Introduction
Hepatocellular carcinoma (HCC) is one of the most
common types of malignant cancer, which is associated with a poor
prognosis. In addition, HCC is the fourth most common cause of
cancer-associated mortality worldwide (1). It was previously estimated that
39,230 new cases of HCC and 27,170 cases of HCC-associated
mortality would occur in the United States in 2016 (1). Due to the highest occurrence of HCC
in East Asia, this estimation should be further increased in this
area (2). Chronic liver diseases,
including hepatitis, fibrosis and cirrhosis, are the primary causes
of HCC. In addition, ~90% of cases of HCC develop from cirrhotic
livers (3), and HCC and cirrhosis
share numerous risk factors, including hepatitis, alcohol
consumption, obesity, diabetes, gender and advanced age (4,5).
These findings suggest that cirrhosis and HCC may share
pathophysiological similarities.
Despite the aforementioned correlations between HCC
and cirrhosis, the detailed mechanisms linking these two diseases
remain to be fully elucidated. Hepatic stellate cells (HSCs) are
primary fibrogenic cells, which are involved in cirrhosis-dependent
carcinogenesis (6). Su et
al (7) indicated that HSCs can
be activated by inflammatory signals and macrophages, which
contribute to fibrogenesis, and therefore may be considered
positive risk factors for HCC. Furthermore, it has been
demonstrated that activated HSCs can foster a conducive environment
to directly support hepatic tumorigenesis via secreting numerous
cytokines, including Wnt ligands, interleukin 6 and growth factors
(8). De Minicis et al
(9) reported that HSCs can be
activated by lipopolysaccharide produced by the intestinal
microflora via the toll-like receptor 4 signaling pathway, which
can upregulate the expression of proinflammatory chemokines or
cytokines, and facilitate the invasion and migration of HCC
(10). Increased extracellular
matrix (ECM) production is another feature of cirrhosis that
contributes to tumorigenesis (11). Previous studies have reported that
increased ECM production may promote the growth, survival and
migration of precancerous cells via integrin signaling (6,12).
Furthermore, increased ECM production may disturb cell signaling by
sequestering growth factors, including interleukin families,
fibroblast growth factor and transforming growth factor (13). This sequestration may serve a role
in the aberration of normal liver cells. However, although these
existing studies have confirmed that the development of HCC is
closely associated with cirrhosis, the detailed mechanisms remain
to be elucidated. Therefore, further investigations into how HCC
develops from cirrhosis are required.
In order to identify the epigenetic alterations and
potential role of DNA methylation markers in HCC and its prognosis,
Villanueva et al (14)
generated a methylation-based prognostic signature based on 304
samples from patients with HCC after surgical resection using a
training-validation scheme. The GSE63898 dataset contains data from
this study, which analyzed whole-genome transcriptome alterations
(14). Villanueva et al
(14) confirmed a high prevalence
of methylation deregulated genes, including Ras association
domain family member 1, APC, WNT signaling pathway regulator
and insulin like growth factor 2 (IGF2), and
identified potential epidrivers, such as ephrin B2 and
septin 9; however, the mechanisms underlying the development
of HCC from cirrhosis were not determined. In order to investigate
the pathogenesis of HCC, the GSE63898 dataset was used to analyze
differences between HCC and cirrhotic liver tissues. The present
study aimed to identify potential target molecules, which may aid
HCC clinical treatment.
Materials and methods
Data acquisition
The GSE63898 expression profile (14), which contained data from HCC and
liver cirrhosis samples, was downloaded from the Gene Expression
Omnibus (www.ncbi.nlm.nih.gov/geo/). The samples were sequenced
using the Affymetrix Human Genome U219 Array [HG-U219] platform. In
this profile, a total of 396 liver tissue samples were collected,
including 228 HCC tissue samples and 168 non-tumor liver adjacent
cirrhotic tissue samples. The research was authorized by the
Institutional Review Boards of the participating centers.
Data preprocessing
The downloaded raw data were preprocessed using the
Robust Multi-array Average method (15,16)
in the Affy package of R (17),
including background correction, normalization and expression
calculation. Gene expression was presented as the mean value of
different probes.
Differentially expressed genes (DEGs)
screening
DEGs between HCC samples and cirrhotic liver samples
were screened by Student's t-test in the Linear Models for
Microarray Data package (18). The
P-value obtained from the Student's t-test was adjusted according
to the Benjamini and Hochberg method (19) and the obtained adjusted P<0.01
was set as the threshold.
Functional and pathway enrichment
analyses
Gene Ontology (GO) is a free database used in
biological process (BP), molecular function and cellular component
analyses (20). Kyoto Encyclopedia
of Genes and Genomes (KEGG) is also a free database specifically
used in pathway analysis (21).
The Database for Annotation, Visualization and Integrated Discovery
(DAVID) is a common online tool used in large-scale gene functional
analyses (22). Therefore, GO
functional and KEGG pathway enrichment analyses of DEGs were
conducted using DAVID with the criterion of P<0.05.
Protein-protein interaction (PPI)
network construction and analysis
According to the required confidence threshold set
(combined score) >0.7, PPIs were analyzed using Search Tool for
the Retrieval of Interacting Genes (23) and the PPI network was constructed
by Cytoscape (24). CytoNCA
(25), which is a plugin of
Cytoscape for PPI network evaluation, was used to perform a
topological analysis, including degree centrality (DC), betweenness
centrality (BC) and closeness centrality (CC), for hub gene
screening (26). Proteins were
defined as nodes, and PPI associations were defined as edges in the
PPI network.
Sub-module analysis
Based on the PPI network, the MCODE plugin of
Cytoscape was used to screen specific bio-functional sub-modules
(27) in the network. KEGG pathway
analysis of these modules was performed using DAVID with the
criterion of P<0.05. Proteins were defined as nodes, and PPI
associations were defined as edges in modules screened from the PPI
network.
Results
DEGs screening
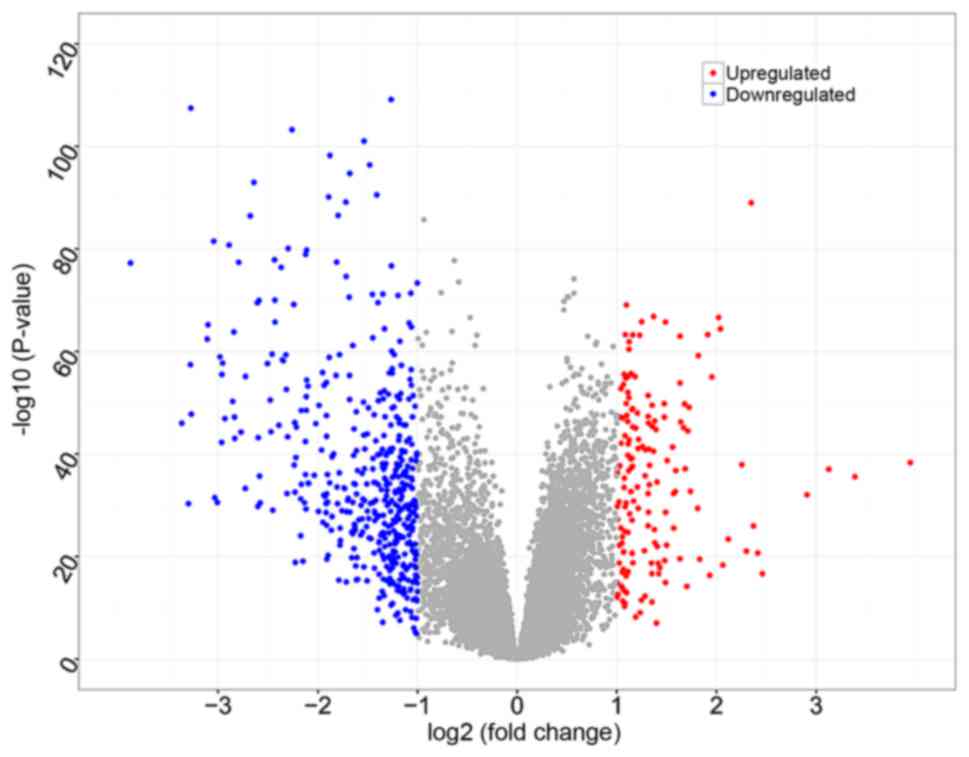
Based on data preprocessing and Student's t-test, a
total of 20,568 genes were identified, and 634 DEGs were identified
between HCC samples and cirrhotic liver samples, of which 165 were
upregulated and 469 were downregulated (Fig. 1).
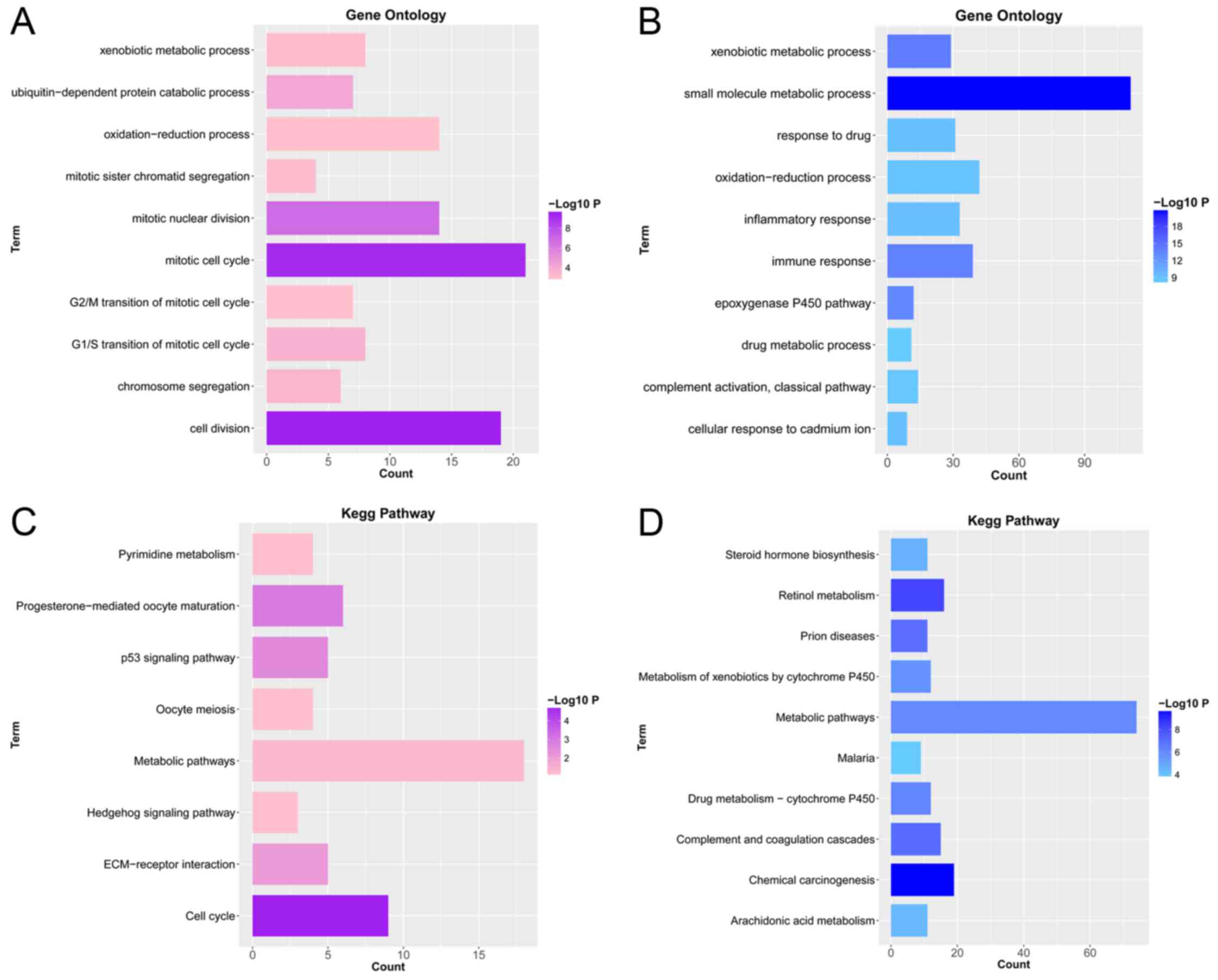
Functional and pathway enrichment
To investigate the biological functions of the DEGs,
GO_BP functional and KEGG pathway enrichment analyses were
conducted. GO_BPs of upregulated DEGs were significantly enriched
in cell division (P=2.91×10−10), mitotic cell cycle
(P=5.57×10−10) and mitotic nuclear division
(P=1.42×10−7), etc. (Fig.
2A). Furthermore, GO_BPs of downregulated DEGs were
significantly enriched in small molecule metabolic process
(P=1.87×10−21), xenobiotic metabolic process
(P=7.15×10−15) and immune response
(P=1.83×10−14), etc. (Fig.
2B). KEGG pathways of upregulated DEGs were significantly
enriched in cell cycle (P=1.67×10−5),
progesterone-mediated oocyte maturation (P=0.001) and p53 signaling
pathway (P=0.003), etc. (Fig. 2C).
In addition, KEGG pathways of downregulated genes were
significantly enriched in chemical carcinogenesis
(P=1.92×10−10), retinol metabolism
(P=4.53×10−9) and complement and coagulation cascades
(P=8.67×10−8), etc. (Fig.
2D).
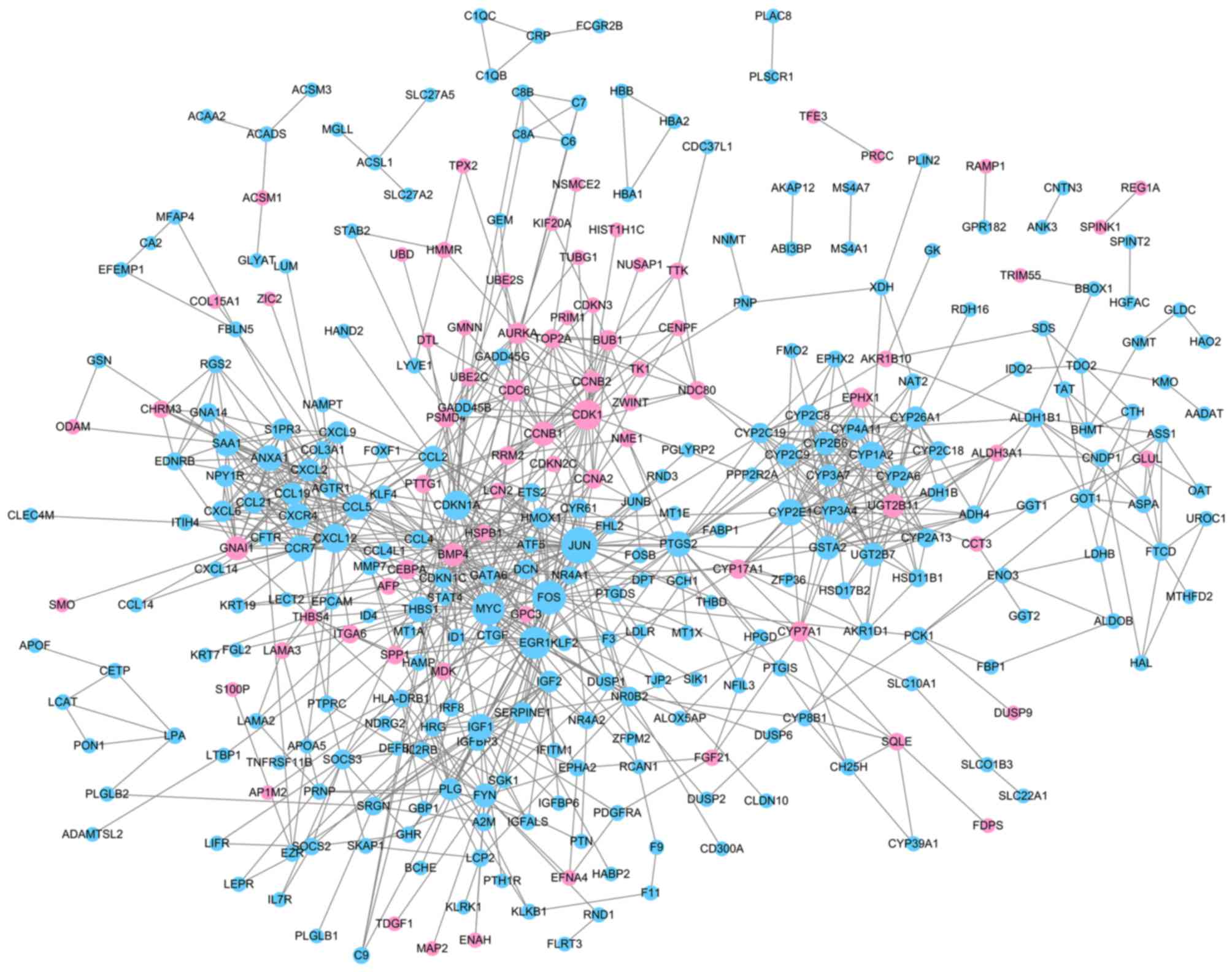
PPI network analysis
With required confidence >0.7, the PPI network
for DEGs was constructed with 324 nodes and 915 edges (Fig. 3, Table
I), including 74 upregulated genes and 250 downregulated genes.
Downregulated Jun proto-oncogene, AP-1 transcription factor subunit
(JUN; degree, 39), Fos proto-oncogene, AP-1 transcription factor
subunit (FOS; degree, 34) and v-myc avian myelocytomatosis viral
oncogene homolog (MYC; degree, 32) had high degrees and were
therefore identified as the hub nodes in the PPI network.
 | Table I.Proteins with a degree ≥20, as
determined by topological analysis of the protein-protein
interaction network. |
Table I.
Proteins with a degree ≥20, as
determined by topological analysis of the protein-protein
interaction network.
| Gene | DC | BC | CC |
|---|
| JUN | 39 | 17,219.9 | 0.0283 |
| FOS | 34 | 19,632.8 | 0.0283 |
| MYC | 32 | 8,693.0 | 0.0282 |
| EGR1 | 31 | 7,531.4 | 0.0282 |
| CDK1 | 27 | 6,022.2 | 0.0280 |
| CDKN1A | 26 | 4,038.7 | 0.0280 |
| CXCL12 | 25 | 3,844.4 | 0.0279 |
| CYP2E1 | 22 | 2,721.3 | 0.0278 |
| CYP1A2 | 22 | 4,144.9 | 0.0276 |
| IGF1 | 21 | 6,096.5 | 0.0280 |
| CCR7 | 21 | 2,227.9 | 0.0277 |
| CYP3A4 | 21 | 1550.2 | 0.0277 |
| PTGS2 | 20 | 6,968.9 | 0.0281 |
| THBS1 | 20 | 5,690.0 | 0.0281 |
| CCL5 | 20 | 3,738.3 | 0.0280 |
| ANXA1 | 20 | 634.9 | 0.0275 |
| SAA1 | 20 | 1,475.6 | 0.0275 |
Sub-module analysis
According to the selection criteria, four
sub-modules were screened and named module 1, 2, 3 and 4 (Table II). In module 1 (Fig. 4A), there were 14 nodes with 91
edges, and according to the KEGG pathway analysis, the genes were
significantly enriched in chemokine signaling pathway
(P=3.14×10−13) and cytokine-cytokine receptor
interaction (P=5.07×10−7), etc. In particular,
downregulated C-X-C motif chemokine ligand 12 (CXCL12; degree, 13),
C-C motif chemokine receptor 7 (CCR7; degree, 13) and C-C motif
chemokine ligand 5 (CCL5; degree, 13) were the three nodes with the
highest degrees in this module. In addition, there were 7 nodes
with 21 edges identified in module 2 (Fig. 4B), and according to the KEGG
pathway analysis, the genes were significantly enriched in
complement and coagulation cascades (P=5.85×10−4) and
p53 signaling pathway (P=0.04). In particular, downregulated IGF1
(degree, 6), plasminogen (degree, 6) and IGF2 (degree, 6) were the
three nodes with the highest degrees in this module. In module 3,
there were 7 nodes with 20 edges (Fig.
4C), and according to the KEGG pathway analysis, the genes were
significantly enriched in retinol metabolism
(P=5.57×10−13) and steroid hormone biosynthesis
(P=1.11×10−5), etc. In particular, downregulated
cytochrome P450 family 1 subfamily A member 2 (degree, 6),
cytochrome P450 family 4 subfamily A member 11 (degree, 6) and
cytochrome P450 family 3 subfamily A member 4 (degree, 6) were the
three nodes with the highest degrees in this module. In addition,
there were 9 nodes with 24 edges in module 4 (Fig. 4D), and according to the KEGG
pathway analysis, the genes were significantly enriched in chemical
carcinogenesis (P=2.31×10−16) and drug metabolism
cytochrome P450 (P=8.13×10−10), etc. In particular,
downregulated cytochrome P450 family 2 subfamily E member 1
(CYP2E1; degree, 6), cytochrome P450 family 2 subfamily C member 9
(CYP2C9; degree, 6) and cytochrome P450 family 2 subfamily A member
6 (CYP2A6; degree, 5) were the three nodes with the highest degrees
in this module.
 | Table II.Top 5 enriched Kyoto Encyclopedia of
Genes and Genomes terms obtained from the subnet module
analysis. |
Table II.
Top 5 enriched Kyoto Encyclopedia of
Genes and Genomes terms obtained from the subnet module
analysis.
| Term | Count | P-value | Proteins |
|---|
| Cluster 1 |
|
|
|
|
Chemokine signaling
pathway | 10 |
3.14×10−13 | CCR7, GNAI1, CXCR4,
CCL21, CXCL2 … |
|
Cytokine-cytokine receptor
interaction | 7 |
5.07×10−7 | CCR7, CXCR4, CCL21,
CXCL9, CCL19 … |
|
Rheumatoid arthritis | 3 | 0.007870 | CXCL6, CCL5,
CXCL12. |
| NF-κB
signaling pathway | 3 | 0.008048 | CCL21, CCL19,
CXCL12. |
|
Leukocyte transendothelial
migration | 3 | 0.014455 | GNAI1, CXCR4,
CXCL12. |
| Cluster 2 |
|
|
|
|
Complement and coagulation
cascades | 3 |
5.85×10−4 | A2M, SERPINE1,
PLG. |
| p53
signaling pathway | 2 | 0.037772 | SERPINE1,
IGF1. |
| Cluster 3 |
|
|
|
| Retinol
metabolism | 7 |
5.57×10−13 | CYP3A4, CYP4A11,
CYP2B6, UGT2B11, CYP26A1 … |
| Steroid
hormone biosynthesis | 4 |
1.11×10−5 | CYP3A4, UGT2B11,
CYP1A2, UGT2B7. |
|
Chemical carcinogenesis | 4 |
2.94×10−5 | CYP3A4, UGT2B11,
CYP1A2, UGT2B7. |
|
Metabolic pathways | 7 |
3.00×10−5 | CYP3A4, CYP4A11,
CYP2B6, UGT2B11, CYP26A1 … |
| Drug
metabolism cytochrome P450 | 3 |
7.30×10−4 | CYP3A4, CYP2B6,
CYP1A2. |
| Cluster 4 |
|
|
|
|
Chemical carcinogenesis | 9 |
2.31×10−16 | GSTA2, CYP3A7,
CYP2C19, CYP2C9, CYP2C18 … |
| Drug
metabolism-cytochrome P450 | 6 |
8.13×10−10 | GSTA2, CYP2C19,
CYP2C9, CYP2C8, CYP2A6 … |
|
Metabolism of xenobiotics
by | 5 |
2.30×10−7 | GSTA2, CYP2C9,
EPHX1, CYP2A6, CYP2E1. |
|
cytochrome P450 |
|
|
|
| Retinol
metabolism | 5 |
4.91×10−7 | CYP3A7, CYP2C9,
CYP2C18, CYP2C8, CYP2A6. |
|
Linoleic acid metabolism | 4 |
3.70×10−6 | CYP2C19, CYP2C9,
CYP2C8, CYP2E1. |
Discussion
Disease-specific differential gene expression
reveals potential alterations associated with disease development.
According to the analysis criteria of the present study, a total of
634 DEGs were identified in HCC versus cirrhotic tissue samples, of
which 165 were upregulated and 469 were downregulated. After GO
functional and KEGG pathway enrichment analyses, a PPI network was
constructed with 324 nodes and 915 edges. In particular, JUN, FOS
and MYC were the hub nodes in this network. Based on the PPI
network, four specific modules were identified. According to the
KEGG analysis results, module 1 was significantly enriched in
chemokine signaling pathway, module 2 was significantly enriched in
complement and coagulation cascades, module 3 was significantly
enriched in retinol metabolism, and module 4 was significantly
enriched in drug metabolism cytochrome P450.
The majority of the nodes in module 1 were
chemokines or their receptors, including CXCL12, CCR7 and CCL5.
Chemokines are cytokines that specifically respond to
proinflammatory stimuli, and are involved in the migration of
immune cells to damaged organs and are associated with HCC
development (28). Chemokines are
small molecules that can be divided into four groups (C, CC, CXC,
and CXC3C) by the motifs of their NH2 terminals
(29). CXCL12, also known as
stromal cell-derived factor 1, is produced by HSCs, biliary
epithelial cells and liver sinusoidal endothelial cells (30), and has a higher expression in
cirrhotic liver tissue and HCC (31). It is well known that the
CXCL12-CXCR4 axis serves an important role in the pathogenesis of
liver diseases, including cirrhosis (32), migration (33), invasion (34) and HCC prognosis (35). However, in the present study, a
significant downregulation was identified in HCC compared with in
cirrhosis. Furthermore, Neve Polimeno et al (36) and Shibuta et al (37) demonstrated that the expression of
CXCL12 is significantly reduced in hepatoma carcinoma cells and HCC
compared with normal controls; however, the detailed mechanism of
this reduction remains unclear. Therefore, it may be hypothesized
that the CXCL12-CXCR4 axis serves various roles in different
conditions of HCC originating from cirrhosis. CCR7 has been
reported to be associated with the clinicopathological parameters
of HCC (38). An in vitro
study indicated that the expression of CCR7 is reduced in HCC cell
lines compared with in the normal liver cell line L-02 (39). A similar result was identified in
the present study; CCR7 expression was downregulated in HCC tissue
samples compared with in cirrhotic tissue samples. Shi et al
(40) reported that CCL1, which is
exclusively expressed in non-hematopoietic stromal cells (41), may serve as a tumor suppressor by
inhibiting CCR7-associated chemotaxis in HCC. Furthermore,
overexpression of CCL21, the ligand of CCR7, in HCC cell lines
presented a potential antitumor effect in a model of HCC (42); however, the detailed function of
CCR7-CCL21 requires further investigation. CCL5 has been reported
to be associated with the inflammatory cirrhosis stages in chronic
liver disease (43). It has been
demonstrated that CCL5 downregulation is able to inhibit the
effects of the human bone marrow stromal cell line HS-5 on Huh-7
cell migration and invasion via the phosphatidylinositol
3-kinase/Akt pathway (44). These
findings suggested that CCL5 may promote the migration and invasion
of Huh-7 cells. In addition, Sadeghi et al (45) reported that CCL5 is upregulated in
the serum of patients with HCC compared with patients with
cirrhosis. Conversely, in the present study, a significant
downregulation in CCL5 was detected in HCC tissue samples compared
with in cirrhotic tissue. Therefore, it may be hypothesized that
CCL5 expression differs between tissue and serum and in
vitro and in vivo. This may be a novel opportunity for
CCL5-targeted therapy.
CYPs belong to a superfamily of enzymes that serve
important roles in the metabolism of procarcinogens, carcinogens
and drugs (46). CYP2E1, CYP2C9
and CYP2A6 were three important CYPs identified in module 4 in the
present study. CYP2E1 is often deficient in HCC cell lines
(47). A significant decrease in
CYP2E1 expression was identified in HCC samples compared with
cirrhotic tissue samples in the present study. In addition,
Kinoshita and Miyata (48)
reported that CYP2E1 is significantly downregulated in HCC liver
tissue. Wu et al (49)
reported that CYP2E1 can be downregulated by resveratrol to
attenuate diethylnitrosamine and 2-acetylaminofluorene-induced
hepatocarcinogenesis in Sprague Dawley rats, thereby suggesting
that CYP2E1 may have an inhibitory effect on HCC carcinogenesis. In
addition, hepatitis B virus (HBV)-x protein can inhibit the
expression of CYP2E1 via downregulating hepatocyte nuclear factor 4
(HNF4), resulting in the promotion of human hepatoma cell growth
(48). Taken together, these
results suggested that CYP2E1 may be a promising target for the
drug-targeted therapy of HCC. CYP2C9 has been reported to be
downregulated in HCC compared with in peri-HCC and normal control
tissues (50); this result was
similar to the findings of the present study. In addition, it has
been reported that CYP2C9 can be suppressed by
hsa-microRNA-128-3P, resulting in the invasion of HCC (51). Myung et al (52) also reported that CYP2C9 is involved
in the sensitization of liver cancer stem cells to anticancer drugs
via the signal transducer and activator of transcription 3
signaling pathway, particularly in advanced stages. However, the
detailed mechanisms of these results remain unclear; therefore,
further study is required. CYP2A6 is another important member of
the CYP family that was downregulated in module 4. Similar to
CYP2C9, CYP2A6 can be regulated by HNF4 and is involved in the
metabolism of nitrosamines and aflatoxin B1 (53). Fushiya et al (54) reported that CYP2A6 is implicated in
the metabolism of 5-fluorouracil, which is commonly used in HCC
clinical chemotherapy. In addition, a previous study indicated that
CYP2A6 was downregulated in HBV- and hepatitis C
virus-infected livers, and in HCC (55); the present study suggested that HCC
samples had a significantly lower expression of CYP2A6
compared with cirrhotic liver samples. Furthermore, a previous
study reported that CYP2A6 activity was decreased in moderate or
severe alcoholic liver diseases, but not in mild severe alcoholic
liver disease (56). Therefore, it
may be hypothesized that CYP2A6 may be negatively correlated with
the stage of HCC development. Further studies are required to
investigate the CYP2A6-associated functions and pathways in
HCC.
Although numerous DEGs with their potential
functions were identified between HCC and cirrhosis samples in an
in silico analysis, there remain limitations to the present
study. For example, in silico analysis of the obtained
results revealed that numerous chemokines and cytokines were
involved in HCC development; however, the involvement of the
identified genes in vitro remains unknown. Therefore,
further experimental verifications of these genes are required.
Furthermore, these analyses were based on subjective criteria;
therefore, some genes, which have lower degrees but important roles
in HCC carcinogenesis, may have been overlooked. Despite these
limitations, these analytical results may provide novel insights
into the mechanism of HCC originating from cirrhosis.
In conclusion, the present study identified a series
of DEGs between HCC and cirrhotic tissue samples. Based on the GO
and KEGG enrichment analyses of DEGs in the PPI network,
chemokines, such as CXCL12, CCR7, CCL5, and cytokines, such as
CYP2E1, CYP2C9, CYP2A6, were identified as two important components
in the process of HCC developing from cirrhosis as they are
associated with the regulation of inflammation, growth and the
invasion of pre-cancerous cells in the liver; thus, they may serve
crucial roles in HCC development. However, these results were
derived from bioinformatics analysis; therefore, the effects of
these genes in HCC in vivo require further verification.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
HSCs
|
hepatic stellate cells
|
|
GO
|
Gene Ontology
|
|
BP
|
biological process
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
DC
|
degree centrality
|
|
BC
|
betweenness centrality
|
|
CC
|
closeness centrality
|
|
CCR7
|
C-C motif chemokine receptor 7
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan Q, He M, Deng X, Wu WK, Zhao L, Tang
J, Wen G, Sun X and Liu Y: Derepression of c-Fos caused by
microRNA-139 down-regulation contributes to the metastasis of human
hepatocellular carcinoma. Cell Biochem Funct. 31:319–324. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Archambeaud I, Auble H, Nahon P, Planche
L, Fallot G, Faroux R, Gournay J, Samuel D, Kury S and Féray C:
Risk factors for hepatocellular carcinoma in caucasian patients
with non-viral cirrhosis: The importance of prior obesity. Liver
Int. 35:1872–1876. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saunders D, Seidel D, Allison M and
Lyratzopoulos G: Systematic review: The association between obesity
and hepatocellular carcinoma-epidemiological evidence. Aliment
Pharmacol Ther. 31:1051–1063. 2010.PubMed/NCBI
|
|
5
|
Naveau S: Body mass index and risk of
liver cirrhosis in middle aged UK women: prospective study.
Gastroentérol Clin Biol. 34:429–430. 2010. View Article : Google Scholar
|
|
6
|
Zhang DY and Friedman SL:
Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology.
56:769–775. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Su TH, Kao JH and Liu CJ: Molecular
mechanism and treatment of viral hepatitis-related liver fibrosis.
Int J Mol Sci. 15:10578–10604. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Friedman SL: Hepatic stellate cells:
Protean, multifunctional, and enigmatic cells of the liver. Physiol
Rev. 88:125–172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De Minicis S, Seki E, Uchinami H, Kluwe J,
Zhang Y, Brenner DA and Schwabe RF: Gene expression profiles during
hepatic stellate cell activation in culture and in vivo.
Gastroenterology. 132:1937–1946. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu WT, Jing YY, Yu GF, Han ZP, Yu DD, Fan
QM, Ye F, Li R, Gao L, Zhao QD, et al: Toll like receptor 4
facilitates invasion and migration as a cancer stem cell marker in
hepatocellular carcinoma. Cancer Lett. 358:136–143. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee SK, Kim MH, Cheong JY, Cho SW, Yang SJ
and Kwack K: Integrin alpha V polymorphisms and haplotypes in a
Korean population are associated with susceptibility to chronic
hepatitis and hepatocellular carcinoma. Liver Int. 29:187–195.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu BH, Wu ZZ and Qin J: Effects of
integrins on laminin chemotaxis by hepatocellular carcinoma cells.
Mol Biol Rep. 37:1665–1670. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vlodavsky I, Miao HQ, Medalion B, Danagher
P and Ron D: Involvement of heparan sulfate and related molecules
in sequestration and growth promoting activity of fibroblast growth
factor. Cancer Metastasis Rev. 15:177–186. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Villanueva A, Portela A, Sayols S,
Battiston C, Hoshida Y, Méndez-González J, Imbeaud S, Letouzé E,
Hernandez-Gea V, Cornella H, et al: DNA methylation-based prognosis
and epidrivers in hepatocellular carcinoma. Hepatology.
61:1945–1956. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smyth GK: limma: Linear models for
microarray data. Springer; New York: pp. 397–420. 2005
|
|
19
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc. 57:289–300. 1995.
|
|
20
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanehisa M and Goto S: KEGG: Κyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ehling J and Tacke F: Role of chemokine
pathways in hepatobiliary cancer. Cancer Lett. 379:173–183. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Charo IF and Ransohoff RM: The many roles
of chemokines and chemokine receptors in inflammation. N Engl J
Med. 354:610–621. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Marra F and Tacke F: Roles for chemokines
in liver disease. Gastroenterology. 147:577–594.e1. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wald O, Pappo O, Safadi R, Dagan-Berger M,
Beider K, Wald H, Franitza S, Weiss I, Avniel S, Boaz P, et al:
Involvement of the CXCL12/CXCR4 pathway in the advanced liver
disease that is associated with hepatitis C virus or hepatitis B
virus. Eur J Immunol. 34:1164–1174. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ghanem I, Riveiro ME, Paradis V, Faivre S,
de Parga PM and Raymond E: Insights on the CXCL12-CXCR4 axis in
hepatocellular carcinoma carcinogenesis. Am J Transl Res.
6:340–352. 2014.PubMed/NCBI
|
|
33
|
Schimanski CC, Bahre R, Gockel I, Müller
A, Frerichs K, Hörner V, Teufel A, Simiantonaki N, Biesterfeld S,
Wehler T, et al: Dissemination of hepatocellular carcinoma is
mediated via chemokine receptor CXCR4. Br J Cancer. 95:210–217.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shah AD, Bouchard MJ and Shieh AC:
Interstitial fluid flow increases hepatocellular carcinoma cell
invasion through CXCR4/CXCL12 and MEK/ERK signaling. PLoS One.
10:e01423372015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiang ZL, Zeng ZC, Tang ZY, Fan J, Zhuang
PY, Liang Y, Tan YS and He J: Chemokine receptor CXCR4 expression
in hepatocellular carcinoma patients increases the risk of bone
metastases and poor survival. BMC Cancer. 9:1762009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Neve Polimeno M, Ierano C, D'Alterio C,
Simona Losito N, Napolitano M, Portella L, Scognamiglio G,
Tatangelo F, Maria Trotta A, Curley S, et al: CXCR4 expression
affects overall survival of HCC patients whereas CXCR7 expression
does not. Cell Mol Immunol. 12:474–482. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shibuta K, Mori M, Shimoda K, Inoue H,
Mitra P and Barnard GF: Regional expression of CXCL12/CXCR4 in
liver and hepatocellular carcinoma and cell-cycle variation during
in vitro differentiation. Jpn J Cancer Res. 93:789–797. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schimanski CC, Bahre R, Gockel I,
Junginger T, Simiantonaki N, Biesterfeld S, Achenbach T, Wehler T,
Galle PR and Moehler M: Chemokine receptor CCR7 enhances
intrahepatic and lymphatic dissemination of human hepatocellular
cancer. Oncol Rep. 16:109–113. 2006.PubMed/NCBI
|
|
39
|
Luo KQ, Shi YN and Peng JC: The effect of
chemokine CC motif ligand 19 on the proliferation and migration of
hepatocellular carcinoma. Tumour Biol. 35:12575–12581. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shi JY, Yang LX, Wang ZC, Wang LY, Zhou J,
Wang XY, Shi GM, Ding ZB, Ke AW, Dai Z, et al: CC chemokine
receptor like 1 functions as a tumor suppressor by impairing
CCR7-related chemotaxis in hepatocellular carcinoma. J Pathol.
235:546–558. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Heinzel K, Benz C and Bleul CC: A silent
chemokine receptor regulates steady-state leukocyte homing in vivo.
Proc Natl Acad Sci USA. 104:pp. 8421–8426. 2007; View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liang CM, Chen L, Hu H, Ma HY, Gao LL, Qin
J and Zhong CP: Chemokines and their receptors play important roles
in the development of hepatocellular carcinoma. World J Hepatol.
7:1390–1402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mohs A, Kuttkat N, Reißing J, Zimmermann
HW, Sonntag R, Proudfoot A, Youssef SA, de Bruin A4, Cubero FJ and
Trautwein C: Functional role of CCL5/RANTES for HCC progression
during chronic liver disease. J Hepatol. 66:743–753. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bai H, Weng Y, Bai S, Jiang Y, Li B, He F,
Zhang R, Yan S, Deng F, Wang J and Shi Q: CCL5 secreted from bone
marrow stromal cells stimulates the migration and invasion of Huh7
hepatocellular carcinoma cells via the PI3K-Akt pathway. Int J
Oncol. 45:333–343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sadeghi M, Lahdou I, Oweira H, Daniel V,
Terness P, Schmidt J, Weiss KH, Longerich T, Schemmer P, Opelz G
and Mehrabi A: Serum levels of chemokines CCL4 and CCL5 in
cirrhotic patients indicate the presence of hepatocellular
carcinoma. Br J Cancer. 113:756–762. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Korobkova EA: Effect of natural
polyphenols on CYP metabolism: implications for diseases. Chem Res
Toxicol. 28:1359–1390. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Puszyk WM, Hlady R, Robertson K, Cabrera R
and Liu C: Epigenetic signatures of alcohol abuse in hepatocellular
carcinoma. FASEB J. 30 1 Suppl:S516.112016.
|
|
48
|
Kinoshita M and Miyata M: Underexpression
of mRNA in human hepatocellular carcinoma focusing on eight loci.
Hepatology. 36:433–438. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu X, Li C, Xing G, Qi X and Ren J:
Resveratrol downregulates Cyp2e1 and attenuates chemically induced
hepatocarcinogenesis in SD rats. J Toxicol Pathol. 26:385–392.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen H, Shen ZY, Xu W, Fan TY, Li J, Lu
YF, Cheng ML and Liu J: Expression of P450 and nuclear receptors in
normal and end-stage Chinese livers. World J Gastroenterol.
20:8681–8690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu D, Green B, Marrone A, Guo Y, Kadlubar
S, Lin D, Fuscoe J, Pogribny I and Ning B: Suppression of CYP2C9 by
microRNA hsa-miR-128-3p in human liver cells and association with
hepatocellular carcinoma. Sci Rep. 5:85342015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Myung SJ, Yoon JH and Yu SJ: STAT3 &
Cytochrome P450 2C9: A novel signaling pathway in liver cancer stem
cells. Biomed Pharmacother. 66:612–616. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jover R, Bort R, Gómez-Lechón MJ and
Castell JV: Cytochrome P450 regulation by hepatocyte nuclear factor
4 in human hepatocytes: A study using adenovirus-mediated antisense
targeting. Hepatology. 33:668–675. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fushiya N, Takagi I, Nishino H, Akizuki S
and Ohnishi A: Genetic polymorphisms of enzymes related to oral
tegafur/uracil therapeutic efficacy in patients with hepatocellular
carcinoma. Anticancer Drugs. 24:617–622. 2013.PubMed/NCBI
|
|
55
|
Iizuka N, Oka M, Hamamoto Y, Mori N,
Tamesa T, Tangoku A, Miyamoto T, Uchimura S, Tamesa T, Tangoku A,
et al: Altered levels of cytochrome p450 genes in hepatitis B or C
virus-infected liver identified by oligonucleotide microarray.
Cancer Genomics Proteomics. 1:53–58. 2004.
|
|
56
|
Sotaniemi EA, Rautio A, Bäckstrom M,
Arvela P and Pelkonen O: CYP3A4 and CYP2A6 activities marked by the
metabolism of lignocaine and coumarin in patients with liver and
kidney diseases and epileptic patients. Br J Clin Pharmacol.
39:71–76. 1995. View Article : Google Scholar : PubMed/NCBI
|