Introduction
Bone is a dynamic tissue that undergoes continuous
repair and remodeling throughout the lifetime of an individual
(1,2). The regulation of bone remodeling
involves a balance between the number of osteoblasts and
osteoclasts, which produce or resorb bone, respectively (3). Osteoblasts, osteocytes and
osteoclasts are specialized cells that orchestrate the process of
bone remodeling. As osteoblasts are highly sensitive to mechanical
stimuli, these cells have the leading role in the bone remodeling
process (4). It has been
previously established that external mechanical stress, such as
physiological levels of compression and tension force, induces the
formation of new bone (5).
Distraction osteogenesis (DO) is a bone regenerative response and
is employed surgically to treat skeletal injuries and deformities
(6). DO leads to the generation of
new bone by applying tensile forces to the skeleton after a
controlled osteotomy has been established (7). However, the mechanism by which the
mechanical load is translated into intracellular signals remains
unclear.
It is established that canonical Wnt signaling has
an important role in the regulation of osteoblastogenesis and bone
formation (8–10). Various studies have demonstrated
that Wnt promotes osteoblast differentiation, proliferation and
mineralization activity. The major component of the Wnt signaling
pathway is β-catenin, which is a transcription factor and a
structural adaptor protein that facilitates the binding of
cadherins to the actin cytoskeleton during cell-cell adhesion
(11,12). Activation of Wnt signaling results
in a loss of cadherin-mediated cell-cell adhesion and increased
cytoplasmic β-catenin (13).
β-catenin acts as a transcription cofactor with T-cell factor
(TCF)/lymphoid enhancer-binding factor (LEF) in the Wnt pathway
(14). Under normal conditions,
β-catenin that is not associated with cadherins undergoes rapid
phosphorylation by glycogen synthase kinase (GSK)-3β, which targets
it for degradation (15). However,
upon phosphorylation of GSK-3β, GSK-3β-mediated phosphorylation of
β-catenin is prevented, which leads to increased cytoplasmic levels
of unphosphorylated β-catenin. Wnt signaling-induced cytoplasmic
accumulation of β-catenin leads to increased entry of β-catenin
into the nucleus and increased heterodimer formation with TCF/LEF
transcription factors to regulate the expression of various Wnt
target genes, such as cyclin D1 (16–19).
Previous studies have demonstrated that
Wnt/β-catenin signaling is involved in the early responses of
osteoblastic cells to load bearing. The current study investigated
the effects of mechanical strain on β-catenin in Saos-2
osteoblastic cells and investigated whether mechanical stimuli
modulated Wnt/β-catenin signaling.
Materials and methods
Saos-2 cell culture
The Saos-2 human osteosarcoma cell line (American
Type Culture Collection, Manassas, VA, USA) was obtained from the
Center Laboratory for Tissue Engineering, College of Stomatology,
The Fourth Military Medical University (Xi'an, China) and used as
osteoblasts (20). The cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
v/v fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.)
at 37°C in a humidified atmosphere of 5% CO2 and 95%
air. After reaching 90% confluence, the cells were detached by
treatment with 10% trypsin-EDTA (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and cultured for 72 h on 6-well
flexible-bottomed plates (type I collagen-coated BioFlex I plates
at 37°C; Flexcell International Corp., Burlington, NC, USA) at
1×105 cells per well. Subsequently, 10% FBS was replaced
with 1% FBS prior to the application of strain force to cells. The
1% FBS provided minimal nutrition for the cells while allowing them
to achieve a quiescent phase (G0). After 24 h starvation, the media
was replaced again with DMEM containing 10% FBS and cells were
incubated for an additional 12 h at 37°C. The plates were
subsequently placed into the Flexcell's FX-4000T tension plus
system (Flexcell International Corp.) in which the 6-well plates
were docked.
Application of strain force
Flexcell's FX-4000T tension plus system (BF-3001U
BioFlex®) was used to generate cyclic tensile strain in
the Saos-2 cells. The instrument comprises a computer system
controller and monitor, a control module that regulates negative
and positive pressure, and a vacuum baseplate and gasket upon which
the flexible-bottomed plates are placed (BioFlex I culture plates).
Control of frequency, strain rate and degree of elongation of the
deformation regimen are achieved by regulating the rate of
evacuation (vacuum level) and rate of air influx to the plate
bottoms. The flexible membrane with the adherent cells were
subjected to cyclic, uniaxial strain at different magnitudes at a
frequency of 1 cycle/sec (Hz) along the long axis. Unstrained
plates served as controls. Strain is the amount of deformation
caused by an applied stress.
MTT analysis
Saos-2 cells were subjected to 0 or 12% elongation
by using Flexcell's FX-4000T tension plus system. At 0, 4, 8 and 12
h following loading, a 20 ml sample of MTT solution (5 g/l
dissolved in PBS) was added to the culture plates and incubated at
37°C for 6 h. The pipetted supernatant was discarded and 150 ml
dimethyl sulfoxide (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) was added to dissolve the blue MTT formazan produced by
mitochondrial succinate dehydrogenase. The absorbance was measured
at 492 nm in a spectrophotometer. The percentage of viable cells
was calculated as the relative ratio of their absorbance to the
control.
Biochemical alkaline phosphatase (ALP)
assay
Biochemical analysis was employed to investigate the
effects of different magnitudes of mechanical strain (as detailed
above) on ALP activity. ALP activity was measured directly on the
monolayer of the cultures. Following medium removal, cells were
washed three times with PBS and shaken for 30 min at 37°C in 1 ml
saline buffer (PBS) containing 10 mM p-nitrophenylphosphate (PNP;
Sigma-Aldrich; Merck KGaA). PNP solution was removed and 1 ml 1 N
NaOH was added to cells. The optical density was measured at 405
nm. The ALP activity values were normalized to the relative number
of viable cells, as determined using the proliferation assay
described above.
Nodule formation and
mineralization
Saos-2 cells were treated with either 0 or 12%
tensile stress for 8 h/day for 7 or 14 days. Mineralized nodule
formation and the degree of mineralization were determined for
osteoblast cultures cultured in 6-well plates using Alizarin Red S
staining. Briefly, medium was aspirated from the wells and cells
were rinsed twice with PBS. Cells were fixed with ice-cold 70% v/v
ethanol for 1 h. Ethanol was removed and the cells were rinsed
twice with deionized water. Cells were subsequently stained with 40
mM Alizarin Red S in deionized water (adjusted to pH 4.2) for 10
min at room temperature. Alizarin Red S solution was removed by
aspiration and the cells were rinsed five times with deionized
water. Water was removed by aspiration and the cells were incubated
in PBS for 15 min at room temperature on an orbital rotator. PBS
was removed and the cells were rinsed once with fresh PBS.
Mineralized Alizarin Red S-positive nodules present in each well
were subsequently counted using light microscopy at magnification,
×40; 5 fields of view were randomly selected per well.
Immunofluorescence
Immediately following treatment with 12% tensile
stress for 0, 4 or 8 h, cells were prepared by washing with PBS and
were immersion-fixed with 4% paraformaldehyde in PBS for 15 min at
room temperature. Cells were subsequently permeabilized with 0.2%
Triton-X-100 in PBS at room temperature for 5 min. Then cells were
blocked by 2% goat serum (OriGene Technologies, Beijing, China) at
37°C for 1 h. Primary antibodies were diluted in PBS/5% bovine
serum albumin (Sigma-Aldrich; Merck KGaA). After rinsing three
times with PBS, cells were incubated overnight at 4°C in a moist
environment with a mixture containing primary antibodies against
β-catenin (rabbit; 1:100; sc-7199; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and anti-E-cadherin (mouse; 1:100; sc-9988; Santa
Cruz Biotechnology, Inc.), followed by incubation with a secondary
antibody mixture containing Alexa Fluor® 594 donkey
anti-mouse IgG (1:500; R37115; Molecular Probes; Thermo Fisher
Scientific, Inc.) and Alexa Fluor® 488 donkey
anti-rabbit IgG (1:500; R37118; Molecular Probes; Thermo Fisher
Scientific, Inc.) for 2 h at room temperature. F-actin was
additionally stained with Alexa Fluor® 350 phalloidin
(A22281; Molecular Probes; Thermo Fisher Scientific, Inc.) at a
concentration of 7.5 U/ml for 30 min at room temperature, which was
followed by washing three times with PBS. The cells were mounted on
a drop of n-propyl gallate in 70% glycerol-PBS and viewed with a
laser scanning confocal microscope (FLUOVIEW; Olympus Corporation,
Tokyo, Japan) at magnification, ×600 using FV1000 Viewer version
1.7a software (Olympus Corporation).
Reverse transcriptase-polymerase chain
reaction (RT-PCR)
Cells were subjected to 0 or 12% tensile stress for
0 or 4 h. Cellular RNA was isolated from each culture using an
RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA), according to the
manufacturer's protocol. To remove contaminating genomic DNA, the
RNA samples were treated with RNase-free DNase I (Qiagen, Inc.) at
37°C for 30 min. Template RNA (1 µl) was added to the individual
tubes containing the master mix, using the Omniscript RT kit
(Qiagen, Inc.). The tubes were centrifuged at 120 × g for 1 min at
room temperature to collect residual liquid from the walls of the
tube. The tubes were incubated for 2 h at 37°C. RNase A (2 µl; 10
mg/ml) was added and the reaction mix incubated for 10 min at 65°C
and then for 5 min at 93°C before being cooled immediately on ice.
Subsequently, 1 µl cDNA mixture was subjected to PCR amplification
using specific primers. The primer sequences used in the current
study are presented in Table I.
Each PCR was performed in a 50 µl mixture containing 1 µl cDNA, 5
µl 10X Qiagen PCR buffer, 10 µl 5X Q-Solution, 1 µl each of
deoxynucleotide triphosphate mix (10 mM), 0.1 µM of each sense and
antisense primer, and 0.5 µl Taq DNA polymerase by using Taq DNA
Polymerase kit (Qiagen, Inc.). The amplification reaction consisted
of initial denaturation at 94°C for 3 min, followed by three-step
cycling that consisted of denaturation at 94°C for 30 sec,
annealing at a temperature optimized for each primer pair for 30
sec and extension at 72°C for 1 min for 30 cycles (optimal number
of cycles determined prior to the experiment). A final extension
step was performed at 72°C for 10 min. The 150 ng amplification
products were electrophoresed on 2% agarose gels and visualized by
ethidium bromide staining. The results were normalized to the mRNA
level of β-actin, a housekeeping enzyme. These experiments were
performed using samples from at least five different cell
preparations and quantification (BioSens Gel Documentation System,
Shanghai Bio-Tech Co., Ltd., Shanghai, China) of mRNA was confirmed
using the same cell sample at least in triplicate.
 | Table I.Primer sequences used in
semi-quantitative reverse transcription-polymerase chain
reaction. |
Table I.
Primer sequences used in
semi-quantitative reverse transcription-polymerase chain
reaction.
|
| Primer |
|
|
|---|
|
|
|
|
|
|---|
| Gene | Forward | Reverse | Size, bp | Annealing
temperature, °C |
|---|
| β-actin |
5′-ATCATGTTTGAGACCTTCAA-3′ |
5′-CATCTCTTGCTCGAAGTCCA-3′ | 212 | 55 |
| β-catenin |
5′-AAGGTCTGAGGAGCAGCTTC-3′ |
5′-TGGACCATAACTGCAGCCTT-3′ | 668 | 55 |
| COX-2 |
5′-TCAAGTCCCTGAGCATCTAC-3′ |
5′-CATTCCTACCACCAGCAACC-3′ | 488 | 58 |
| Cyclin D1 |
5′-GAGACCATCCCCCTGACGGC-3′ |
5′-CTCTTCCTCCTCCTCGGCGGC-3′ | 484 | 56 |
| c-fos |
5′-ACCAGTCCGGACCTGCAGTGG-3′ |
5′-GCGGCATTTGGCTGCAGCCAT-3′ | 261 | 57 |
| c-Jun |
5′-CCCCTGTCCCCCATCGACATG-3′ |
5′-TTGCAACTGCGTTAGCAT-3′ | 267 | 55 |
Reporter gene assay
Cells were switched to media containing 0.5% FBS and
1% penicillin/streptomycin and transfected using FuGENE 3
transfection reagent (Roche Diagnostics, Indianapolis, IN, USA) 500
µl TOPflash (wild-type TCF) or FOPflash (mutant TCF) luciferase
reporter plasmids (300 ng/ml) (Upstate Biotechnology; EMD
Millipore, Billerica, MA, USA) were co-transfected into Saos-2
cells (~1×106) with 500 µl Renilla luciferase
marker plasmid (5 ng/ml) (Promega Corporation, Madison, WI, USA) at
37°C for 48 h. The cells were subjected to 0 or 12% tensile stress
for 1 h. Cells were subsequently incubated for 4 h in 12 ml DMEM +
0.1% FBS and 1% penicillin/streptomycin, after which the cells were
processed for Dual Luciferase Assays (Dual-Luciferase®
Reporter Assay System; Promega Corporation). Results were
calculated by standardizing to Renilla luciferase counts and
subtracting the mutant (FOPflash) luciferase counts from the
wild-type (TOPflash) counts.
Western blotting, immunoprecipitation
and immunoblotting
Cells that were subjected to 0 or 12% tensile stress
for 1 h were cultured on flex membranes. Following treatment, cells
were washed three times with ice-cold PBS and placed on ice. Cells
were lysed with 500 µl ice-cold lysis buffer at pH 7.4 (50 mM
HEPES, 5 mM EDTA and 50 mM NaCl), 1% Triton-X-100, protease
inhibitors (10 µg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride
and 10 µg/ml leupeptin) and phosphatase inhibitors (50 mM sodium
fluoride, 1 mM sodium orthovanadate and 10 mM sodium
pyrophosphate). Solubilized proteins were centrifuged at 14,000 × g
in a microfuge (4°C) for 15 min and supernatants were stored at
−80°C. Extracted proteins were quantified by the Bradford assay.
Nuclear extract was also prepared: The cell homogenates were
centrifuged at 450 × g for 5 min at 4°C. The supernatant was
discarded, and 200 µl of hypotonic buffer containing 10 mM HEPES, 1
mM EDTA, 1 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 20 µg/ml
aprotinin, 0.5% Nonidet P-40, 4 µg/ml leupeptin and 0.2 mM
phenylmethanesulfonyl fluoride were added to the cell pellet.
Following centrifugation at 7,200 × g for 15 min at 4°C, the
supernatant, which contained the membranes and the cytosolic
fraction, was discarded. The pellets, consisting of crude nuclei,
were suspended for 15 min at 4°C in 20 µl of a hypertonic solution
(20 mM HEPES, 420 mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.2
mM PMSF, 0.2 mM EDTA and 25% glycerol) containing protease
inhibitor. The samples were centrifuged at 16,000 × g for 30 min at
4°C. The supernatant, which contained nuclear proteins, was stored
at −80°C until use.
For immunoprecipitation, 600–700 µg/µl cell lysates
were incubated with E-cadherin (1:200; sc-9988; Santa Cruz
Biotechnology, Inc.) or β-catenin (1:200; sc-7199; Santa Cruz
Biotechnology, Inc.) antibodies overnight at 4°C and subsequently
incubated with 25 ml protein A-G agarose beads (Beyotime Institute
of Biotechnology, Shanghai, China) for 1 h at 4°C with gentle
rocking. The beads were washed four times with 500 ml lysis buffer
and proteins were solubilized in Laemmli sample buffer (sc-286962;
Santa Cruz Biotechnology, Inc.) containing 2-mercaptoethanol.
Immunoprecipitated samples or samples prepared directly after cell
lysis were separated using 10% SDS-PAGE and transferred onto
nitrocellulose membranes. The amount of protein was 70 µg per lane.
Membranes were blocked overnight at 4°C with PBS containing 5%
non-fat dry milk and 0.1% Tween-20. The blots were incubated
overnight at 4°C with primary antibodies against phosphorylated
(p)-GSK-3β (1:100; sc-373800), GSK-3β (1:100; sc-377213; both Santa
Cruz Biotechnology, Inc.), β-actin (1:4,000; A2228; Sigma, St.
Louis, MO, USA), β-catenin (1:100; sc-7199), lamin B (1:100;
374015) and E-cadherin (1:100; sc-9988; all Santa Cruz
Biotechnology, Inc.) in PBS containing 1% non-fat dry milk and 0.1%
Tween-20. Following incubation with secondary antibodies (1:2,000;
7074 and 7076; anti-rabbit or anti-mouse IgG antibody conjugated to
horseradish peroxidase; Cell Signaling Technology; Danvers, MA,
USA) for 1 h at 37°C in PBS containing 1% non-fat dry milk and 0.1%
Tween-20, proteins were detected by chemiluminescence (SuperSignal™
West Femto Trial kit; Thermo Fisher Scientific, Inc.). and protein
expression were analysed by Bandscan version 5.0 software (Glyko,
Inc., Novato, CA, USA).
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analysis was processed by SPSS version 16.0 (SPSS,
Inc., Chicago, IL, USA). Data in Fig.
1 were subjected to repeated analysis of variance and multiple
comparisons were made by Tukeys method. Student's t-test was used
to analyze differences between two groups. P<0.05 was considered
to indicate a statistically significant difference.
Results
The effects of cyclic tensile strain
on proliferation, ALP activity and mineralization nodule formation
in Saos-2 osteoblastic cells
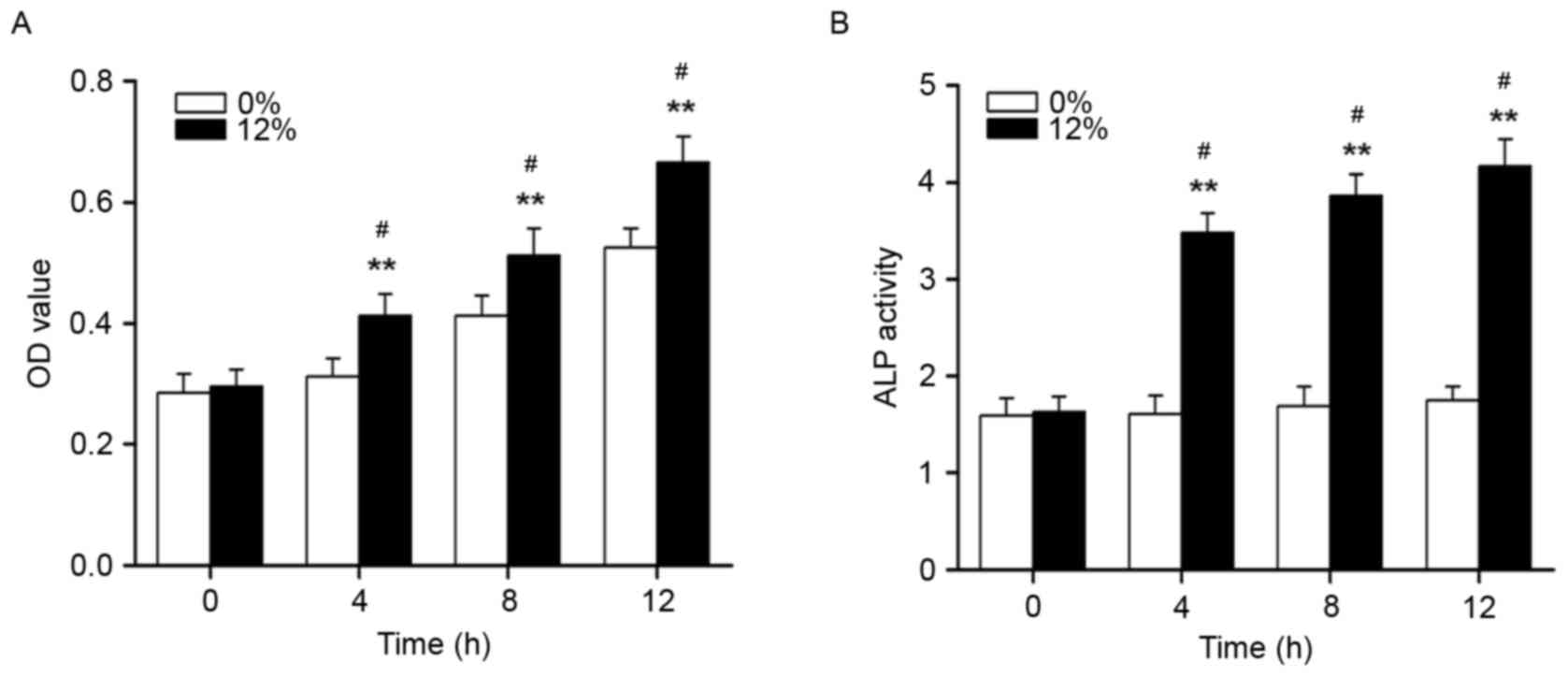
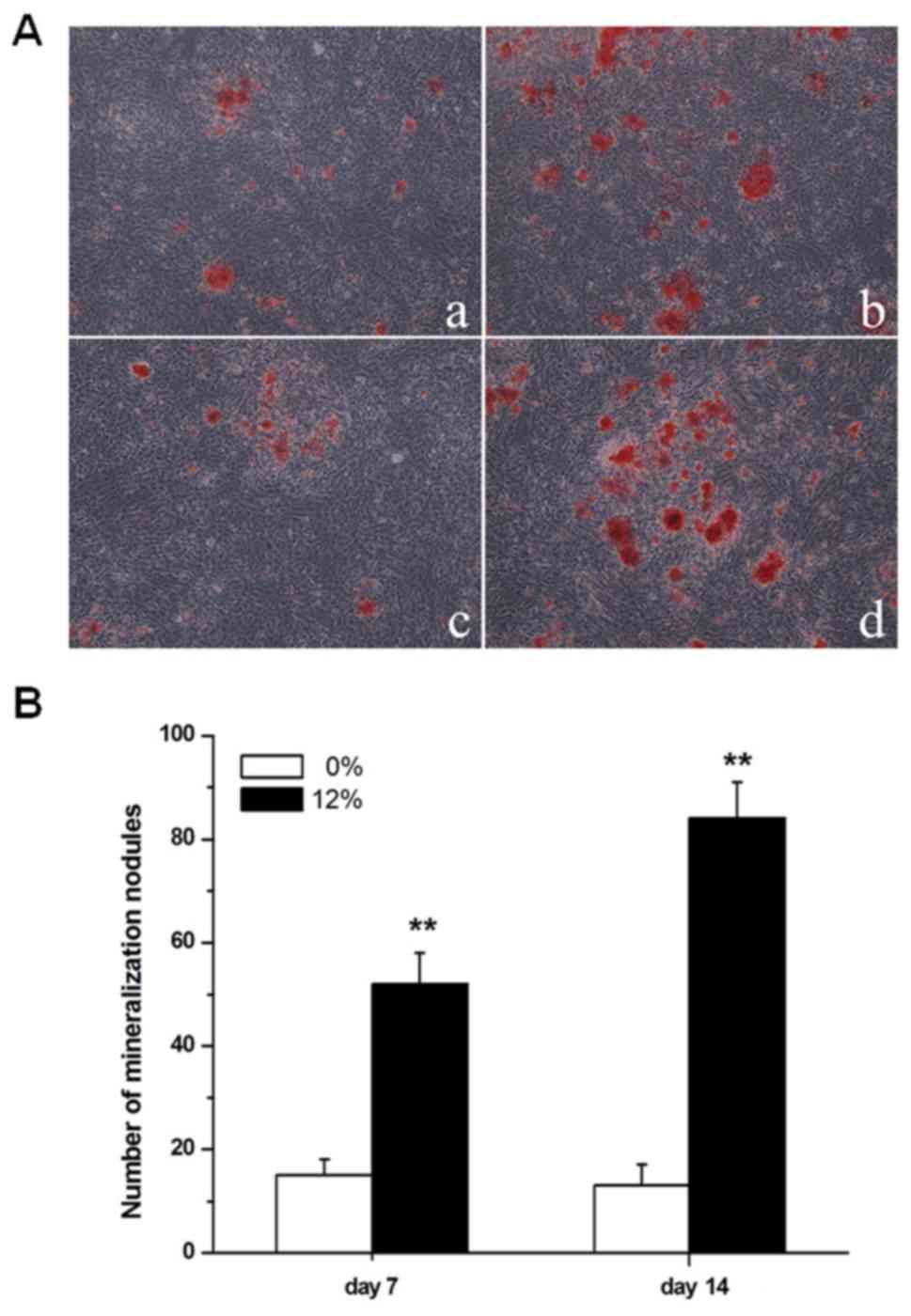
As demonstrated in Fig.
1, the proliferation and ALP activity of Saos-2 osteoblastic
cells were significantly increased following mechanical strain
treatment at 4, 8 and 12 h (P<0.01). The formation of
mineralization nodules in Saos-2 osteoblastic cells was also
promoted by a 12% mechanical strain elongation rate, compared with
unstrained cells (P<0.01; Fig.
2).
The effects of cyclic tensile strain
application on β-catenin expression in Saos-2 cells
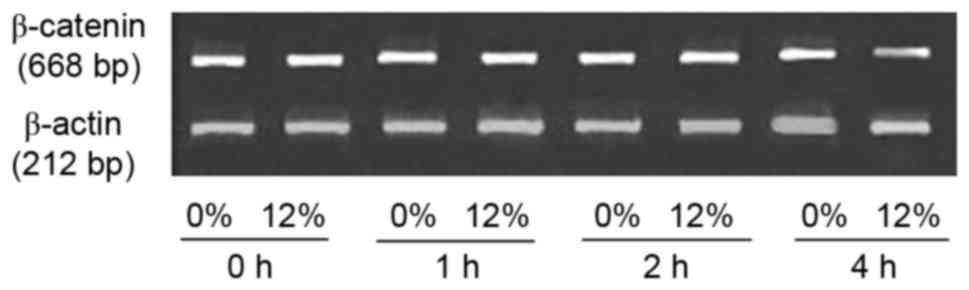
The expression level of β-catenin mRNA was measured
using RT-PCR (Fig. 3). No marked
differences in the expression level of β-catenin mRNA were observed
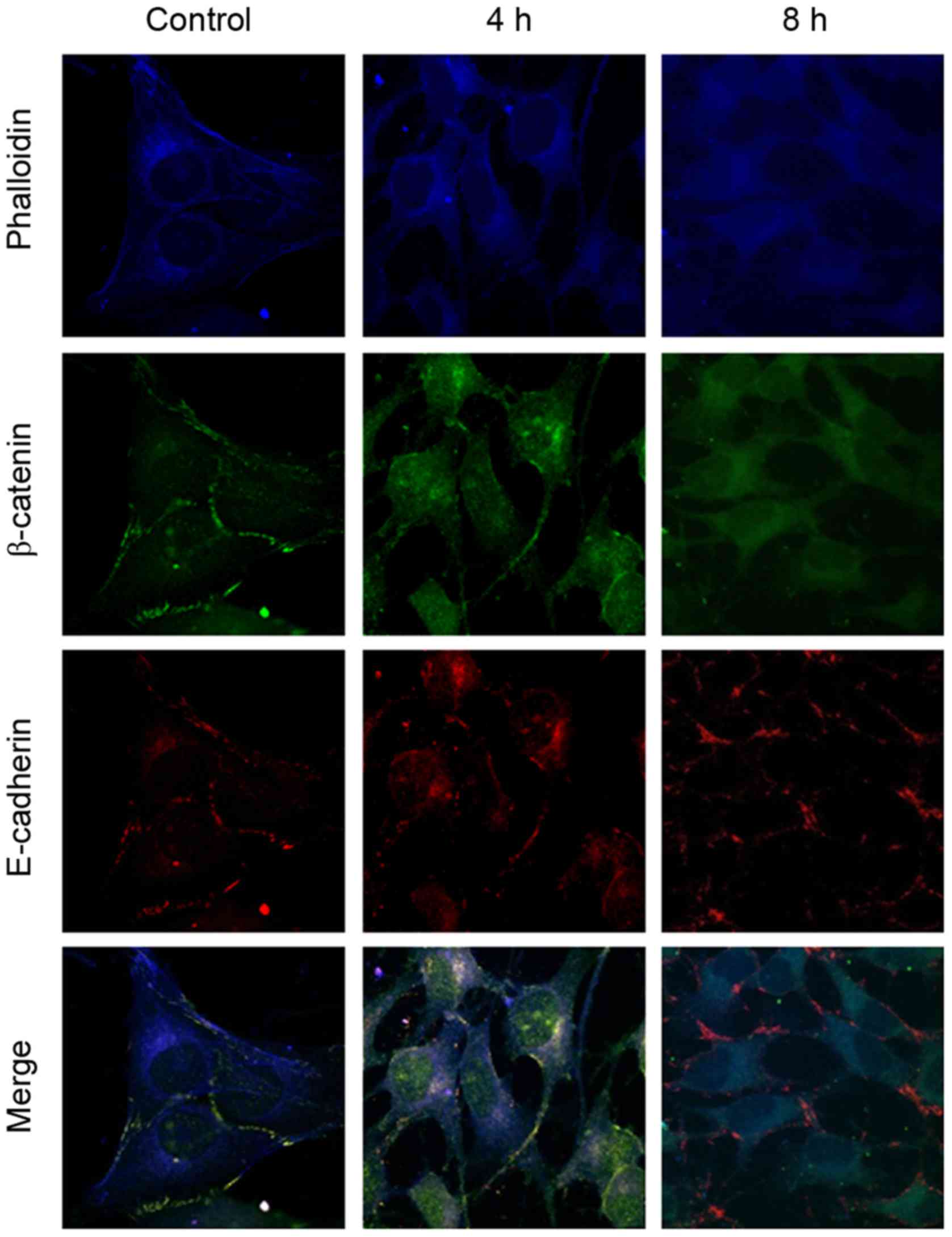
between the 0 and 12% tensile stress groups. The immunofluorescent
staining results demonstrated that β-catenin and E-cadherin were
detected prior to (control; Fig.
4) and following (4 and 8 h; Fig.
4) 12% tensile stress. β-catenin was localized primarily on the
cytomembrane prior to stress, while it was observed in the
cytoplasm and nucleus following stress. However, E-cadherin
remained on the cytomembrane throughout. Co-localization of
β-catenin and E-cadherin was observed prior to stress; however,
co-localization was not observed following 12% tensile stress for 8
h (Fig. 4).
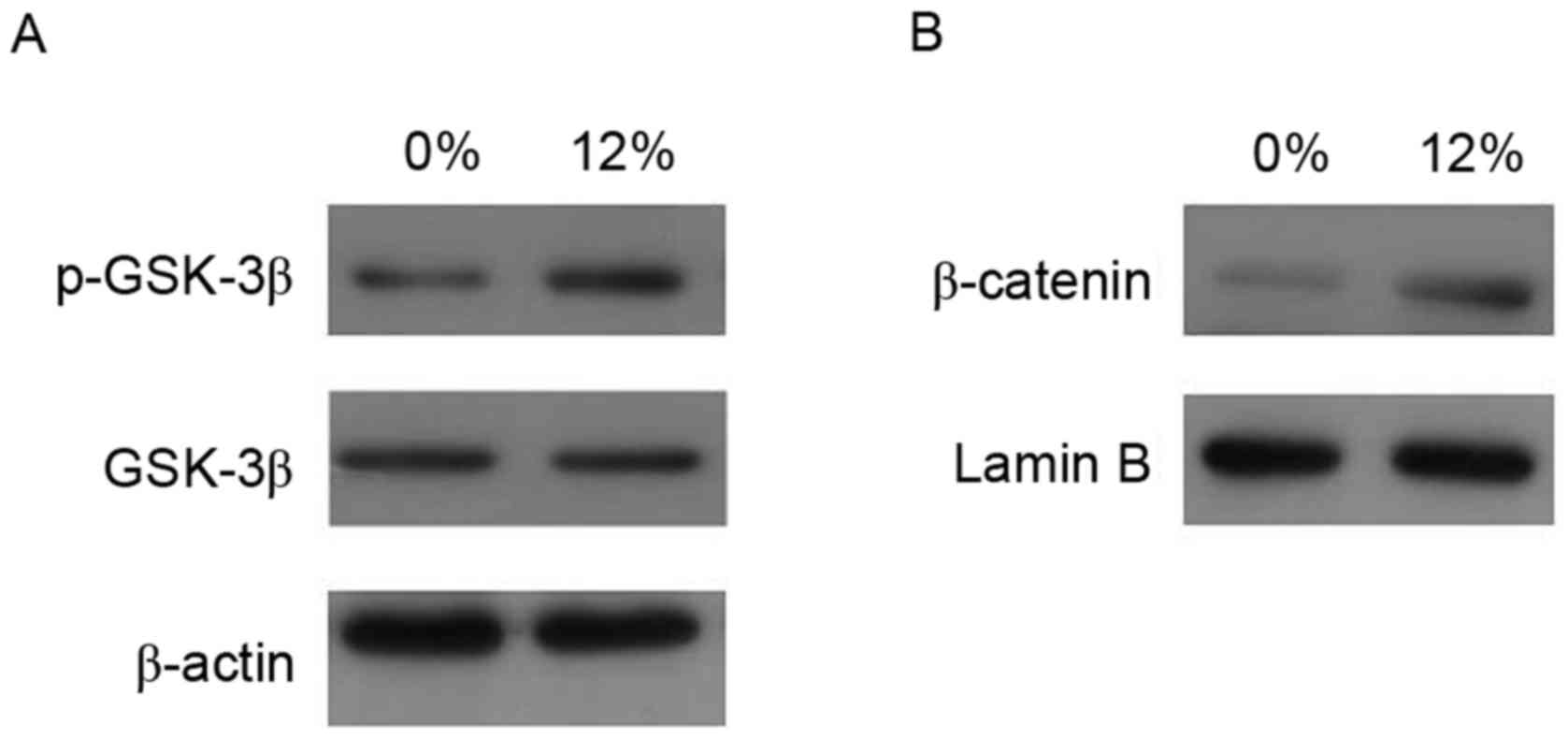
GSK-3β phosphorylates β-catenin and
subsequently facilitates the degradation of β-catenin
The present study further investigated whether
mechanical strain influences GSK-3β activity (Fig. 5). The results demonstrated that 12%
tensile strain increased the cytosolic phosphorylation of GSK-3β
expression, while it had no effect on GSK-3β levels. In addition,
nuclear β-catenin levels were upregulated by tensile stress.
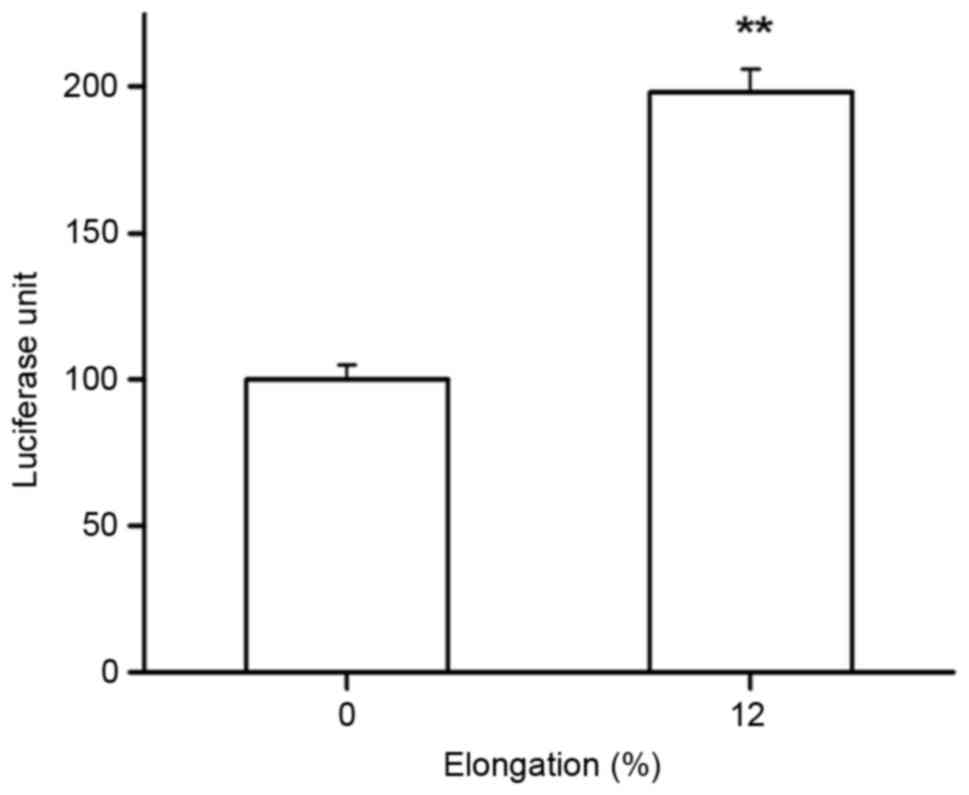
The effects of cyclic tensile strain
application on TCF reporter gene activity
In the current study, TCF reporter gene
transcriptional activity was also determined in Saos-2 cells, and
the results demonstrated a significant increase in TCF reporter
gene activity compared with the control group following 12% tensile
for 1 h (P<0.05; Fig. 6).
Therefore, the Wnt/β-catenin pathway may be functionally activated
under tensile stress.
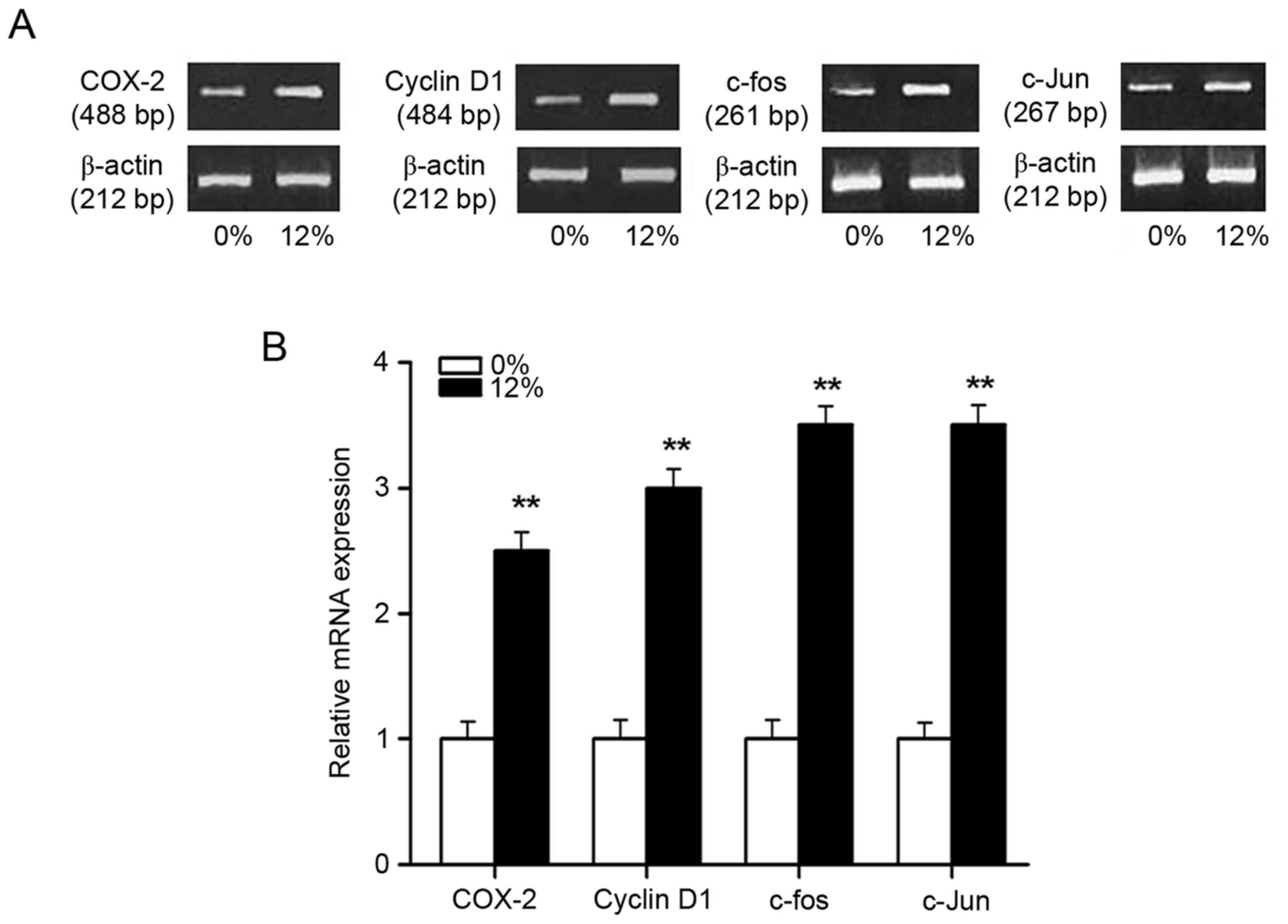
The effects of cyclic tensile strain
application on mRNA expression of genes downstream of the
Wnt/β-catenin pathway
The mRNA expression of downstream genes (COX-2,
c-fos, c-Jun and cyclin D1) of the Wnt/β-catenin pathway was
investigated by RT-PCR following 0 or 12% tensile stress for 4 h
(Fig. 7). The results demonstrated
a significant increase in the mRNA transcript levels of these genes
in the 12% tensile stress group compared with the control group
(P<0.01; Fig. 7).
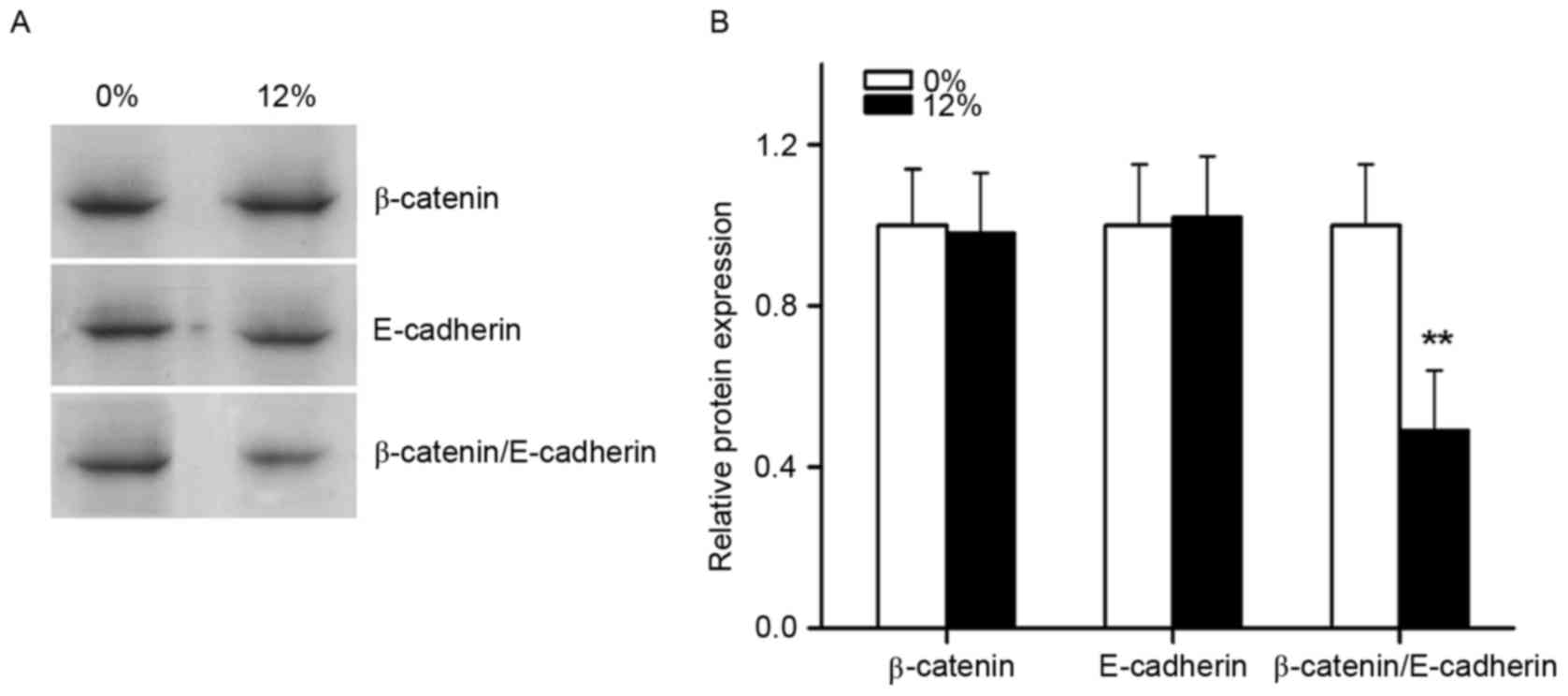
The effects of cyclic tensile strain
application on binding of the β-catenin/E-cadherin complex
Immunoprecipitation was performed to observe the
effects of tensile stress on the binding of the
β-catenin/E-cadherin complex (Fig.
8). Immunoblot assays demonstrated that β-catenin and
E-cadherin protein levels in the experimental groups were unchanged
following 12% tensile stress for 1 h. However, immunoprecipitation
assays demonstrated a significant decrease in the binding of
β-catenin to E-cadherin.
Discussion
Numerous in vitro and in vivo
experiments have reported that mechanical loading promoted
osteoblast proliferation and increased the transcription of various
bone-associated genes (4–6). However, the molecular mechanism by
which mechanical signals are transduced in osteoblasts remains
unclear. Previous studies have indicated that the integrin/focal
adhesion kinase pathway and calcium ion channels are involved in
the signal transduction process (21,22).
Robinson et al (23)
demonstrated that target gene expression in the Wnt/β-catenin
pathway was upregulated by the application of a four-point bending
load to the long bones of low-density lipoprotein receptor related
protein (Lrp) 5 transgenic mice, indicating that mechanical loading
may activate the Wnt/β-catenin signaling pathway.
The Wnt/β-catenin signaling pathway has become a
major research focus in developmental biology. Mammalian Wnt
proteins bind to the receptor complex of the Lrp5/Lrp6
extracellular region and the Frizzled receptor. Subsequently,
through a series of cytoplasmic protein interactions, β-catenin
accumulates in the cytoplasm, translocates to the nucleus and
functions with transcription factors, TCF and LEF-1, to activate
bone-associated target gene transcription. This pathway is referred
to as the classical Wnt/β-catenin pathway, and it has an important
role in bone formation (24,25).
Loss-of-function mutations in Lrp5 resulted in osteoblast
dysfunction and subsequent osteoporosis (25). Furthermore, Boyden et al
(26) demonstrated that a mutation
at position 171 in the Lrp5 protein led to the total loss of
dickkopf Wnt signaling pathway inhibitor 1 antagonism in the Wnt
signaling pathway in vitro, which resulted in in bone
sclerosis. In addition, Bain et al (10) reported that increased expression
and activity of the early osteoblast differentiation marker ALP was
induced when β-catenin was overexpressed in C3H1OT1/2 cells or when
lithium chloride was added to enhance the expression of endogenous
β-catenin. These previous reports indicated that β-catenin has an
important role in the proliferation and differentiation of
osteoblast precursor cells and osteoblasts. The present study
applied 12% cyclical tensile stress to osteoblasts and also
demonstrated that the proliferation, ALP activity and
mineralization nodule formation of Saos-2 osteoblastic cells were
promoted. Subsequently, the present study investigated the
alterations in the Wnt/β-catenin signaling pathway following
tensile stress application in osteoblasts in vitro.
Previous studies have demonstrated that
intracellular β-catenin serves a dual function in cell-cell
adhesion structures and as a regulation factor of downstream gene
expression. Under normal circumstances, β-catenin forms a complex
with cadherins in the cell membrane, while free β-catenin is
rapidly degraded via phosphorylation by the complex of GSK-3β, axin
and adenomatous polyposis coli in the cytoplasm. However, if GSK-3β
becomes inactivated by phosphorylation, β-catenin is not degraded.
As a result, β-catenin accumulates in the cytoplasm and triggers
signal transduction, followed by translocation into the nucleus to
promote target gene transcription by binding to transcription
factors. Therefore, the nuclear translocation of β-catenin into is
a prerequisite for the activation of the classical Wnt/β-catenin
pathway (13,27). The present study demonstrated that
the phosphorylation of GSK-3β and nuclear β-catenin expression was
significantly enhanced following tensile stress. Nuclear β-catenin
was more apparent in the 4 and 8 h tensile stress groups compared
with the control group. These results indicated that 12% cyclical
tensile stress altered the localization of β-catenin in the cell,
which may trigger β-catenin-mediated Wnt signal transduction and
maintain the activation of this signaling pathway for an extended
period of time. Furthermore, β-catenin mRNA and protein expression
levels were also measured in the current study, however, no marked
differences between the experimental and control groups were
observed, indicating that tensile stress did not increase the level
of β-catenin transcription or its protein synthesis. Instead,
tensile stress primarily resulted in alterations in the spatial
localization of β-catenin and promoted its translocation into the
nucleus. In the current study, 12% tensile stress was selected as
this value falls within the tolerable physiological range for bone.
In the study by Norvell et al (28), β-catenin nuclear translocation was
observed in primary rat osteoblasts after a 10 dynes/cm2
fluid shear stress was applied, and this translocation was
maintained at a relatively high level up to 6 h after loading.
Furthermore, it was reported by Case et al (29) that transient nuclear translocation
of β-catenin occurred in CIMC-4 cells after a 2% stretch force was
applied, however, this event was decreased at 1 h. The difference
between these results may be attributed to the different loading
systems, force values and cell types used.
In the present study, the results also demonstrated
that, following transfection of the TCF reporter gene into
osteoblasts, the activity of the nuclear transcription factor TCF
was significantly enhanced following a 12% cyclical stretch. The
TCF/LEF family is a well-defined group of Wnt/β-catenin downstream
target molecules. Unlike other traditional transcription factors,
TCF/LEF cannot activate transcription alone. Instead, they rely on
the binding and activation of β-catenin. TCF/LEF mutually bind to
transcription inhibitors, when the Wnt/β-catenin pathway remains
inactive. However, upon activation of the pathway, nuclear
β-catenin facilitates Wnt/β-catenin target gene expression by
replacing the inhibitory factors and binding to the N-terminal
region of TCF/LEF to enhance its interaction with chromatin and
restore transcriptional activity. In the study by Chou et al
(30), impaired bone formation was
observed in TCF-4 deficient mice, indicating that the activation
and expression of this gene is closely associated with bone
formation. The results of the TCF luciferase reporter gene assay in
the present study were consistent with the immunofluorescence
results, which demonstrated nuclear localization of β-catenin
following 12% tensile stress. Together, these results demonstrate
that nuclear translocation and activation of β-catenin due to
stretching may further enhance the transcriptional activity of TCF
and lead to the functional activation of the Wnt/β-catenin
signaling pathway.
To further confirm and observe the activation of the
Wnt/β-catenin signaling pathway following stretch loading, the
present study investigated the expression of downstream target
genes, including COX-2, c-fos, c-Jun and cyclin D1. The mRNA
transcript levels of COX-2, c-fos, c-Jun and cyclin D1 were
significantly enhanced after 4 h stretch loading, as revealed by
RT-PCR analysis. Norvell et al (28) reported that the expression of COX-2
was enhanced by the activation of the Wnt/β-catenin pathway by
GSK-3β inhibitors. Additionally, the function of COX-2 in bone
tissue reconstruction following force loading was also confirmed.
This indicated that COX-2 is an important target gene in the
Wnt/β-catenin pathway and its expression may be triggered by the
activation of the Wnt/β-catenin pathway by tensile stress. Robinson
et al (23) reported that,
after Lrp5 G171V transgenic mice were treated with a GSK-3β
inhibitor or stress loading, the expression of the Wnt/β-catenin
target genes, including c-fos, c-Jun and cyclin D1, in the two
groups was higher compared with the control, although no
significant differences were observed between the two experimental
groups. This is consistent with the present study's experimental
results from cells after stretch loading in vitro. Together,
these results indicate that stretch loading may activate the
Wnt/β-catenin pathway and trigger the expression of downstream
target genes that are closely associated with bone formation.
In addition, the current study investigated the
mechanism by which tensile stress activated the Wnt/β-catenin
signaling pathway in osteoblasts. Previous studies have
demonstrated that mechanical loading promote osteoblast
proliferation and differentiation (4,5,31),
however, the mechanism by which osteoblasts convert stimulating
mechanical signals to biochemical signals is unknown. The results
of the current study demonstrated that the levels of β-catenin and
E-cadherin were not significantly altered, however, the binding
level of β-catenin with E-cadherin was significantly reduced
following tensile stress. It is established that cadherins are a
cell-cell adhesion mediation and signal transduction family, and
they are comprised of three different types, including E-cadherin,
N-cadherin and P-cadherin, that are differentially distributed in
different tissues. The cytoplasmic region of cadherin molecules is
highly conserved and is connected to the cytoskeleton. Acting as a
transmembrane calcium-dependent adhesion molecule in various cell
types, including osteoblasts, it binds directly to β-catenin and
forms a complex in the cell membrane, restricting β-catenin to the
cell membrane and preventing it from translocating into the nucleus
to function in the regulation of downstream gene expression.
Adkison et al (32)
demonstrated that the amount of vascular endothelial cadherin
binding to β-catenin was significantly reduced after 34% stretch
loading was applied to mouse pulmonary artery endothelial cells for
24 h. In addition, the present study demonstrated that the binding
level of E-cadherin with β-catenin was significantly reduced and
the amount of β-catenin that translocated into the nucleus was
increased after 12% tensile stress in osteoblasts for 1 h.
Therefore, we hypothesize that this may be due to the structural
deformation of the cell membrane and cytoskeleton under tensile
stress, which disrupts the β-catenin and E-cadherin complex and
causes the release of β-catenin from the complex. As a result,
β-catenin accumulates in the cytoplasm and translocates into the
nucleus. This is followed by binding to and activation of the
nuclear transcription factor TCF to promote the expression of the
downstream bone formation-associated target genes COX-2, c-fos,
c-Jun and cyclin D1, thus facilitating the conversion of mechanical
stimuli to chemical signals.
In conclusion, the results of the current study
demonstrated that 12% tensile stress activated the Wnt/β-catenin
signaling pathway and promoted the expression of downstream target
genes, including COX-2, c-fos, c-Jun and cyclin D1, and that
β-catenin had a pivotal role in this process. The disassociation of
the β-catenin/E-cadherin complex in the osteoblast membrane under
stretch loading and the subsequent translocation of β-catenin into
the nucleus may be an intrinsic mechanical signal transduction
mechanism. Together, these observations provide novel insight in
order to obtain a deeper understanding of the molecular mechanisms
of mechanical signal transduction and bone reconstruction.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 31070836 and
31200706).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FFL was mainly responsible for the design of the
experiment, cell culture, statistical analysis and the writing of
the article. BZ was responsible for the MTT, immunofluorescence and
ALP assays. JHC was responsible for RT-PCR, reporter gene assay,
western blot analysis and immunoprecipitation. FLC was responsible
for the design of the experiment and revising the paper. YD and XF
were responsible for the design of the experiment.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing
interests.
References
|
1
|
Karlsson MK, Johnell O and Obrant KJ: Is
bone mineral density advantage maintained long-term in previous
weight lifters? Calcif Tissue Int. 57:325–328. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lemaire T, Capiez-Lernout E, Kaiser J,
Naili S and Sansalone V: What is the importance of multiphysical
phenomena in bone remodelling signals expression? A multiscale
perspective. J Mech Behav Biomed Mater. 4:909–920. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim JY, Kim BI, Jue SS, Park JH and Shin
JW: Localization of osteopontin and osterix in periodontal tissue
during orthodontic tooth movement in rats. Angle Orthod.
82:107–114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eriksen EF: Cellular mechanisms of bone
remodeling. Rev Endocr Metab Disord. 11:219–227. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wallace JM, Rajachar RM, Allen MR,
Bloomfield SA, Robey PG, Young MF and Kohn DH: Exercise-induced
changes in the cortical bone of growing mice are bone-and
gender-specific. Bone. 40:1120–1127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liedert A, Kaspar D, Blakytny R, Claes L
and Ignatius A: Signal transduction pathways involved in
mechanotransduction in bone cells. Biochem Biophys Res Commun.
349:1–5. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rawadi G, Vayssière B, Dunn F, Baron R and
Roman-Roman S: BMP-2 controls alkaline phosphatase expression and
osteoblast mineralization by a Wnt autocrine loop. J Bone Miner
Res. 18:1842–1853. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jackson A, Vayssière B, Garcia T, Newell
W, Baron R, Roman-Roman S and Rawadi G: Gene array analysis of
Wnt-regulated genes in C3H10T1/2 cells. Bone. 36:585–598. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Caetano-Lopes J, Canhão H and Fonseca JE:
Osteoblasts and bone formation. Acta Reumatol Port. 32:103–110.
2007.PubMed/NCBI
|
|
10
|
Bain G, Müller T, Wang X and Papkoff J:
Activated beta-catenin induces osteoblast differentiation of
C3H10T1/2 cells and participates in BMP2 mediated signal
transduction. Biochem Biophys Res Commun. 301:84–91. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gottardi CJ and Gumbiner BM: Adhesion
signaling: How beta-catenin interacts with its partners. Curr Biol.
11:R792–R794. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nelson WJ and Nusse R: Convergence of Wnt,
beta-catenin, and cadherin pathways. Science. 303:1483–1487. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jamora C and Fuchs E: Intercellular
adhesion, signalling and the cytoskeleton. Nat Cell Biol.
4:E101–E108. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smith E and Frenkel B: Glucocorticoids
inhibit the transcriptional activity of LEF/TCF in differentiating
osteoblasts in a glycogen synthase kinase-3beta-dependent and
-independent manner. J Biol Chem. 280:2388–2394. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sakamoto I, Kishida S, Fukui A, Kishida M,
Yamamoto H, Hino S, Michiue T, Takada S, Asashima M and Kikuchi A:
A novel beta-catenin-binding protein inhibits
beta-catenin-dependent Tcf activation and axis formation. J Biol
Chem. 275:32871–32878. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kulkarni NH, Onyia JE, Zeng Q, Tian X, Liu
M, Halladay DL, Frolik CA, Engler T, Wei T, Kriauciunas A, et al:
Orally bioavailable GSK-3alpha/beta dual inhibitor increases
markers of cellular differentiation in vitro and bone mass in vivo.
J Bone Miner Res. 21:910–920. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kato M, Patel MS, Levasseur R, Lobov I,
Chang BH, Glass DA II, Hartmann C, Li L, Hwang TH, Brayton CF, et
al: Cbfa1-independent decrease in osteoblast proliferation,
osteopenia, and persistent embryonic eye vascularization in mice
deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 157:303–314.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vaes BL, Dechering KJ, van Someren EP,
Hendriks JM, van de Ven CJ, Feijen A, Mummery CL, Reinders MJ,
Olijve W, van Zoelen EJ and Steegenga WT: Microarray analysis
reveals expression regulation of Wnt antagonists in differentiating
osteoblasts. Bone. 36:803–811. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Glass DA II and Karsenty G: Molecular
bases of the regulation of bone remodeling by the canonical Wnt
signaling pathway. Curr Top Dev Biol. 73:43–84. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Reyes L, Eiler-McManis E, Rodrigues PH,
Chadda AS, Wallet SM, Bélanger M, Barrett AG, Alvarez S, Akin D,
Dunn WA Jr and Progulske-Fox A: Deletion of lipoprotein PG0717 in
Porphyromonas gingivalis W83 reduces gingipain activity and alters
trafficking in and response by host cells. PLoS One. 8:e742302013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan Y, Sun H, Gong Y, Yan Z, Zhang X, Guo
Y and Wang Y: Mechanical strain promotes osteoblastic
differentiation through integrin-β1-mediated β-catenin signaling.
Int J Mol Med. 38:594–600. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang H, Sun W, Ma J, Pan Y, Wang L and
Zhang W: Polycystin-1 mediates mechanical strain-induced
osteoblastic mechanoresponses via potentiation of intracellular
calcium and Akt/β-catenin pathway. PLoS One. 9:e917302014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robinson JA, Chatterjee-Kishore M,
Yaworsky PJ, Cullen DM, Zhao W, Li C, Kharode Y, Sauter L, Babij P,
Brown EL, et al: Wnt/beta-catenin signaling is a normal
physiological response to mechanical loading in bone. J Biol Chem.
281:31720–31728. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Manolagas SC and Almeida M: Gone with the
Wnts: Beta-catenin, T-cell factor, forkhead box O, and oxidative
stress in age-dependent diseases of bone, lipid, and glucose
metabolism. Mol Endocrinol. 21:2605–2614. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao C, Li Y, Wang X, Zou S, Hu J and Luo
E: The effect of uniaxial mechanical stretch on Wnt/β-catenin
pathway in bone mesenchymal stem cells. J Craniofac Surg.
28:113–117. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Boyden LM, Mao J, Belsky J, Mitzner L,
Farhi A, Mitnick MA, Wu D, Insogna K and Lifton RP: High bone
density due to a mutation in LDL-receptor-related protein 5. N Engl
J Med. 346:1513–1521. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Roura S, Miravet S, Piedra J, de Herreros
García A and Duñach M: Regulation of E-cadherin/Catenin association
by tyrosine phosphorylation. J Biol Chem. 274:36734–36740. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Norvell SM, Alvarez M, Bidwell JP and
Pavalko FM: Fluid shear stress induces beta-catenin signaling in
osteoblasts. Calcif Tissue Int. 75:396–404. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Case N, Ma M, Sen B, Xie Z, Gross TS and
Rubin J: Beta-catenin levels influence rapid mechanical responses
in osteoblasts. J Biol Chem. 283:29196–29205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chou HY, Howng SL, Cheng TS, Hsiao YL,
Lieu AS, Loh JK, Hwang SL, Lin CC, Hsu CM, Wang C, et al: GSKIP is
homologous to the Axin GSK3beta interaction domain and functions as
a negative regulator of GSK3 beta. Biochemistry. 45:11379–11389.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brady RT, O'Brien FJ and Hoey DA:
Mechanically stimulated bone cells secrete paracrine factors that
regulate osteoprogenitor recruitment, proliferation, and
differentiation. Biochem Biophys Res Commun. 459:118–123. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Adkison JB, Miller GT, Weber DS, Miyahara
T, Ballard ST, Frost JR and Parker JC: Differential responses of
pulmonary endothelial phenotypes to cyclical stretch. Microvasc
Res. 71:175–184. 2006. View Article : Google Scholar : PubMed/NCBI
|