Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common malignant disease and the third leading cause of
cancer-related death worldwide (1). Most patients with HCC are diagnosed
at a later stage of disease, leading to poor prognosis with a
5-year survival rate of less than 16% (2). However, the 5-year survival rate
increases to more than 50% for HCC detected at early stages
(2). Therefore, searching for
biomarkers for detecting early-stage HCC is critical in improvement
of the overall prognosis of HCC.
At present, imaging techniques and determination of
α-fetoprotein (AFP) levels are widely used as screening tools.
Because of the high cost and radiation exposure, or insufficient
sensitivity and specificity, imaging techniques have limitations in
the screening of early HCC (3,4). AFP
is currently the main serum biomarker used in the diagnosis of HCC,
with low sensitivity of 46–59% and specificity of 87–93% for
detection of early HCC (5). Other
biomarkers reported in recent years include lens culinaris
agglutinin-reactive fraction of AFP (AFP-L3),
des-γ-carboxyprothrombin (DCP), squamous cell carcinoma antigen
(SCCA), and golgi protein 73 (GP73) (6–10),
with area under the curves (AUCs) of 0.67 to 0.77, sensitivities of
28.6–70.9%, and specificities of 74.9–92.7% for early-stage HCC
(9–13).
In recent years, tumor-associated antigens (TAAs)
with promising diagnostic value for tumors at early stages,
including HCC, have been identified (14). Among them, serological autoantibody
to centromere protein F (CENP-F) has been recognized to have
potential value in the detection of early HCC (14,15).
CENP-F is a kinetochore protein of 3,210 amino acids that plays a
role in centromere formation and kinetochore organization during
mitosis (16–21). Using protein microarray to evaluate
the diagnostic value of CENP-F autoantibody in a large HCC cohort,
our previous study showed that CENP-F antibody had better
sensitivity for the detection of early-stage HCC compared with AFP
and that combined autoantibody to CENP-F with AFP further improved
the diagnostic capability for early HCC (22). However, much less has been known
regarding the dominant epitopes of CENP-F antigen, and the exact
regions corresponding to the dominant peptides of CENP-F antigen
remain to be explored.
In the present study, we aimed to screen and
evaluate potential dominant epitope peptides in the full-length
CENP-F antigen protein with the aim of obtaining novel CENP-F
antigens and improving the early diagnosis of HCC.
Materials and methods
Study population
Screening group: for screening of antigens with the
best serodiagnostic performance among eight individual antigens we
collected serum samples including 47 cases of HCC (38 men and 9
women aged 44–80 years, with a median age of 57.0 years) and 48
healthy controls (21 men and 27 women aged 20–59 years, with a
median age of 46.0 years). Validation group: For validation of the
serodiagnostic performance of the selected antigen dominant
epitopes we collected another set of 405 serum samples, including
153 cases of HCC with AFP data available (127 men and 26 women aged
0–81 years, with a median age of 57.0 years), of which 70 cases
were early-stage HCC (57 men and 13 women aged 28–80 years, with a
median age of 56.5 years); 126 cases of liver cirrhosis (95 men and
31 women aged 27–73 years, with a median age of 51.0 years); and
126 healthy controls (65 men and 61 women aged 20–70 years, with a
median age of 47.0 years). Clinical characteristics of the samples
are shown in Table I.
 | Table I.Study population characteristics. |
Table I.
Study population characteristics.
| Characteristics | HCC (N=153) | LC (N=126) | HC (N=126) |
|---|
| Age (mean ± SD) | 56.8±11.2 | 50.0±9.7 | 46.9±8.5 |
| Sex (n, %) |
|
|
|
| Male | 127 (83.0) | 95 (75.4) | 65 (51.6) |
|
Female | 26 (17.0) | 31 (24.6) | 61 (48.4) |
| HBV infection (n,
%) |
|
|
|
| HBV
(+) | 107 (69.9) | 101 (80.2) | 0 (0.0) |
| HBV
(−) | 44 (28.8) | 25 (19.8) | 126 (100.0) |
|
Missing | 2 (1.3) | 0 (0.0) | 0 (0.0) |
| HCV infection (n,
%) |
|
|
|
| HCV
(+) | 13 (8.5) | 0 (0.0) | 0 (0.0) |
| HCV
(−) | 138 (90.2) | 126 (100.0) | 126 (100.0) |
|
Missing | 2 (1.3) | 0 (0.0) | 0 (0.0) |
| TNM tumor stage (n,
%) |
|
|
|
| I | 70 (45.8) | – | – |
|
>I | 83 (54.2) | – | – |
| Child-Pugh (n,
%) |
|
|
|
| A | 83 (54.2) | 57 (45.2) | – |
| B | 23 (15.0) | 33 (26.2) | – |
| C | 14 (9.2) | 23 (18.3) | – |
|
Missing | 33 (21.6) | 13 (10.3) | – |
| AFP |
|
|
|
| ≥20
ng/ml | 81 (52.9) | 22 (17.5) | 0 (0.0) |
| <20
ng/ml | 72 (47.1) | 104 (82.5) | 126 (100.0) |
| AST (U/l) | 54.2
(11.0–659.5) | 35.2
(13.9–393.7) | – |
| ALT (U/l) | 43.3 (8.2–837.4) | 30.6
(8.2–1328.0) | – |
| ALB (g/l) | 35.9 (22.1–63.4) | 36.2 (3.9–49.4) | – |
| TBIL (µmol/l) | 32.5 (6.9–945.0) | 23.9 (5.1–475.1) | – |
| DBIL (µmol/l) | 6.3 (0.9–243.0) | 5.6 (1.0–221.7) | – |
All samples were obtained from the Cancer Hospital,
Chinese Academy of Medical Science, Beijing, China; Beijing Youan
Hospital, Capital Medical University, Beijing, China; and Beijing
Friendship Hospital, Capital Medical University (Beijing, China)
from November 2013 to December 2016.
A diagnosis of HCC was based on the guideline for
diagnosis and treatment of primary HCC (2012 version, China).
Early-stage HCC was defined as a tumor at TMN stage I. Diagnosis of
LC was based on ultrasound, computed tomography (CT), or magnetic
resonance imaging (MRI) characteristics, laboratory indexes, and
histopathology (3). Healthy
controls were healthy examiners with normal liver biochemistry, no
history of liver disease, and no malignant disease.
All serum samples were stored at −80°C until
testing. The study protocol was approved by the Clinical Research
Ethics Committee of Beijing Friendship Hospital, Capital Medical
University (Beijing, China).
Bioinformatics analysis of dominant
epitope peptides of CENP-F
Candidate dominant epitopes of CENP-F protein were
predicted using BioSun version 3.0 software developed by the Center
of Computational Biology, Beijing Institute of Basic Medical
Sciences (Beijing, China). Based on the epitope curve, peptides
containing the dominant CENP-F epitopes with highest peak values
were selected as the target peptides.
Construction, expression, and
purification of recombinant proteins
Coding sequences of each dominant antigen peptide of
eight single antigens (121–220 a.a, 335–416 a.a, 1100–1265 a.a,
1670–1791 a.a, 1759–2093 a.a, 2075–2210 a.a, 2485–2592 a.a,
2808–2960 a.a) with GST or His tags were chemically synthesized and
inserted into the prokaryotic expression plasmid pET-6P with GST or
6-His tags (constructed in house) using specific endonuclease
restriction sites BamHI and XhoI. The recombinant plasmids were
transformed into Escherichia coli BL21 or BL21 (DE3) and the
fusion proteins were expressed following induction with 0.1 M
isopropyl β-D-thiogalactoside at 16°C for 12 h. The soluble
expression of recombinant proteins were purified by affinity
chromatography using GST-Sefinose resin or His-Sefinose resin
(Sangon Biotech Co., Ltd., Shanghai, China). The purity of fusion
proteins was analyzed by sodium alt-polyacrylamide gel
electrophoresis (SDS-PAGE) and Gel-Pro Analyzer version 3.1.00.00
(Media Cybernetics, Inc., Silver Spring, MD, USA) and the protein
concentration was determined by the bicinchoninic acid (BCA) method
(Pierce, Rockford, IL, USA).
Enzyme-linked immunosorbent assay
(ELISA) for screening and evaluation of dominant peptides
For ELISA, 96-well microplates (Nunc A/S, Roskilde,
Denmark) were coated with individual antigens at 5 µg/ml (100
µl/well) in coating buffer (0.05 M carbonate/bicarbonate, pH 9.6)
and incubated at 4°C overnight. The plates were washed once with
phosphate-buffered saline (PBS) containing 0.05% Tween-20 and
blocked by the addition of 200 µl of 10% newborn bovine serum (Life
Technologies, Burlington, ON, Canada) and incubation at 37°C for 2
h. Next, 100 µl of standard serum (in-house preparation) diluted
1:4 (1,000 µg/ml), 1:8 (500 µg/ml), 1:16 (250 µg/ml), 1:32 (125
µg/ml), 1:64 (62.5 µg/ml), 1:128 (31.25 µg/ml), 1:256 (15.625
µg/ml), and 1:512 (7.8125 µg/ml) in 10% newborn bovine serum or 100
µl of patient serum diluted 1:11 in PBS containing 10% newborn
bovine serum was added to the wells and incubated for 1 h at 37°C.
The plates were washed five times and then 100 µl of a 1:8,000
dilution of rabbit anti-human IgG-peroxidase antibody
(Sigma-Aldrich, St. Louis, MO, USA) was added and incubated for 30
min at 37°C, followed by addition of 100 µl TMB HRP-Substrate
(Beijing Solarbio Science & Technology Co., Ltd., Beijing,
China) and incubation for 10 min at 37°C. The reaction was stopped
by addition of 50 µl stop solution (Beijing Solarbio Science &
Technology Co., Ltd.) and absorbance was immediately read at 450 nm
or 630 nm using a microplate reader SpectraMax M3 (Molecular
Devices, LLC, Sunnyvale, CA, USA).
Statistical analysis
All statistical analyses were performed using SPSS
(version 23.0; IBM Corp., Armonk, NY, USA) and GraphPad Prism
(version 6.0c; GraphPad Software, Inc., La Jolla, CA, USA).
Receiver operating characteristic (ROC) curves were plotted and the
following diagnosis-related indicators, including sensitivity,
specificity and AUCs along with 95% confidence intervals [95%
confidence intervals (CIs)], were used to evaluate the diagnostic
performance of individual biomarkers. The respective optimal
cut-off values of individual biomarkers in detecting HCCs were
determined by the Youden's index (sensitivity + specificity-1). We
used the Chi-square (χ2) test to evaluate the correlation between
the level of auto-antibody to CENP-F and TNM stage of HCC.
In addition, we further evaluated the diagnostic
potential of multiple biomarkers using logistic regression models.
The predicted probabilities were used to conduct ROC analyses, and
diagnosis-related indicators were therefore calculated and
reported.
Results
Prediction of peptides containing
dominant epitopes of CENP-F
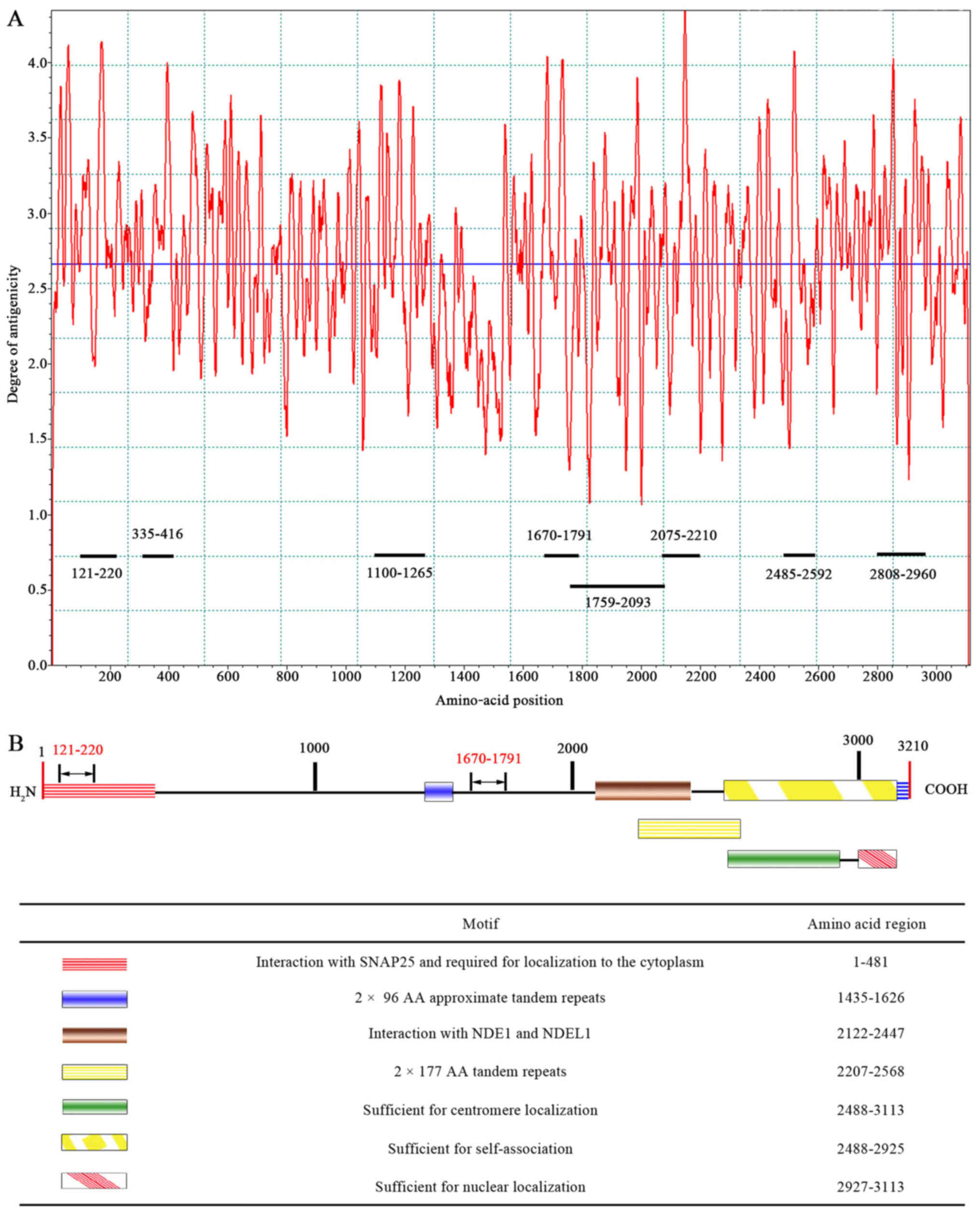
Based on the epitope curve (Fig. 1A), eight peptides containing CENP-F
dominant epitopes with higher peak values for each antigen were
determined as follows: 121–220 a.a peptide, 335–416 a.a peptide,
1100–1265 a.a peptide, 1670–1791 a.a peptide, 1759–2093 a.a
peptide, 2075–2210 a.a peptide, 2485–2592 a.a peptide, and
2808–2960 a.a peptide. The eight peptides covered all functional
domains except for the domain responsible for 2×96 AA approximate
tandem repeats (Fig. 1B).
Recombinant proteins of the eight predicted dominant peptides were
prepared, including 1759–2093 a.a peptide with his tag, and other
seven peptides with GST tag (Fig.
2).
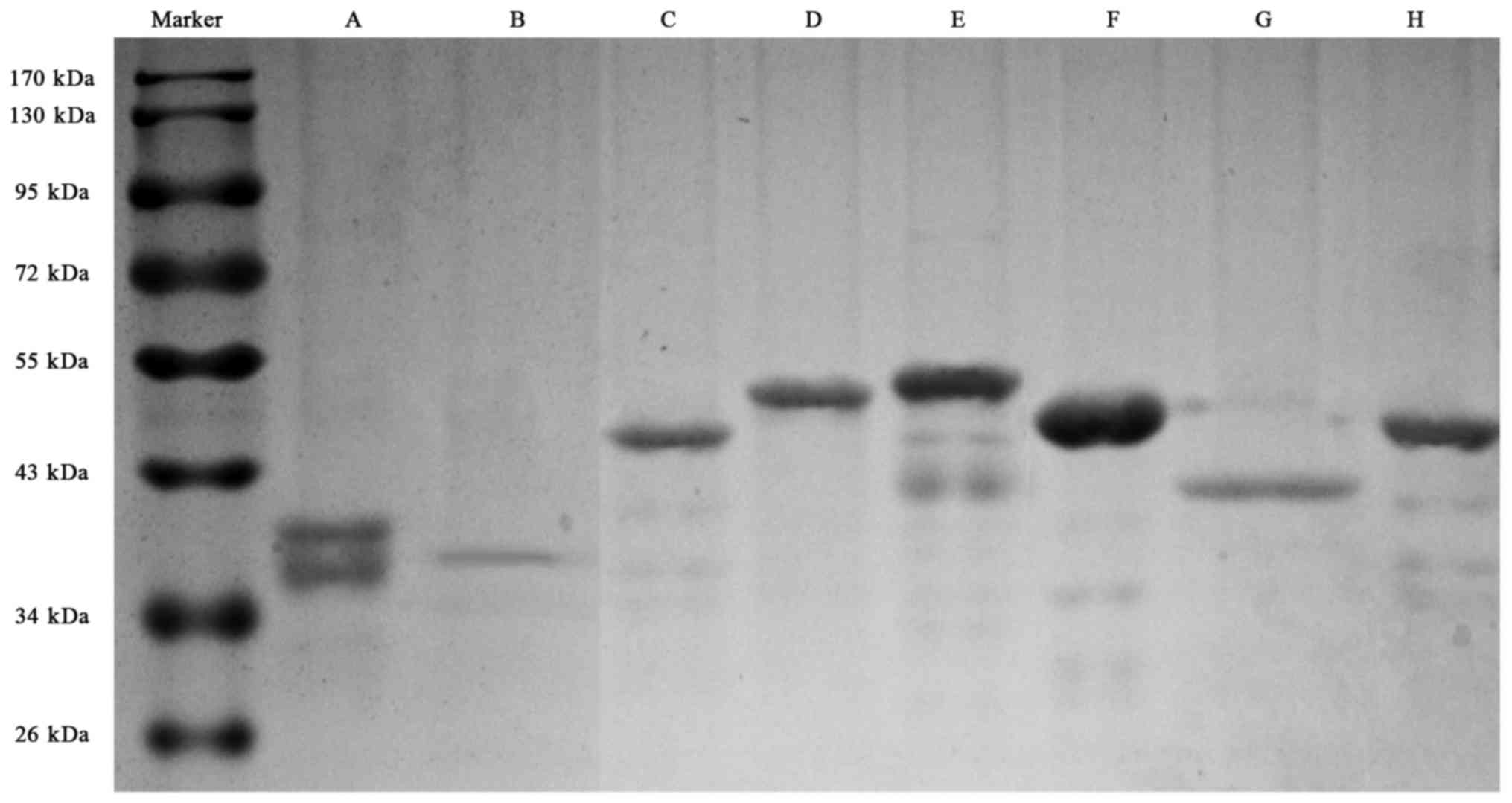 | Figure 2.SDS-PAGE electrophoresis for the eight
recombinant fragments of CENP-F protein. (A), 121–220 a.a with GST
tag, with a degraded band (with CENP-F antigenicity); (B), 335–416
a.a with GST tag; (C), 1759–2093 a.a with his tag; (D), 1100–1265
a.a with GST tag; E, 1670–1791 a.a with GST tag; F, 2075–2210 a.a
with GST tag; G, 2485–2592 a.a with GST tag; H, 2808–2960 a.a with
GST tag. SDS-PAGE, sodium alt-polyacrylamide gel electrophoresis;
CENP-F, centromere protein F; a.a, amino acid. |
Serological reactivity of the eight
individual antigen peptides
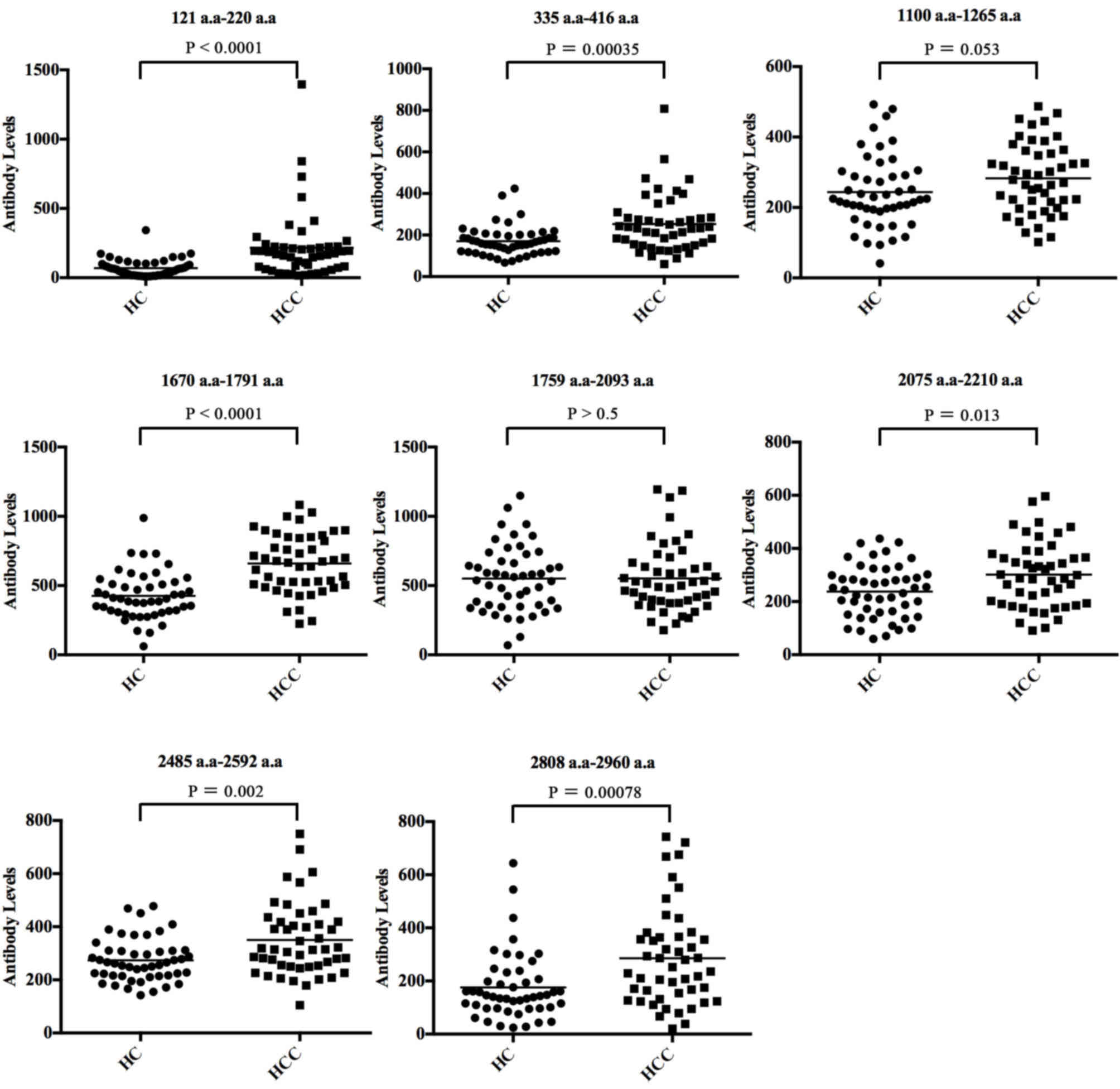
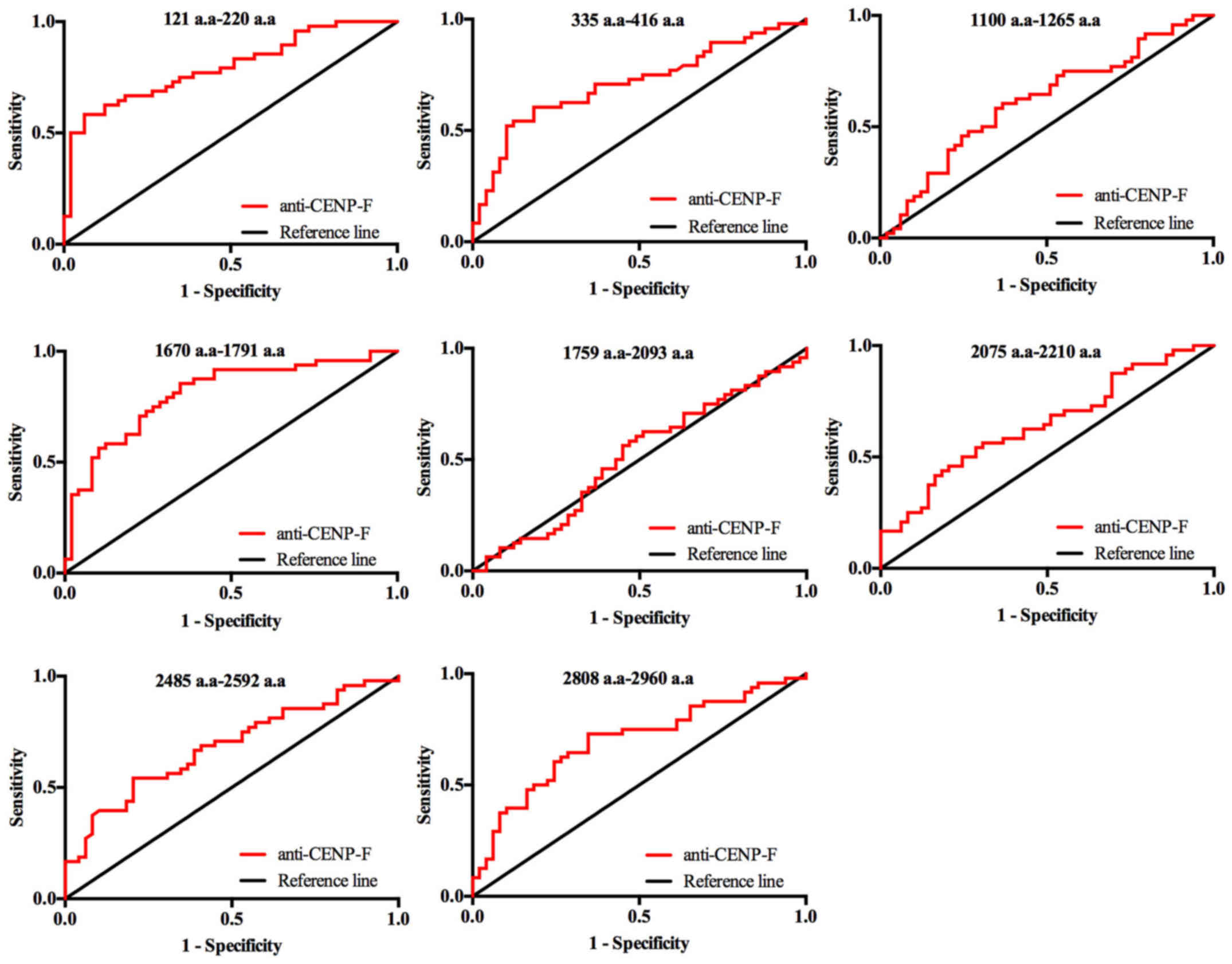
We used an indirect ELISA to evaluate the diagnostic
performance of the individual peptides by the analysis of
anti-CENP-F level in serum of 47 cases of HCC and 48 healthy
control. The AUC value and sensitivities and specificities of the
eight individual antigen peptides are shown in Table II. The scatter plots showing the
eight antigens of CENP-F between HCC and HC groups are presented in
Fig. 3, and the ROC curves of the
eight individual CENP-F antigens in discriminating between HCC and
healthy controls (HC) are shown in Fig. 4. The sensitivities of the eight
individual peptide antigens ranged from 25.50 to 85.40%, and the
specificities ranged from 59.20 to 93.30%. Among them, two peptides
of CENP-F, 121–220 a.a and 1670–1791 a.a, had the highest
diagnostic value for HCC with AUC values of 0.795 and 0.809,
respectively. Specificity was better for peptide 121–220 a.a
(93.9%) but sensitivity was better for peptide 1670–1791 a.a
(Figs. 3 and 4, Table
II).
 | Table II.Diagnostic performance of the eight
predicted dominant peptides of CENP-F antigen evaluated by ELISA
analysis of samples of screening group. |
Table II.
Diagnostic performance of the eight
predicted dominant peptides of CENP-F antigen evaluated by ELISA
analysis of samples of screening group.
| Dominant
peptides | AUC value | 95% CI | Sensitivity
(%) | Specificity
(%) | Cut-off value |
|---|
| 121–220 a.a | 0.795 | 0.706–0.884 | 58.3 | 93.9 | 154 |
| 335–416 a.a | 0.711 | 0.606–0.815 | 54.2 | 87.8 | 225 |
| 1100–1265 a.a | 0.614 | 0.501–0.727 | 60.4 | 63.3 | 250 |
| 1670–1791 a.a | 0.809 | 0.721–0.896 | 85.4 | 65.3 | 458 |
| 1759–2093 a.a | 0.513 | 0.397–0.629 | 25.0 | 85.7 | 793 |
| 2075–2210 a.a | 0.630 | 0.520–0.740 | 22.9 | 98.0 | 441 |
| 2485–2592 a.a | 0.682 | 0.576–0.788 | 68.8 | 59.2 | 276 |
| 2808–2960 a.a | 0.656 | 0.547–0.766 | 72.9 | 59.2 | 163 |
Validation of the diagnostic
performance of the 121–220 a.a and 1670–1791 a.a antigen
peptides
A total of 405 serum samples from HCC, LC, and HC
were used to evaluate the diagnostic value of the two antigen
peptides 121–220 a.a and 1670–1791 a.a, as well as the effect of
combined antigen peptide with AFP, for the detection of HCC.
As shown in Table
III, the 121–220 a.a peptide gave results consistent with the
data obtained in the screening group; the AUC value for the
discrimination of HCC from HC was 0.749 (95% CI, 0.692–0.807) with
sensitivity of 68% and specificity of 72.2%, and the AUC for
discrimination of early HCC from HC was 0.743 (95% CI, 0.674–0.812)
with sensitivity of 68.6 and specificity of 72.2%, but there was no
significant difference between HCC or early HCC and LC. However,
the 1670–1791 a.a peptide showed lower performance compared with
the results in the screening group; the AUC value for the
discrimination of HCC from HC was 0.628 (95% CI, 0.563–0.693) with
sensitivity of 62.1% and specificity of 61.9%, and the AUC for
discrimination of early HCC from HC was 0.584 (95% CI, 0.500–0.668)
with sensitivity of 72.9% and specificity of 50.0%, with no
significant difference between HCC or early HCC and LC.
 | Table III.Diagnostic value of 121–220 a.a and
1670–1791 a.a dominant peptides of CENP-F evaluated by ELISA
analysis of samples of validation group. |
Table III.
Diagnostic value of 121–220 a.a and
1670–1791 a.a dominant peptides of CENP-F evaluated by ELISA
analysis of samples of validation group.
| Dominant
peptides | Cases | AUC value | 95% CI | Sensitivity
(%) | Specificity
(%) |
|---|
|
| HCC vs. HC |
|
|
|
|
| 121–220 a.a |
| 0.749 | 0.692–0.807 | 68.0 | 72.2 |
| 1670–1791 a.a |
| 0.628 | 0.563–0.693 | 62.1 | 61.9 |
|
| HCC vs. LC |
|
|
|
|
| 121–220 a.a |
| 0.559 | 0.491–0.627 | 51.0 | 61.9 |
| 1670–1791 a.a |
| 0.541 | 0.474–0.609 | 68.0 | 43.7 |
|
| Early-stage HCC vs.
HC |
|
|
|
|
| 121–220 a.a |
| 0.743 | 0.674–0.812 | 68.6 | 72.2 |
| 1670–1791 a.a |
| 0.584 | 0.500–0.668 | 72.9 | 50.0 |
|
| Early-stage HCC vs.
LC |
|
|
|
|
| 121–220 a.a |
| 0.465 | 0.383–0.546 | 92.9 | 15.1 |
| 1670–1791 a.a |
| 0.518 | 0.435–0.602 | 75.7 | 38.1 |
Specifically, the combination of AFP, for which a
serum level greater than 20 ng/ml was defined as positive, and
autoantibody to 121–220 a.a dominant peptide of CENP-F antigen
improved the ability to distinguish HCC from the healthy controls,
with the AUC (95% CI), sensitivity, specificity of 0.875 (95% CI,
0.835–0.914), 75.2 and 84.9%, respectively, better than AFP solely
(Table IV). Meanwhile, improved
diagnostic performance for detection of early-stage HCC was also
observed for the combination, with AUC of 0.84, higher than AFP
(0.72) solely (Table IV).
 | Table IV.Diagnostic value of the combination
of AFP and 121–220 a.a or 1670–1791 a.a dominant peptides of CENP-F
antigen. |
Table IV.
Diagnostic value of the combination
of AFP and 121–220 a.a or 1670–1791 a.a dominant peptides of CENP-F
antigen.
| Dominant
peptides | Cases | AUC value | 95% CI | Sensitivity
(%) | Specificity
(%) |
|---|
|
| HCC vs. HC |
|
|
|
|
| 121–220 a.a +
AFP |
| 0.875 | 0.835–0.914 | 75.2 | 84.9 |
| 1670–1791 a.a +
AFP |
| 0.827 | 0.779–0.876 | 63.4 | 93.7 |
| AFP |
| 0.768 | 0.712–0.824 | 53.6 | 100.0 |
|
| HCC vs. LC |
|
|
|
|
| 121–220 a.a +
AFP |
| 0.702 | 0.641–0.763 | 53.6 | 82.5 |
| 1670–1791 a.a +
AFP |
| 0.700 | 0.638–0.761 | 55.6 | 82.5 |
| AFP |
| 0.681 | 0.618–0.744 | 53.6 | 82.5 |
|
| Early-stage HCC vs.
HC |
|
|
|
|
| 121–220 a.a +
AFP |
| 0.840 | 0.781–0.899 | 81.4 | 72.2 |
| 1670–1791 a.a +
AFP |
| 0.779 | 0.706–0.852 | 51.4 | 93.7 |
| AFP |
| 0.721 | 0.639–0.804 | 44.3 | 100.0 |
|
| Early-stage HCC vs.
LC |
|
|
|
|
| 121–220 a.a +
AFP |
| 0.626 | 0.543–0.709 | 42.9 | 84.9 |
| 1670–1791 a.a +
AFP |
| 0.639 | 0.557–0.721 | 48.6 | 80.2 |
| AFP |
| 0.634 | 0.383–0.546 | 44.3 | 82.5 |
Discussion
Autoantibody to CENP-F has been recognized as a
potential serological biomarker for the early diagnosis of HCC
(22). As CENP-F is a high
molecular weight protein of 3210 a.a, the immunogenicity of CENP-F
antigen is critical for the sensitivity and specificity of
detection of autoantibody to CENP-F. In the present study, we
screened the predominant epitopes within the full-length protein by
bioinformatics analysis followed by ELISA detection, and selected
two peptides (121–220 a.a and 1670–1791 a.a) for further clinical
evaluation in a large cohort of HCC cases. Among the eight peptides
of CENP-F tested, peptide 121–220 a.a demonstrated the best
diagnostic value.
The CENP-F protein contains several motifs including
tandem repeats that are sufficient for centromere, cytoplasm, or
nuclear localization and for self-association (21) (Fig.
1B). Rattner et al (23) reported that the C-terminal end of
CENP-F is especially antigenic; however, the exact regions were not
determined. In other studies, Welner et al (24) evaluated the 1882–2153 a.a peptide
of CENP-F antigen by indirect ELISA using overlapping 20-mer
peptides of CENP-F spanning the amino acid sequence from 1882 to
2153 and two independent monoclonal antibodies to CENP-F in serum
samples and found several peptides with potentially good
immunogenicity. They further showed that approximately 50% of
patients who were clinically tested for antinuclear antibody (ANA)
and expressed antibodies to CENP-F were diagnosed with various
kinds of cancer, confirming that such antibodies may function as
circulating tumor markers.
In the present study we first predicted antigen
epitope peptides and screened dominant epitopes through
bioinformatics analysis, identifying eight candidate epitopes of
CENP-F peptides. In subsequent evaluation of the serological
responses of these eight antigens by indirect ELISA, two of the
candidate epitopes showed better diagnostic value for HCC; the
121–220 a.a peptide of CENP-F had good specificity whereas the
1670–1791 a.a peptide had better sensitivity (Table III) suggesting that combined use
of both peptides of CENP-F in further studies would enhance both
the sensitivity and the specificity. The 121–220 a.a peptide is
located in the N-terminal of the CENP-F protein, which is quite
different from the antigen reported previously, whereas the
1670–1791 a.a peptide is located close to the 1882–2153 a.a peptide
of CENP-F reported in other studies (23,24).
Finally, we conducted clinical evaluation in a large cohort of
cases to validate the diagnostic value of the two candidate
epitopes. Our results confirmed the promising diagnostic value of
the 121–220 a.a peptide of CENP-F in the detection of early HCC
(Table III); however, the
1670–1791 a.a peptide had lower diagnostic performance in the
validation group compared with the screening group suggesting that
further study with more cases is essential to understand the
diagnostic value of this peptide of CENP-F.
CENP-F has already been reported as a potential
biomarker for early-stage HCC (15,22).
Through high-throughput microarray analysis in large-scale cohorts
of HCC and early-stage HCC cases, our previous study confirmed the
diagnostic performance of anti-CENP-F in the detection of early
HCC. In the present study, we evaluated the clinical significance
of the 121–220 a.a peptide of CENP-F by indirect ELISA in 405 serum
samples including patients with early-stage HCC, advanced HCC, and
LC. The results showed that anti-CENP-F antibody had promising
diagnostic performance in the detection of HCC and, moreover, could
complement AFP leading to improved diagnosis of HCC or early HCC.
However, the results also revealed the limited value of anti-CENP-F
antibody in the discrimination of HCC and LC, consistent with our
previous studies (22).
According to the ROC curve, we defined cases with
antibody level of more than 125 ng/ml as positive for auto-antibody
to CENP-F (121–220 a.a). The results showed that 48 of 70 HCC cases
(68.6%) with TNM stage I, and 56 of 83 HCC cases (67.5%) with TNM
stage II or III, are positive for auto-antibody to CENP-F
(P=0.512), suggesting CENP-F auto-antibody level is not related
with the stage of HCC. However, the CENP-F had the highest
prevalence of autoantibody positivity in HCC cases with TNM stage I
implies that auto-antibody to CENP-F may have value in detection of
early HCC.
There are some other limitations in our study.
Although two candidate epitope peptides of CENP-F were identified,
the exact structure of the antigen and the underlining mechanism
remain unknown. In addition, the numbers of early-stage HCC cases
were still limited, and further studies with a larger sample sizes
would further warrant the findings in our study. Finally, as the
pathology analysis of liver biopsy is the golden standard for
diagnosis of HCC, but in the present study only a part of HCC cases
had pathology results. In our future study, we will use more
pathologically confirmed HCC cases to further evaluate the
diagnostic value of the CENP-F antibody.
In conclusion, through bioinformatics analysis and
clinical evaluation, we identified the 121–220 a.a peptide as the
peptide with highest immunogenicity in the CENP-F antigen. It also
showed promising diagnostic value in detecting early-stage HCC and
could therefore be a complement to AFP in early diagnosis of
HCC.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81071973 and
81602032), and Capital Foundation of Medical Developments
(2016-2-2025) and State Key Projects Specialized on Infectious
Diseases (2017ZX10201201-007-002).
References
|
1
|
Venook AP, Papandreou C, Furuse J and de
Guevara LL: The incidence and epidemiology of hepatocellular
carcinoma: A global and regional perspective. Oncologist. 15 Suppl
4:S5–S13. 2010. View Article : Google Scholar
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Colli A, Fraquelli M, Casazza G, Massironi
S, Colucci A, Conte D and Duca P: Accuracy of ultrasonography,
spiral CT, magnetic resonance and alpha-fetoprotein in diagnosing
hepatocellular carcinoma: A systematic review. Am J Gastroenterol.
101:513–523. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Poon D, Anderson BO, Chen LT, Tanaka K,
Lau WY, Van Cutsem E, Singh H, Chow WC, Ooi LL, Chow P, et al:
Management of hepatocellular carcinoma in Asia: Consensus statement
from the Asian oncology summit 2009. Lancet Oncol. 10:1111–1118.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marrero JA, Feng Z, Wang Y, Nguyen MH,
Befeler AS, Roberts LR, Reddy KR, Harnois D, Llovet JM, Normolle D,
et al: Alpha-fetoprotein, des-gamma carboxyprothrombin, and
lectin-bound alpha-fetoprotein in early hepatocellular carcinoma.
Gastroenterology. 137:110–118. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Z, Zhang Y, Wang Y, Xu L and Xu W:
Alpha-fetoprotein-L3 and Golgi protein 73 may serve as candidate
biomarkers for diagnosing alpha-fetoprotein-negative hepatocellular
carcinoma. Onco Targets Ther. 9:123–129. 2016.PubMed/NCBI
|
|
7
|
Ji J, Wang H, Li Y, Zheng L, Yin Y, Zou Z,
Zhou F, Zhou W, Shen F and Gao C: Diagnostic evaluation of
Des-gamma-carboxy prothrombin versus α-fetoprotein for hepatitis B
virus-related hepatocellular carcinoma in China: A large-scale,
multicentre study. PLoS One. 11:e01532272016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang J, Shao C, Zhou Q, Zhu Y, Zhu J and
Tu C: Diagnostic accuracy of serum squamous cell carcinoma antigen
and squamous cell carcinoma antigen-immunoglobulin M for
hepatocellular carcinoma: A meta-analysis. Mol Clin Oncol.
3:1165–1171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shen Q, Fan J, Yang XR, Tan Y, Zhao W, Xu
Y, Wang N, Niu Y, Wu Z, Zhou J, et al: Serum DKK1 as a protein
biomarker for the diagnosis of hepatocellular carcinoma: A
large-scale, multicentre study. Lancet Oncol. 13:817–826. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lim TS, Kim DY, Han KH, Kim HS, Shin SH,
Jung KS, Kim BK, Kim SU, Park JY and Ahn SH: Combined use of AFP,
PIVKA-II and AFP-L3 as tumor markers enhances diagnostic accuracy
for hepatocellular carcinoma in cirrhotic patients. Scand J
Gastroenterol. 51:344–353. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu J, Wang ZJ, Chen LH and Dong WZ:
Diagnostic value of serum squamous cell carcinoma antigen for
hepatocellular carcinoma: A systematic review and meta-analysis.
Scand J Clin Lab Invest. 77:8–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giannelli G, Fransvea E, Trerotoli P,
Beaugrand M, Marinosci F, Lupo L, Nkontchou G, Dentico P and
Antonaci S: Clinical validation of combined serological biomarkers
for improved hepatocellular carcinoma diagnosis in 961 patients.
Clin Chim Acta. 383:147–152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marrero JA, Romano PR, Nikolaeva O, Steel
L, Mehta A, Fimmel CJ, Comunale MA, D'Amelio A, Lok AS and Block
TM: GP73, a resident Golgi glycoprotein, is a novel serum marker
for hepatocellular carcinoma. J Hepatol. 43:1007–1012. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hong Y: Autoantibodies against
tumor-associated antigens for detection of hepatocellular
carcinoma. World J Hepatol. 7:1581–1585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang JY, Zhu W, Imai H, Kiyosawa K, Chan
EK and Tan EM: De-novo humoral immune responses to
cancer-associated autoantigens during transition from chronic liver
disease to hepatocellular carcinoma. Clin Exp Immunol. 125:3–9.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dai Y, Liu L, Zeng T, Zhu YH, Li J, Chen
L, Li Y, Yuan YF, Ma S and Guan XY: Characterization of the
oncogenic function of centromere protein F in hepatocellular
carcinoma. Biochem Biophys Res Commun. 436:711–718. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu X, Mancini MA, Chang KH, Liu CY, Chen
CF, Shan B, Jones D, Yang-Feng TL and Lee WH: Characterization of a
novel 350-kilodalton nuclear phosphoprotein that is specifically
involved in mitotic-phase progression. Mol Cell Biol. 15:5017–5029.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma L, Zhao X and Zhu X: Mitosin/CENP-F in
mitosis, transcriptional control and differentiation. J Biomed Sci.
13:205–213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Varis A, Salmela AL and Kallio MJ: Cenp-F
(mitosin) is more than a mitotic marker. Chromosoma. 115:288–295.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu X, Ding L and Pei G: Carboxyl terminus
of mitosin is sufficient to confer spindle pole localization. J
Cell Biochem. 66:441–449. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
The national center for biotechnology
information database. http://www/uniprot.org/uniprot/P49454
|
|
22
|
Hong Y, Long J, Li H, Chen S, Liu Q, Zhang
B, He X, Wang Y, Li H, Li Y, et al: An analysis of immunoreactive
signatures in early stage hepatocellular carcinoma. EBioMedicine.
2:438–446. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rattner JB, Rees J, Whitehead CM, Casiano
CA, Tan EM, Humbel RL, Conrad K and Fritzler MJ: High frequency of
neoplasia in patients with autoantibodies to centromere protein
CENP-F. Clin Invest Med. 20:308–319. 1997.PubMed/NCBI
|
|
24
|
Welner S, Trier NH, Houen G and Hansen PR:
Identification and mapping of a linear epitope of centromere
protein F using monoclonal antibodies. J Pept Sci. 19:95–101. 2013.
View Article : Google Scholar : PubMed/NCBI
|