Introduction
Idiopathic pulmonary fibrosis (IPF) is a form of
chronic progressive interstitial pneumonia that continues to be
associated with high morbidity and mortality, despite the fact that
new anti-fibrotic drugs have recently been added to our quiver
(1).
The exact cause of the disease remains unknown,
however there is a strong association with aging and smoking,
similarly to lung cancer (LC) (2,3).
Aggregating mutational burden, increased epigenetic gene silencing
through aberrant DNA methylation patterns and telomere dysfunction
frame the probable underlying correlation (4–8). A
deeper view of the pathophysiology of the two diseases reveals
several similarities and common pathways involved (4,5,9–11).
Crucial cellular mechanisms known to be involved in cell
proliferation, resistance to apoptosis and epithelial-mesenchymal
transition, are activated in both LC and pulmonary fibrosis
(12–14). Preneoplastic lesions (atypia,
metaplasia, dysplasia), microsatellite instability and loss of
heterozygosity in genes that underlie the tumorigenesis have also
been reported in lung fibrosis tissue (15).
Interestingly, LC is a common comorbidity among
patients with IPF with major impact on their survival. Increasing
data support the hypothesis that LC occurs secondarily on the
ground of fibrosis rather than as a preceding finding. In the
majority of patients, LC arise as nodular lesions in the peripheral
area of fibrosis, mainly at the lower areas of the lungs with
squamous cell and adenocarcinoma being the prevalent histological
types (12,16).
MicroRNAs have emphatically emerged as major
regulators of gene expression through epigenetic and
post-transcriptional mechanisms. MicroRNA profiling studies in LC
have currently proposed several microRNAs as potential epigenetic
biomarkers in the diagnostic procedure. Similarly, in IPF several
oncomirs are consistently deregulated, such as the miR-29 family
(17). The miR-29 family is
usually downregulated in affected tissues and plays a crucial role
in fibrogenesis in multiple organs including lung (18), liver (19) and kidney (20). Among the targets of miR-29 are
collagens, enzymes required for collagen synthesis (21,22),
matrix metalloproteases and DNA methylation enzymes such as DNA
methyltransferase (DNMT)3A/B (23). The miR-29 family can be an
effective regulator of tumorigenesis and cancer progression by
targeting multiple tumor-related pathways as cell proliferation,
cell cycle, apoptosis and metastasis as well as affecting the
epigenetic and immune regulation (24).
Additionally, miR-185 is increasingly recognized as
an oncomir commonly deregulated in IPF and LC (25). Downregulation of miR-185 has been
demonstrated in IPF lung tissue (26) and in several malignancies including
LC, glioma, hepatocellular and breast cancer (27). miR-185 supplementation inhibits
cell growth and proliferation by directly targeting AKT1 in
NSCLC28, through DNMT1 targeting, leading to the PTEN upregulation
and inhibition of AKT phosphorylation (29).
Interestingly, the expression of miR-29 and miR-185
is downregulated by TGFb (18,25,30).
Furthermore, there are emerging data suggesting an overlap of
miR-29a and miR-185 targets; miR-185 appears to regulate collagens
(26,30,31),
while miR-29a seems to regulate AKT1 (32) and AKT2 (33). Recently, both miR-185 and miR-29a
were downregulated in IPF bronchoalveolar lavage (BAL) cells
compared to controls (25).
BAL is becoming an attractive, minimally invasive
tool to study alveolar macrophages and other immune cells, as well
as other lung resident cells that may derive from adjacent lesions.
In essence BAL could be considered a form of liquid biopsy in LC
and possibly in IPF34. In this study, the expression of miR-29a and
miR-185 was compared in IPF and LC. Well-established targets of
miR-29a and recently emerged miR-185 targets such as DNMTs (DNMT1
and DNMT3b), AKT1/AKT2 and COL1A1 were analyzed. Furthermore,
microRNA and target gene expression was evaluated in the LC group
according to LC type and endobronchial findings, including, the
presence of endobronchial lesion(s), the side of the BAL procedure
relative to the tumor site and the cytology findings for malignant
cells.
In our study, similar levels of miR-29a, miR-185 and
their targets AKT1, AKT2 and DNMT3b were found in the two diseases.
By contrast, COL1A1 mRNA levels were increased in IPF suggesting a
disease-specific mRNA signature. Importantly, DNMT1 was
downregulated in the LC group and its expression was further
reduced in the presence of increasing malignant burden as indicated
by the endobronchial findings further suggesting an LC-specific
signature.
Materials and methods
Human subjects
Patients were classified as ever smokers and
non-smokers. Pulmonary function tests (PFTs), performed within 1
month of CT, included forced expiratory volume in 1 sec
(FEV1), forced vital capacity (FVC), and diffusing
capacity for carbon monoxide (DLCO) corrected for
hemoglobin concentration, expressed as percentages of the predicted
normal values. A total of 89 subjects were enrolled in this study,
comprising patients with IPF (n=57) and patients with LC (n=32)
with the majority being diagnosed as NSCLC (n=29). All subjects
were recruited from the Department of Thoracic Medicine, University
Hospital of Heraklion (Crete, Greece) between December 2013 and
July 2017. The study was approved by the Ethics Committee of the
University Hospital of Heraklion (IRB no. 17030/12-12-2013 for IPF
patients and Reg. no. 140/4-2-2015 for LC patients). All the
patients provided informed consent in written form.
IPF group: The IPF patients were evaluated with
complete PFTs, performed within 1 month of CT, including
spirometry, measurement of lung volumes and diffusion capacity.
Spirometry, lung volumes using the helium-dilution technique and
DLCO (corrected for haemoglobin) using the single breath
technique were performed using a computerized system (Jaeger 2.12;
MasterLab, Würzburg, Germany). Predicted values were obtained from
the standardized lung function testing of the European Coal and
Steel Community, Luxembourg (1993). PFTs, FEV1, FVC, and
DLCO corrected for haemoglobin concentration, and were
expressed as percentages of the predicted normal values. The
diagnosis of IPF was based on open or video-assisted thoracoscopic
biopsy, with all the biopsies reviewed by the same two
histopathologists, or using ATS/ERS clinical and high resolution
computed tomography (HRCT) criteria (35). In accordance with the
aforementioned criteria, any known cause of pulmonary fibrosis,
such as a systemic connective tissue disorder, was excluded by
immunologic screening and rheumatologic clinical evaluation
(35). All the IPF patients were
newly diagnosed and had not received previous treatment.
LC group: The diagnosis of LC patients and the exact
histological type of malignancy were based on lung biopsy proceeded
through bronchoscopy, CT-guided biopsy by invasive radiologists or
open lung biopsy. The LC patients were all
chemo/radio/immunotherapy naive patients and their diagnostic
management and staging were based on the International Association
for the Study of Lung Cancer (36). The group of LC patients was further
divided into subgroups associated with endobronchial findings
evaluated during bronchoscopy and following BAL cytology
evaluation. Thus, LC patients were assigned into groups according
to the side of the BAL procedure relative to the side of the
malignant lesion as it was depicted in the CT scan with a group
termed ‘same side’ (SS) and a group of ‘opposite side’ (OS), if
this separation was feasible, as patients with mediastinal
lymphadenopathy could not fit in any of those groups. Secondly, the
patients were separated in two groups according to the presence or
absence of malignant cells in the BAL following cytology
evaluation, termed ‘negative cytology’ (NC) and ‘positive cytology’
(PC) respectively. A third option for the cytology test was
‘suspective for malignancy’. Finally, the patients were separated
according to the physician's observations during bronchoscopy for
obvious endobronchial lesions with a group termed ‘endobronchial
lesion’ (OEL) and ‘no endobronchial lesion’ (NOEL). PFTs for LC
patients were not included in the studies.
BAL cell isolation and determination
of cellular composition
BAL was obtained from all the patients, as
previously described (25). Cells
(1–1.5 million) were homogenised in TriReagent™ (MBL) for total RNA
analysis, followed by storage at −80°C. Differential cell
population count was analysed following May-Grünwald Giemsa
staining of cell cytospins, as previously described (25).
MicroRNA and mRNA expression
analyses
Total RNA was isolated as previously described
(25) using the mirVana™ miRNA
isolation kit (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). For the analysis of microRNA expression levels, 10 ng of
total RNA were used in reverse transcriptase and quantitative PCR
reactions using the TaqMan™ microRNA assays (Life Technologies;
Thermo Fisher Scientific, Inc.) and 7500 Fast Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). For gene
expression analyses, 500 ng of total RNA were treated with DNAfree
(Ambion; Thermo Fisher Scientific, Inc.) for genomic DNA
contamination removal, followed by first-strand cDNA synthesis
using Maxima RT™ (Fermentas; Thermo Fisher Scientific, Inc.) and
qPCR analysis using Maxima SYBR-Green qPCRmix (Fermentas; Thermo
Fisher Scientific, Inc.) on Mx3005P qPCR system (Agilent
Technologies, Inc., Santa Clara, CA, USA). Probe and primer
sequences are summarized in Table
II. RNU19 levels were used as endogenous controls for the
normalization of microRNA expression levels in BAL samples. GAPDH
levels were used as endogenous control for the normalization of
mRNA expression levels in BAL samples. Relative expression values
per sample for each microRNA or mRNA assay were calculated by the
2−ΔΔCq method using as calibrator sample the average of
DCT values.
 | Table II.IDs of the TaqMan microRNA assays and
primer sequences used for the quantification of microRNAs and
mRNAs, respectively. |
Table II.
IDs of the TaqMan microRNA assays and
primer sequences used for the quantification of microRNAs and
mRNAs, respectively.
| A, |
|---|
|
|---|
| MicroRNA assay
name | MicroRNA assay
ID |
|---|
| RNU19 | 001003 |
| hsa-miR-185 | 002271 |
| hsa-miR-29a | 002112 |
|
| B, |
|
| Gene
name | Primer
sequences |
|
| GAPDH | F:
agccacatcgctcagaca |
|
| R:
ccaatacgaccaaatccgtt |
| COL1A1 | F:
gggattccctggacctaaag |
|
| R:
ggaacacctcgctctcca |
| AKT1 | F:
gcagcacgtgtacgagaaga |
|
| R:
ggtgtcagtctccgacgtg |
| AKT2 | F:
ctcacacagtcaccgagagc |
|
| R:
tgggtctggaaggcatactt |
| DNMT1 | F:
tttctgatgaaaaagacgaggat |
|
| R:
tttctccgttggttctttgg |
| DNMT3b | F:
agagggacatctcacggttc |
|
| R:
ggttgccccagaagtatcg |
Statistical analysis
Analysis of expression values with lung function
tests and BAL cell population percentages was performed Prism 6
software. Group comparisons were made by analysis of variance, log
transformation and Mann-Whitney test. Student's t-test, Wilcoxon
rank-sum test, or Chi-square test were also used for comparisons as
appropriate. P<0.05 was considered statistically
significant.
Results
Patient demographics
Demographic data and PFTs are summarised in Table I. IPF patients were older than the
patients in the LC group by 4.3 (±4.6) years, while LC patients
were heavier smokers as could be expected. The majority of LC was
NSCLC (29/32), with the rest being SCLC. The group of LC patients
was subdivided according to the endobronchial findings as described
in materials and methods. Fourteen LC patients were assigned to the
group SS and 9 to OS according to the site of broncoscopy. Fourteen
LC patients had no malignant cells detected in their BAL and were
assigned to the NC group and 7 to the PC. Nineteen patients had
obvious OEL and 7 were assigned to NOEL.
 | Table I.Patient demographics. |
Table I.
Patient demographics.
| Demographics | LC (32) | IPF (n=57) | P-value |
|---|
| Age | 67.7 (±11.9) | 72 (±7.3) | 0.01 |
| Sex (M/F) | 26/6 | 46/11 | NS |
| Pyrs | 75 (47.5–100) | 30 (6–45) | 0.001 |
| Non
smoker/smoker | 2/26 | 13/43 | NS |
| PFTs | LC | IPF |
|
| FVC, % | ND | 78.6 (±17,6) | NA |
| FEV1,
% | ND | 84.8 (±19.3) | NA |
|
FEV1/FVC, % | ND | 84.21
(79.8–89) | NA |
| TLC | ND | 75.5
(62.1–83.4) | NA |
| TLCO/SB | ND | 51.3 (±16.8) | NA |
| KCO | ND | 88 (±23.7) | NA |
| BAL, % | LC | IPF |
|
| Macrophages | 86.2
(70.3–93.3) | 80.38 (66–88) | NS |
| Lymphocytes | 5.8 (3.6–11.1) | 6.4 (3.5–12.7) | NS |
| Neutrophils | 3 (1.3–6.3) | 5.8 (2.4–12.9) | NS |
| Eosinophils | 0 (0–1.5) | 1.1 (0.4–2.6) | 0.01 |
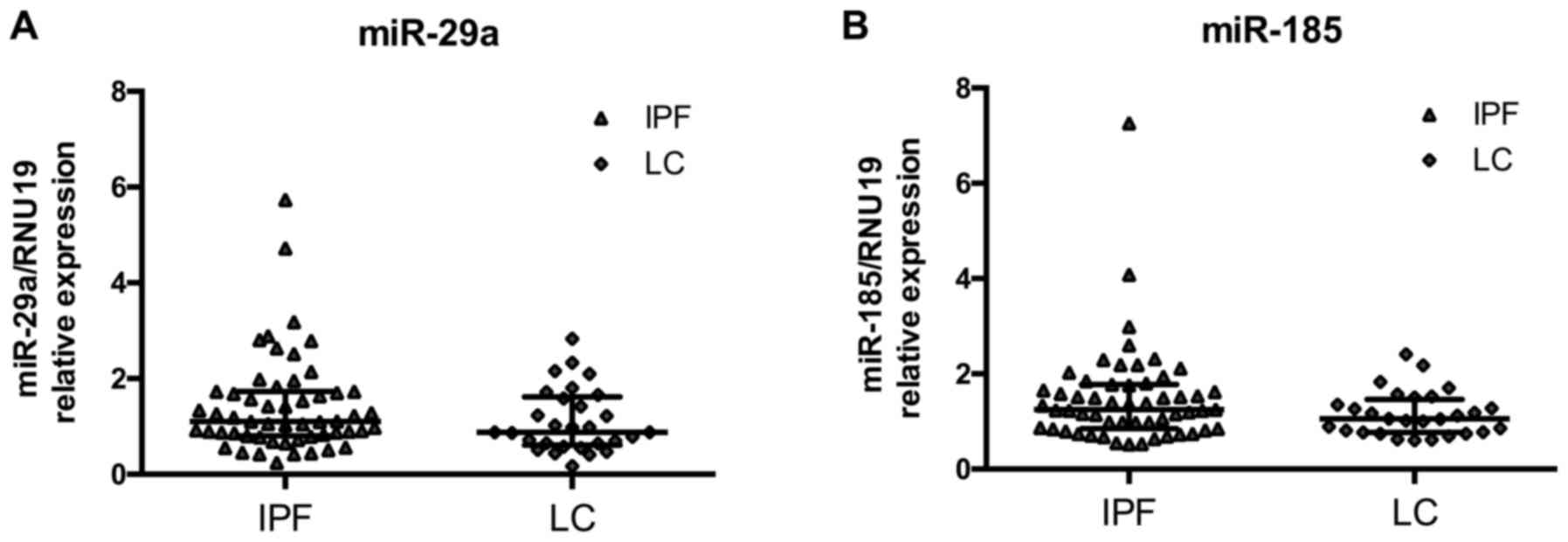
miR-185 and miR-29a levels are similar
between IPF and LC
MicroRNA expression levels in BAL cells were
measured by RT-qPCR and normalized using small nucleolar RNA RNU19.
We have previously shown that both microRNAs were significantly
downregulated in IPF relative to controls (25). The downregulation of the two
microRNAs was also noted for LC since the expression of miR-29a and
miR-185 did not differ between IPF and LC patients (Fig. 1A and B). miR-29a and miR-185
expression levels were significantly correlated within the IPF
group (Spearman's R 0.81, P=6e−14) and the LC group
(Spearman's R 0.71, P=3.6e−5).
DNMTs/AKT/miR-29a/miR-185 axis in IPF
and LC
DNMT1 and DNMT3b are common targets of miR-185 and
miR-29 in IPF and LC. mRNA expression levels in BAL cells were
measured by RT-qPCR and normalized using GAPDH. DNMT1 levels
previously measured in IPF relative to controls showed no
differences. Notably, DNMT1 levels were significantly lower in LC
patients compared to IPF patients. (Fig. 2A; Table III). No differences were detected
in the levels of DNMT3b between the two diseases (Table III). No difference was detected
in the levels of AKT1 and AKT2 between the two diseases (Table III).
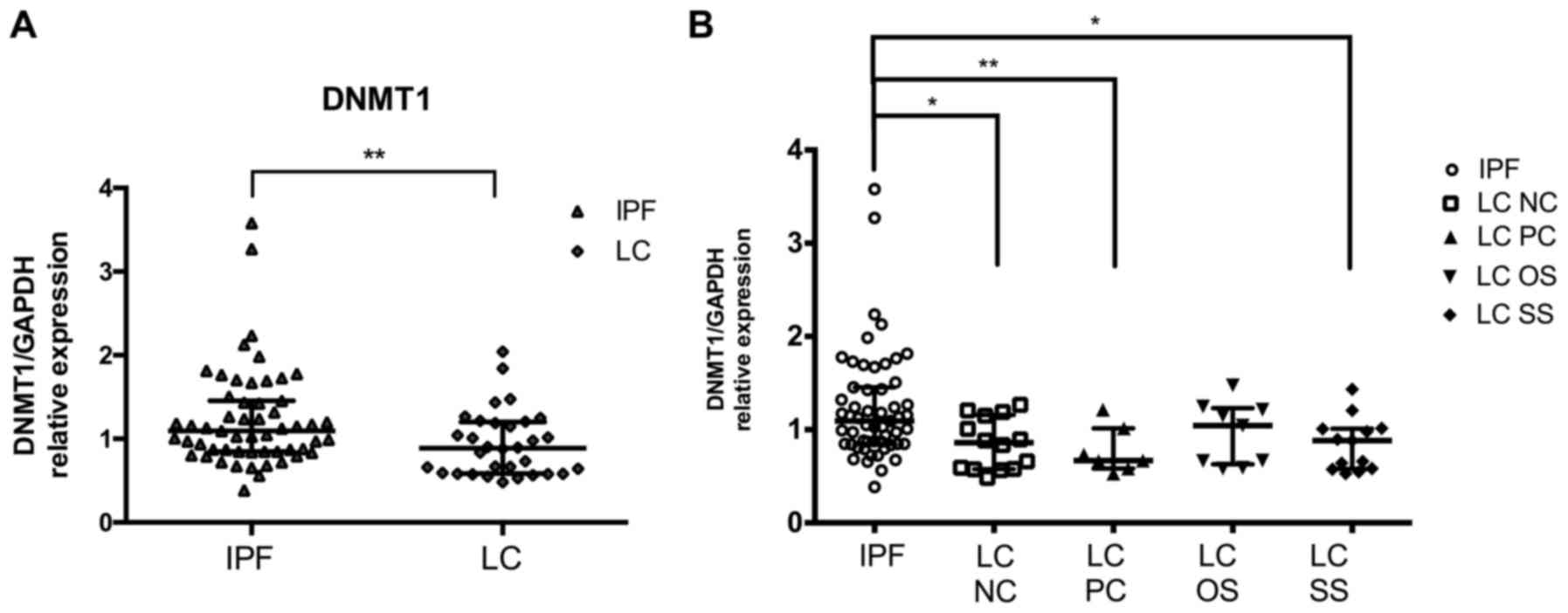 | Figure 2.DNMT1 levels in LC patients compared
to IPF patients. DNMT1 mRNA levels in BAL cells as measured by
RT-qPCR and normalised using GAPDH (A) in LC group and (B) in LC
subgroups NC, PC, OS and SS as compared to IPF. *P<0.05,
**P<0.01 of Mann-Whitney test. DNMT, DNA methyltransferase; LC,
lung cancer; IPF, idiopathic pulmonary fibrosis; BAL,
bronchoalveolar lavage; NC, negative cytology; PC, positive
cytology; OS, opposite side; SS, same side. |
 | Table III.microRNA and gene expression levels
in IPF and LC group and subgroups. |
Table III.
microRNA and gene expression levels
in IPF and LC group and subgroups.
|
| IPF (n=56) | LC (n=32) | LC-SS (n=14) | LC-OS (n=9) | LC-NC (n=14) | LC-PC (n=7) | LC-OEL (n=19) | LC -NOEL (n=7) |
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Variables | Median
(25–75%) | Median
(25–75%) |
P-valuea | Median
(25–75%) |
P-valuea | Median
(25–75%) |
P-valuea | Median
(25–75%) |
P-valuea | Median
(25–75%) |
P-valuea | Median
(25–75%) |
P-valuea | Median
(25–75%) |
P-valuea |
|---|
| miR-185 | 1.25 | 1.1 | NS | 1.05 | NS | 0.74 | NS | 1.05 | NS | 1 | NS | 1.06 | NS | 1.01 | NS |
|
| (0.85–1.78) | (0.77–1.47) |
| (0.80–1.33) |
| (0.62–1.37) |
| (0.62–1.27) |
| (0.77–1.71) |
| (0.74–1.53) |
| (0.62–1.17) |
|
| miR-29a | 1.10 | 0.9 | NS | 0.93 | NS | 0.79 | NS | 0.88 | NS | 1.02 | NS | 0.87 | NS | 0.99 | NS |
|
| (0.80–1.73) | (0.61–1.62) |
| (0.69–1.70) |
| (0.47–1.50) |
| (0.65–1.58) |
| (0.72–2.16) |
| (0.65–1.72) |
| (0.51–1.21) |
|
| DNMT1 | 1.10 | 0.9 | 0.01 | 0.89 | 0.013 | 1.04 | NS | 0.86 | 0.009 | 0.67 | 0.007 | 1.01 | NS | 0.66 | NS |
|
| (0.85–1.45) | (0.58–1.20) |
| (0.58–1.06) |
| (0.63–1.23) |
| (0.58–1.16) |
| (0.58–1.01) |
| (0.64–1.21) |
| (0.59–1.25) |
|
| DNMT3b | 1.03 | 1.0 | NS | 1.00 | NS | 0.82 | NS | 0.92 | NS | 1.15 | NS | 1.08 | NS | 0.66 | NS |
|
| (0.69–1.43) | (0.61–1.47) |
| (0.70–1.61) |
| (0.58–1.66) |
| (0.62–1.61) |
| (0.58–1.65) |
| (0.70–1.65) |
| (0.59–1.06) |
|
| AKT1 | 0.78 | 0.7 | NS | 0.69 | NS | 0.64 | NS | 0.72 | NS | 0.64 | NS | 0.67 | NS | 0.69 | NS |
|
| (0.53–1.22) | (0.41–1.15) |
| (0.39–1.31) |
| (0.47–1.22) |
| (0.56–1.36) |
| (0.24–0.97) |
| (0.38–1.60) |
| (0.56–0.88) |
|
| AKT2 | 0.96 | 0.94 | NS | 0.91 | NS | 1.11 | NS | 0.94 | NS | 0.89 | NS | 0.91 | NS | 1.11 | NS |
|
| (0.75–1.30) | (0.69–1.48) |
| (0.60–1.37) |
| (0.69–1.48) |
| (0.74–1.51) |
| (0.54–1.50) |
| (0.65–1.50) |
| (0.89–1.33) |
|
| COL1A1 | 0.70 | 0.19 | 0.001 | 0.22 | 0.017 | 0.35 | NS | 0.22 | 0.017 | 0.17 | 0.048 | 0.14 | 0.0005 | 0.35 | NS |
|
| (0.24–2.61) | (0.08–1.00) |
| (0.05–1.03) |
| (0.13–0.94) |
| (0.11–0.78) |
| (0.04–0.98) |
| (0.05–0.98) |
| (0.21–1.30) |
|
Effect of malignant burden on DNMT1
levels in BAL cells compared to IPF
Comparing IPF patients with the LC group in detail,
further reduced levels of DNMT1 were detected in the samples where
the BAL procedure was performed at the side of the lesion (SS) as
opposed to the lesion free side (OS). Moreover, LC patients with
positive BAL cytology results (PC) had a more pronounced reduction
in DNMT1 levels than those without the presence cells with
malignant features (NC), when compared with IPF (Fig. 2B). Regarding the histological type
of LC, the more obvious reduction in DNMT1 mRNA levels compared
with IPF was found in NSLC type.
COL1A1/miR-29a/miR-185 axis in IPF and
LC
Next we analysed miR-29a specific target COL1A1 by
RT-qPCR and normalized using GAPDH. Our previous result showed that
IPF BAL cells express significantly higher levels of COL1A1 mRNA
than controls. In this study, we observed that the levels of COL1A1
mRNA were significantly lower in LC patients compared with IPF
patients (Fig. 3A; Table III).
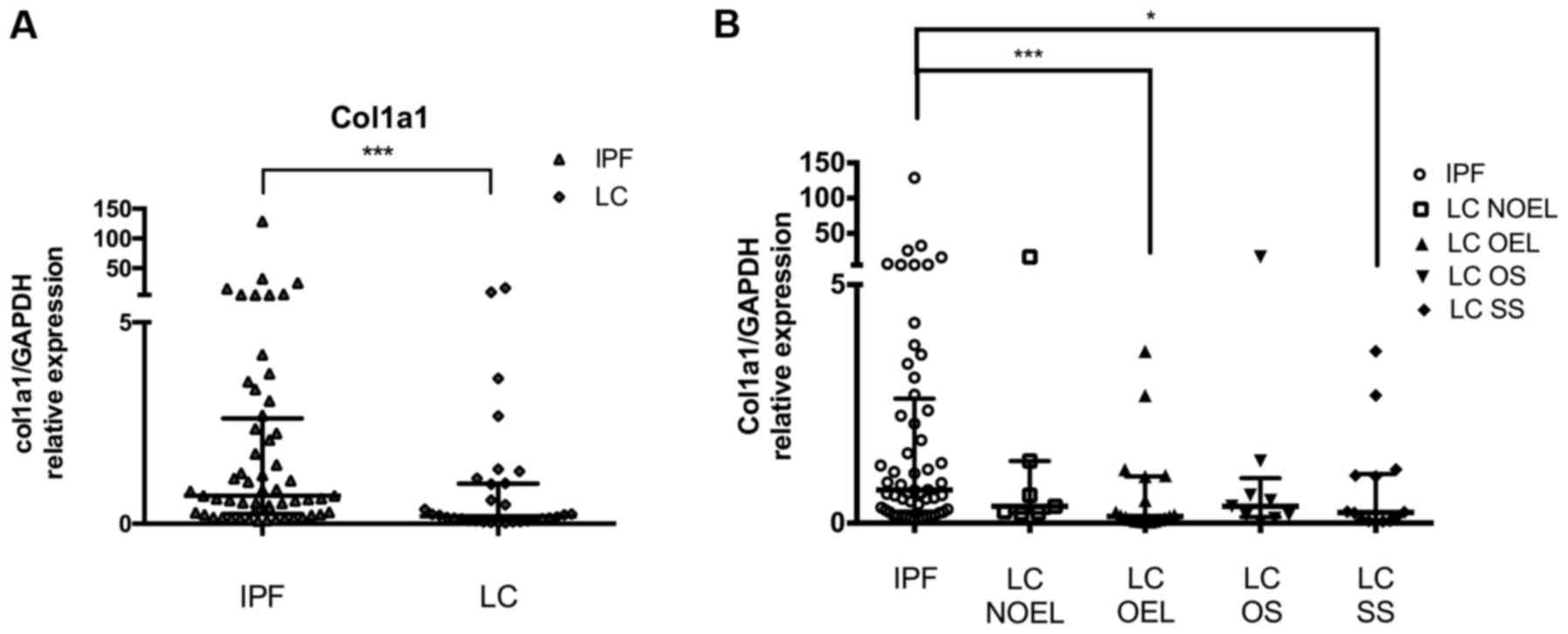 | Figure 3.COL1A1 levels in LC patients compared
to IPF patients. COL1A1 mRNA levels in BAL cells as measured by
RT-qPCR and normalised using GAPDH (A) in the LC group and (B) in
the LC subgroups NOEL, OEL, OS and SS as compared to IPF.
*P<0.05, ***P<0.001, Mann-Whitney test. LC, lung cancer; IPF,
idiopathic pulmonary fibrosis; BAL, bronchoalveolar lavage; NOEL,
no endobronchial lesion; OEL, endobronchial lesion; OS, opposite
side; SS, same side. |
Effect of malignant burden on COL1A1
levels in BAL cells compared to IPF
The mRNA levels of COL1A1 were markedly reduced in
LC patients compared with IPF overall as it was noted previously.
Assorting LC patients depending on the side the BAL procedure was
performed and comparing them with IPF, further reduced levels of
COL1A1 was detected in LC patients when the BAL procedure was done
ipsilaterally of the lesion than contralaterally. The presence of
an OEL during the bronchoscope signified more clearly reduced
levels of COL1A1 (compared with IPF) than in the case of NOEL
(Fig. 3A; Table III). Patients with the
histological type of SCLC had COL1A1 levels similar with IPF
levels, opposing the reduced levels in the NSCLC patients
subgroup.
Discussion
This is a BAL study of the expression of major
epigenetic molecules, miR-29a and miR-185, and their common targets
DNMT1 and DNMT3b, involved in the DNA methylation process. The
downregulation of these molecules has been previously connected
with IPF and LC. The relationship between the lethal diseases which
often coexist is an active field of research as common pathogenic
pathways emerge, with major therapeutic implications. Our cardinal
findings were: i) Both miR-185 and miR-29a were comparably
expressed in IPF and LC BAL samples; ii) no direct correlation with
miR-29a or miR-185 and their targets was observed, albeit DNMT1
downregulation was characteristic of LC and COL1A1 upregulation was
representative of IPF BAL samples; and iii) in LC the malignant
burden affected both DNMT1 and COL1A1 expression.
Similar levels of miR-29a and miR-185 were detected
between IPF and LC in BAL cells while our previous findings showed
that both miR-29a and miR-185 were downregulated in IPF relative to
controls (25). The downregulation
of miR-29a and miR-185 in LC tissue specimens has been previously
established; however, to the best of our knowledge, this is the
first study on LC BAL cells. Our results suggest that miR-185 is a
novel common microRNA deregulation in IPF and LC next to previously
identified microRNAs such as miR-29a. miR-29a and miR-185
expression showed a significant association in both IPF and LC BAL
samples. A possible common regulation of the expression of the two
microRNAs may be related to the increased levels of TGFb in both
IPF and LC BAL (37,38). Activation of TGF-β signaling and
excessive accumulation of ECM proteins are observed in IPF and LC,
highlighting a common molecular mechanism in both diseases that is
directly linked to both microRNAs (39,40).
Of note, miRNA based-therapeutic strategies are already under
evaluation for their use in several malignancies (41) and that imposes the need for
in-depth study of similarities and differences between IPF and LC,
focusing on key molecules involved in the multifarious function of
alveolar macrophages.
For this reason, we examined the expression of a
common target of miR-29a and miR-185 induced by TGFb and central
fibrosis mediator collagen 1a in LC in comparison to IPF. Our
previous results supported that the downregulation of miR-29a in
IPF is associated with the overexpression of COL1A1 gene in
BAL cells, confirming the active role of the miR-29a/COL1A1 pathway
in AMs and lung tissue, while the expression profile of COL1A1 in
LC has not been yet clarified. COL1A1 may be involved in
carcinogenesis as aberrant expression levels were revealed in
several malignancies including hepatocellular carcinoma (42), NSCLC tissue (43) and in malignant gastric tissue
(44). Our findings, however,
showed significantly increased levels of COL1A1 in IPF relative to
LC in BAL cells that establishes the characteristic fibrotic
profile of BAL cells in IPF, which appears to be lacking in LC BAL
cells. The further reduced levels of COL1A1 accordingly with
increased malignant burden cannot thus far be interpreted and
further functional analysis of AMs in LC is needed.
A second pathway/axis affected by both miR-29a and
miR-185 is the expression of DNMTs. An important concept currently
put forward for the pathogenesis of IPF as in cancer is that common
risk factors such as aging, cigarette smoke and environmental
effects induce errors in the maintenance of the methylation marks
of the genome creating aberrant DNA methylation patterns and
accelerating the aging of our epigenome (45,46).
Global methylation profile in IPF lung tissue was different
compared to the controls and partially similar to cancer (6,7).
Furthermore, an increased expression of the DNMTs was observed in
both cancer and IPF lung tissue studies (7,47),
leading to site-specific hypermethylation and gene silencing.
We have previously reported that DNMT1 mRNA levels
in the BAL cells of IPF patients were similar to controls (25). In the current study, we observed a
significant reduction in the mRNA levels of DNMT1 in LC BAL cells
while, DNMT3b levels were similar in IPF and LC. DNMT1, in contrast
to DNMT3, appears to function in cooperation with DNA damage repair
pathways in order to maintain genomic stability and ablation or
reduction of DNMT1 promotes mutagenic events (48), microsatellite instability and
chromosomal translocations (49).
Notably, the samples obtained near the malignant lesion or with
positive malignant cell cytology results showed a more pronounced
reduction of DNMT1 expression suggesting that reduced DNMT1 levels
were associated with increased malignant burden.
LC is a common and prognostically determinant
comorbidity among IPF patients. BAL procedure is a less invasive
and harmless tool for revealing new, disease specific biomarkers
regarding LC in IPF patients, for screening, risk stratification
and diagnostic purposes, verging the promising concept of cancer
liquid biopsy. It would be of great interest to study the
expression profile of those miRNAs and their targets also in
patients who simultaneously suffer from IPF and LC. MicroRNA
based-therapeutic strategies are already under evaluation for their
use in several malignancies (41)
and IPF (50) and would greatly
benefit from in depth study of similarities and differences between
IPF and LC, focusing on key molecules involved in the multifarious
function of alveolar macrophages.
Tissue specimens and cell lines are the predominant
material of research; however, the role of BAL as a minimally
invasive tool to study alveolar macrophages and their implication
in pathogenesis continually recovers ground. In IPF alveolar
macrophages deriving from monocytes recruited to the injured lungs
are actively involved in the fibrotic process (51). Altered alveolar macrophage function
in patients with LC has been recorded under the influence of
tumor-associated polarizing events such as mediators and hypoxic
tissue damage (52). Our study
provides some insight with respect to common alveolar macrophage
function in the two groups as demonstrated by the commonly reduced
expression of miR-29a and miR-185. Further research is needed in
order to identify BAL biomarkers for high risk patients and more
targeted therapies.
Acknowledgements
The authors would like to thank the staff of the
broncoscopy unit and the members of the pathology department of the
University Hospital of Heraklion.
Funding
The project was funded by a grant from the Hellenic
Thoracic Society (HTS 2012).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
EB, ET and KMA conceived and designed the study. EB,
ET, CK and TG performed the experiments. EB, ET, EV, GM and AT
collected patient data. EB and ET performed statistical analysis.
EB, ET and KMA wrote the manuscript. DAS, NT and KMA critically
reviewed and edited the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the University Hospital of Heraklion (IRB no. 17030/12-12-2013 for
IPF patients and Reg. no. 140/4-2-2015 for LC patients). All the
patients provided informed consent in written form.
Consent for publication
Not applicable.
Competing interests
Demetrios A. Spandidos is the Editor-in-Chief for
the journal, but had no personal involvement in the reviewing
process, or any influence in terms of adjudicating on the final
decision, for this article.
Glossary
Abbreviations
Abbreviations:
|
DNMT
|
DNA methyltransferase
|
|
IPF
|
idiopathic pulmonary fibrosis
|
|
LC
|
lung cancer
|
|
HRCT
|
high resolution computed
tomography
|
|
FEV1
|
forced expiratory volume in 1 sec
|
|
FVC
|
forced vital capacity
|
|
DLCO
|
diffusing capacity for carbon
monoxide
|
|
BAL
|
bronchoalveolar lavage
|
|
PFTs
|
pulmonary function tests
|
References
|
1
|
Richeldi L, Collard HR and Jones MG:
Idiopathic pulmonary fibrosis. Lancet. 389:1941–1952. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Antoniou KM, Hansell DM, Rubens MB, Marten
K, Desai SR, Siafakas NM, Nicholson AG, du Bois RM and Wells AU:
Idiopathic pulmonary fibrosis: Outcome in relation to smoking
status. Am J Respir Crit Care Med. 177:190–194. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Selman M and Pardo A: Revealing the
pathogenic and aging-related mechanisms of the enigmatic idiopathic
pulmonary fibrosis. an integral model. Am J Respir Crit Care Med.
189:1161–1172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Giopanou I, Arendt KAM and Stathopoulos
GT: Lung carcinogenesis and fibrosis taken together: Just
coincidence? Curr Opin Pulm Med. 23:290–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Antoniou KM, Tomassetti S, Tsitoura E and
Vancheri C: Idiopathic pulmonary fibrosis and lung cancer: A
clinical and pathogenesis update. Curr Opin Pulm Med. 21:626–633.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Duruisseaux M and Esteller M: Lung cancer
epigenetics: From knowledge to applications. Semin Cancer Biol
S1044-579X(17)30166-9. 2017. View Article : Google Scholar
|
|
7
|
Sanders YY, Ambalavanan N, Halloran B,
Zhang X, Liu H, Crossman DK, Bray M, Zhang K, Thannickal VJ and
Hagood JS: Altered DNA methylation profile in idiopathic pulmonary
fibrosis. Am J Respir Crit Care Med. 186:525–535. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Antoniou KM, Samara KD, Lasithiotaki I,
Margaritopoulos GA, Soufla G, Lambiri I, Giannarakis I, Drositis I,
Spandidos DA and Siafakas NM: Differential telomerase expression in
idiopathic pulmonary fibrosis and non-small cell lung cancer. Oncol
Rep. 30:2617–2624. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sarchianaki E, Derdas SP, Ntaoukakis M,
Vakonaki E, Lagoudaki ED, Lasithiotaki I, Sarchianaki A,
Koutsopoulos A, Symvoulakis EK, Spandidos DA, et al: Detection and
genotype analysis of human papillomavirus in non-small cell lung
cancer patients. Tumour Biol. 35:3203–3209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Samara KD, Antoniou KM, Karagiannis K,
Margaritopoulos G, Lasithiotaki I, Koutala E and Siafakas NM:
Expression profiles of Toll-like receptors in non-small cell lung
cancer and idiopathic pulmonary fibrosis. Int J Oncol.
40:1397–1404. 2012.PubMed/NCBI
|
|
12
|
Vancheri C: Idiopathic pulmonary fibrosis
and cancer: Do they really look similar? BMC Med. 13:2202015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Antoniou KM, Lasithiotaki I, Symvoulakis
E, Derdas SP, Psaraki A, Spandidos DA, Stathopoulos EN, Siafakas NM
and Sourvinos G: Molecular pathological findings of Merkel cell
polyomavirus in lung cancer: A possible etiopathogenetic link? Int
J Cancer. 133:3016–3017. 2013.PubMed/NCBI
|
|
14
|
Lasithiotaki I, Antoniou KM, Derdas SP,
Sarchianaki E, Symvoulakis EK, Psaraki A, Spandidos DA,
Stathopoulos EN, Siafakas NM and Sourvinos G: The presence of
Merkel cell polyomavirus is associated with deregulated expression
of BRAF and Bcl-2 genes in non-small cell lung cancer. Int J
Cancer. 133:604–611. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vassilakis DA, Sourvinos G, Spandidos DA,
Siafakas NM and Bouros D: Frequent genetic alterations at the
microsatellite level in cytologic sputum samples of patients with
idiopathic pulmonary fibrosis. Am J Respir Crit Care Med.
162:1115–1119. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tomassetti S, Gurioli C, Ryu JH, Decker
PA, Ravaglia C, Tantalocco P, Buccioli M, Piciucchi S, Sverzellati
N, Dubini A, et al: The impact of lung cancer on survival of
idiopathic pulmonary fibrosis. Chest. 147:157–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mizuno K, Mataki H, Seki N, Kumamoto T,
Kamikawaji K and Inoue H: MicroRNAs in non-small cell lung cancer
and idiopathic pulmonary fibrosis. J Hum Genet. 62:57–65. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cushing L, Kuang PP, Qian J, Shao F, Wu J,
Little F, Thannickal VJ, Cardoso WV and Lü J: miR-29 is a major
regulator of genes associated with pulmonary fibrosis. Am J Respir
Cell Mol Biol. 45:287–294. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roderburg C, Urban GW, Bettermann K, Vucur
M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi
M, et al: Micro-RNA profiling reveals a role for miR-29 in human
and murine liver fibrosis. Hepatology. 53:209–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Patel V and Noureddine L: MicroRNAs and
fibrosis. Curr Opin Nephrol Hypertens. 21:410–416. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luna C, Li G, Qiu J, Epstein DL and
Gonzalez P: Role of miR-29b on the regulation of the extracellular
matrix in human trabecular meshwork cells under chronic oxidative
stress. Mol Vis. 15:2488–2497. 2009.PubMed/NCBI
|
|
22
|
Bian EB, Li J and Zhao B: miR-29, a
potential therapeutic target for liver fibrosis. Gene. 544:259–260.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cicchini C, de Nonno V, Battistelli C,
Cozzolino AM, De Santis Puzzonia M, Ciafrè SA, Brocker C, Gonzalez
FJ, Amicone L and Tripodi M: Epigenetic control of EMT/MET
dynamics: HNF4α impacts DNMT3s through miRs-29. Biochim Biophys
Acta. 1849:919–929. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Zhang X, Li H, Yu J and Ren X: The
role of miRNA-29 family in cancer. Eur J Cell Biol. 92:123–128.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsitoura E, Wells AU, Karagiannis K,
Lasithiotaki I, Vasarmidi E, Bibaki E, Koutoulaki C, Sato H,
Spandidos DA, Siafakas NM, et al: MiR-185/AKT and miR-29a/collagen
1a pathways are activated in IPF BAL cells. Oncotarget.
7:74569–74581. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lei GS, Kline HL, Lee CH, Wilkes DS and
Zhang C: Regulation of collagen V expression and
epithelial-mesenchymal transition by miR-185 and miR-186 during
idiopathic pulmonary fibrosis. Am J Pathol. 186:2310–2316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu ZJ, Lu LG, Tao KZ, Chen DF, Xia Q, Weng
JJ, Zhu F, Wang XP and Zheng P: MicroRNA-185 suppresses growth and
invasion of colon cancer cells through inhibition of the hypoxia
inducible factor-2α pathway in vitro and in vivo. Mol
Med Rep. 10:2401–2408. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li S, Ma Y, Hou X, Liu Y, Li K, Xu S and
Wang J: MiR-185 acts as a tumor suppressor by targeting AKT1 in
non-small cell lung cancer cells. Int J Clin Exp Pathol.
8:11854–11862. 2015.PubMed/NCBI
|
|
29
|
Qadir XV, Han C, Lu D, Zhang J and Wu T:
miR-185 inhibits hepatocellular carcinoma growth by targeting the
DNMT1/PTEN/Akt pathway. Am J Pathol. 184:2355–2364. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding X, Yu C, Liu Y, Yan S, Li W, Wang D,
Sun L, Han Y, Li M, Zhang S, et al: Chronic obstructive sleep apnea
accelerates pulmonary remodeling via TGF-β/miR-185/CoLA1 signaling
in a canine model. Oncotarget. 7:57545–57555. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiao K, Luo X, Wang X and Gao Z: MicroRNA
185 regulates transforming growth factor β1 and collagen 1 in
hypertrophic scar fibroblasts. Mol Med Rep. 15:1489–1496. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tang B, Li X, Ren Y, Wang J, Xu D, Hang Y,
Zhou T, Li F and Wang L: MicroRNA-29a regulates lipopolysaccharide
(LPS)-induced inflammatory responses in murine macrophages through
the Akt1/ NF-κB pathway. Exp Cell Res. 360:74–80. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li M, Li H, Liu X, Xu D and Wang F:
MicroRNA-29b regulates TGF-β1-mediated epithelial-mesenchymal
transition of retinal pigment epithelial cells by targeting AKT2.
Exp Cell Res. 345:115–124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park S, Hur JY, Lee KY, Lee JC, Rho JK,
Shin SH and Choi CM: Assessment of EGFR mutation status using
cell-free DNA from bronchoalveolar lavage fluid. Clin Chem Lab Med.
55:1489–1495. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Raghu G: Idiopathic pulmonary fibrosis:
Guidelines for diagnosis and clinical management have advanced from
consensus-based in 2000 to evidence-based in 2011. Eur Respir J.
37:743–746. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 World Health Organization
Classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen Z, Xu Z, Sun S, Yu Y, Lv D, Cao C and
Deng Z: TGF-β1, IL-6, and TNF-α in bronchoalveolar lavage fluid:
Useful markers for lung cancer? Sci Rep. 4:55952014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kumar RK, O'Grady R, Maronese SE and
Wilson MR: Epithelial cell-derived transforming growth factor-beta
in bleomycin-induced pulmonary injury. Int J Exp Pathol. 77:99–107.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fernandez IE and Eickelberg O: The impact
of TGF-β on lung fibrosis: From targeting to biomarkers. Proc Am
Thorac Soc. 9:pp. 111–116. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Eser PO and Jänne PA: TGFβ pathway
inhibition in the treatment of non-small cell lung cancer.
Pharmacol Ther. Nov 10–2017.(Epub ahead of print). PubMed/NCBI
|
|
41
|
Shah MY, Ferrajoli A, Sood AK,
Lopez-Berestein G and Calin GA: microRNA therapeutics in cancer -
An emerging concept. EBioMedicine. 12:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hayashi M, Nomoto S, Hishida M, Inokawa Y,
Kanda M, Okamura Y, Nishikawa Y, Tanaka C, Kobayashi D, Yamada S,
et al: Identification of the collagen type 1 α 1 gene (COL1A1) as a
candidate survival-related factor associated with hepatocellular
carcinoma. BMC Cancer. 14:1082014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Oleksiewicz U, Liloglou T, Tasopoulou KM,
Daskoulidou N, Gosney JR, Field JK and Xinarianos G: COL1A1,
PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell
lung cancer. J Cancer Res Clin Oncol. 143:1133–1141. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li J, Ding Y and Li A: Identification of
COL1A1 and COL1A2 as candidate prognostic factors in gastric
cancer. World J Surg Oncol. 14:2972016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Selman M and Pardo A: Stochastic
age-related epigenetic drift in the pathogenesis of idiopathic
pulmonary fibrosis. Am J Respir Crit Care Med. 190:1328–1330. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Langevin SM, Pinney SM, Leung YK and Ho
SM: Does epigenetic drift contribute to age-related increases in
breast cancer risk? Epigenomics. 6:367–369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT
and Wang YC: Alteration of DNA methyltransferases contributes to
5′CpG methylation and poor prognosis in lung cancer. Lung Cancer.
55:205–213. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jin B and Robertson KD: DNA
methyltransferases, DNA damage repair, and cancer. Adv Exp Med
Biol. 754:3–29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen RZ, Pettersson U, Beard C,
Jackson-Grusby L and Jaenisch R: DNA hypomethylation leads to
elevated mutation rates. Nature. 395:89–93. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cushing L, Kuang P and Lü J: The role of
miR-29 in pulmonary fibrosis. Biochem Cell Biol. 93:109–118. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Misharin AV, Morales-Nebreda L, Reyfman
PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, Chen CI, Anekalla
KR, Joshi N, Williams KJN, et al: Monocyte-derived alveolar
macrophages drive lung fibrosis and persist in the lung over the
life span. J Exp Med. 214:2387–2404. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Almatroodi SA, McDonald CF and Pouniotis
DS: Alveolar macrophage polarisation in lung cancer. Lung Cancer
Int. 2014:7210872014. View Article : Google Scholar : PubMed/NCBI
|