:e17882015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cunningham D, Atkin W, Lenz HJ, Lynch HT,
Minsky B, Nordlinger B and Starling N: Colorectal cancer. Lancet.
375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu W, Zhang Z, Zhang Y, Chen X, Guo S,
Lei Y, Xu Y, Ji C, Bi Z and Wang K: HMGB1-mediated autophagy
modulates sensitivity of colorectal cancer cells to oxaliplatin via
MEK/ERK signaling pathway. Cancer Biol Ther. 16:511–517. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hayashi-Nishino M, Fujita N, Noda T,
Yamaguchi A, Yoshimori T and Yamamoto A: A subdomain of the
endoplasmic reticulum forms a cradle for autophagosome formation.
Nat Cell Biol. 11:1433–1437. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mizushima N: Autophagy: process and
function. Genes Dev. 21:2861–2873. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Galluzzi L, Morselli E, Vicencio JM, Kepp
O, Joza N, Tajeddine N and Kroemer G: Life, death and burial:
Multifaceted impact of autophagy. Biochem Soc Trans. 36:786–790.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rubinsztein DC, Codogno P and Levine B:
Autophagy modulation as a potential therapeutic target for diverse
diseases. Nat Rev Drug Discov. 11:709–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou WH, Tang F, Xu J, Wu X, Yang SB, Feng
ZY, Ding YG, Wan XB, Guan Z, Li HG, et al: Low expression of Beclin
1, associated with high Bcl-xL, predicts a malignant phenotype and
poor prognosis of gastric cancer. Autophagy. 8:389–400. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang M, Zhao H, Guo L, Zhang Q, Zhao L,
Bai S, Zhang M, Xu S, Wang F, Wang X and Zhao B: Autophagy-based
survival prognosis in human colorectal carcinoma. Oncotarget.
6:7084–7103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo GF, Jiang WQ, Zhang B, Cai YC, Xu RH,
Chen XX, Wang F and Xia LP: Autophagy-related proteins Beclin-1 and
LC3 predict cetuximab efficacy in advanced colorectal cancer. World
J Gastroenterol. 17:4779–4786. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Troncoso R, Vicencio JM, Parra V,
Nemchenko A, Kawashima Y, Del Campo A, Toro B, Battiprolu PK,
Aranguiz P, Chiong M, et al: Energy-preserving effects of IGF-1
antagonize starvation-induced cardiac autophagy. Cardiovasc Res.
93:320–329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ikeda H, Shiojima I, Ozasa Y, Yoshida M,
Holzenberger M, Kahn CR, Walsh K, Igarashi T, Abel ED and Komuro I:
Interaction of myocardial insulin receptor and IGF receptor
signaling in exercise-induced cardiac hypertrophy. J Mol Cell
Cardiol. 47:664–675. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sekharam M, Nasir A, Kaiser HE and Coppola
D: Insulin-like growth factor 1 receptor activates c-SRC and
modifies transformation and motility of colon cancer in vitro.
Anticancer Res. 23:1517–1524. 2003.PubMed/NCBI
|
|
14
|
Yao W, Dai W, Jiang L, Lay EY, Zhong Z,
Ritchie RO, Li X, Ke H and Lane NE: Sclerostin-antibody treatment
of glucocorticoid-induced osteoporosis maintained bone mass and
strength. Osteoporos Int. 27:283–294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nature Protoc.
3:1101–1108. 2008. View Article : Google Scholar
|
|
16
|
Lyu Q, Tou F, Su H, Wu X, Chen X and Zheng
Z: The natural product peiminine represses colorectal carcinoma
tumor growth by inducing autophagic cell death. Biochem Biophys Res
Commun. 462:38–45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin CW, Jan MS and Kuo JH: Exploring
MicroRNA Expression Profiles Related to the mTOR Signaling Pathway
in Mouse Embryonic Fibroblast Cells Treated with Polyethylenimine.
Mol Pharm. 12:2858–2868. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sobolewska A, Gajewska M, Zarzynska J,
Gajkowska B and Motyl T: IGF-I, EGF, and sex steroids regulate
autophagy in bovine mammary epithelial cells via the mTOR pathway.
Eur J Cell Biol. 88:117–130. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schwartz-Roberts JL, Cook KL, Chen C,
Shajahan-Haq AN, Axelrod M, Warri A, Riggins RB, Jin L, Haddad BR,
Kallakury BV, Baumann WT and Clarke R: Interferon regulatory
factor-1 signaling regulates the switch between autophagy and
apoptosis to determine breast cancer cell fate. Cancer Res.
75:1046–1055. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD,
Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M
and Ohsumi Y: A unified nomenclature for yeast autophagy-related
genes. Dev Cell. 5:539–545. 2013. View Article : Google Scholar
|
|
21
|
Liang XH, Jackson S, Seaman M, Brown K,
Kempkes B, Hibshoosh H and Levine B: Induction of autophagy and
inhibition of tumorigenesis by beclin 1. Nature. 402:672–676. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Betin VM and Lane JD: Caspase cleavage of
Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial
targeting and apoptosis. J Cell Sci. 122:2554–2566. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yousefi S, Perozzo R, Schmid I, Ziemiecki
A, Schaffner T, Scapozza L, Brunner T and Simon HU:
Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis.
Nat Cell Biol. 8:1124–1132. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nikoletopoulou V, Markaki M, Palikaras K
and Tavernarakis N: Crosstalk between apoptosis, necrosis and
autophagy. Biochim Biophys Acta. 1833:3448–3459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jia G, Cheng G, Gangahar DM and Agrawal
DK: Insulin-like growth factor-1 and TNF-alpha regulate autophagy
through c-jun N-terminal kinase and Akt pathways in human
atherosclerotic vascular smooth cells. Immunol Cell Biol.
84:448–454. 2006. View Article : Google Scholar : PubMed/NCBI
|