Introduction
Sepsis is a form of systemic inflammation caused by
an infection, and multiple organ failure is known to occur when
this condition intensifies. Clinical studies demonstrated that the
central nervous system might be one of the first organs affected by
sepsis. Sepsis-associated encephalopathy (SAE), characterized by
diffuse cerebral dysfunction, is secondary to sepsis and is related
to increased morbidity and mortality (1,2).
Clinical symptoms of SAE are delirium, fluctuating changes in
mental status, lack of attention, and disorganized thinking.
Encephalopathy of variable severity occurs in 9–71% of septic
patients, and those with central nervous disorders have the worst
prognosis in terms of cognitive and motor function. In addition, it
has been reported that the mortality rate of septic patients with
SAE is approximately twice that of septic patients without SAE;
moreover, the mortality rate increases to 63% when patients with
SAE present with a Glasgow Coma Scale (GCS) value of 3–8 (1,2).
Although several mechanisms including inflammation or the
disturbance of neurotransmission disturbance, have been proposed,
the precise mechanisms responsible for sepsis-induced cognitive
impairment have not been fully elucidated.
The kynurenine (KYN) pathway is well known to be a
major mechanism of tryptophan catabolism, and it is activated
during neuroinflammation during several neurodegenerative diseases
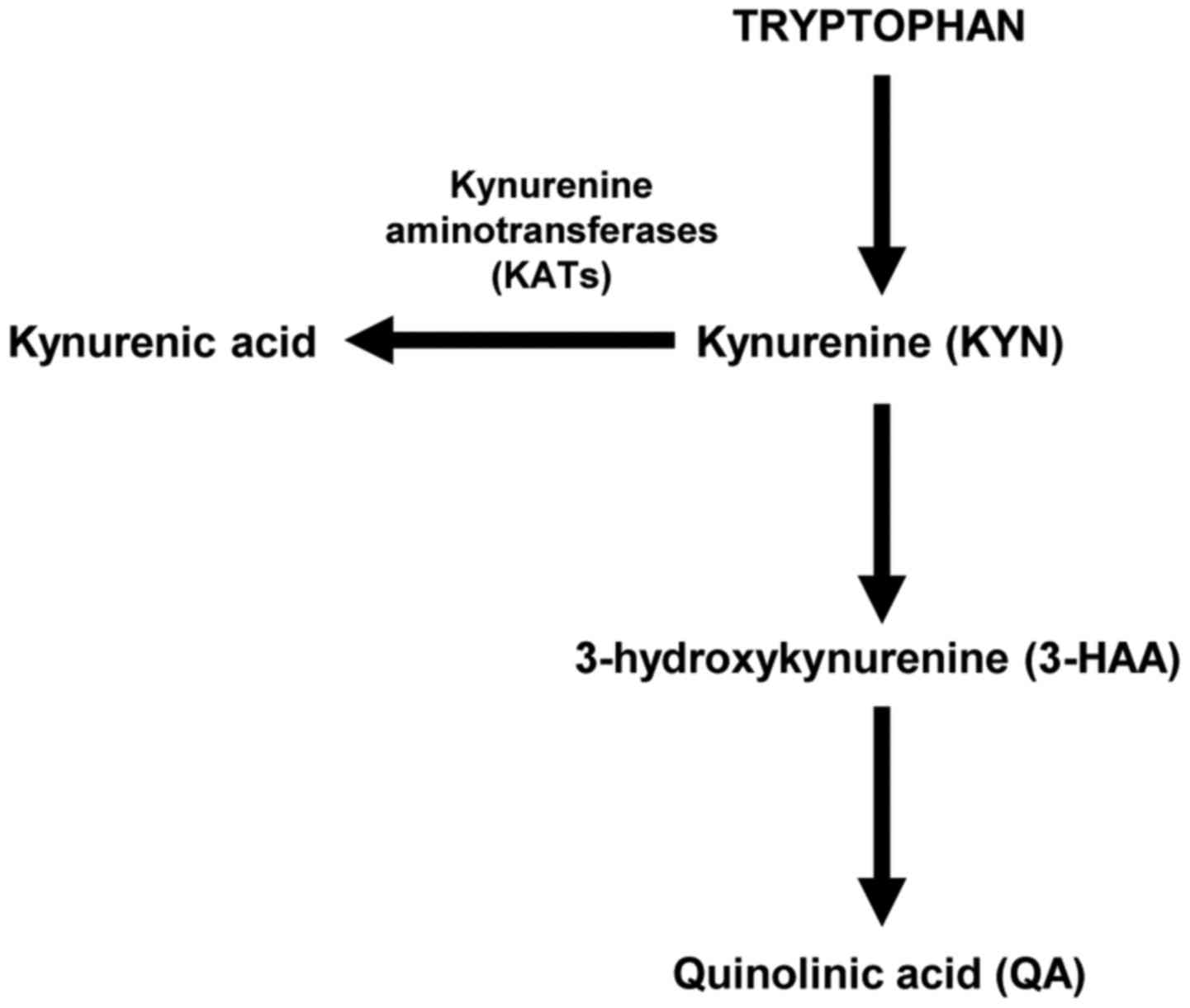
(3,4) such as SAE (5). Tryptophan is converted to KYN, and
KYN is converted into three intermediates, quinolinic acid (QA),
3-hydroxykynurenine (3-HAA), and kynurenic acid (KYNA) via two
different pathways (Fig. 1). 3-HAA
and QA are neurotoxic, whereas KYNA is neuroprotective. The
relative balance between the two branches of this pathway might
play an important role in the development of neuroinflammation. The
conversion to KYNA is catalyzed by kynurenine aminotransferases
(KATs), which have been detected in the brain and peripheral
tissues such as the skeletal muscle (6,7).
Recent studies showed that the peroxisome proliferator-activated
receptor (PPAR)-PPAR coactivator-1 (PGC-1) pathway induces skeletal
muscle KAT expression during exercise (7,8). The
analysis of PGC-1α1 skeletal muscle-specific transgenic mice showed
that increased expression of skeletal muscle KATs induced KYN
metabolism. Synthesis of KYNA was enhanced and the accumulation of
KYN was reduced, thereby protecting against stress-induced
depression (7).
Among several hundreds of E3 ubiquitin ligases,
synoviolin (Syvn1), identified from the cDNA of rheumatoid synovial
cells, is the only known regulator of PGC-1β ubiquitination
(9). We recently demonstrated that
Syvn1 interacts with PGC-1β and induces its degradation (9). Global elimination of Syvn1 in
post-neonatal mice is associated with weight loss and reduced white
adipose tissue through enhanced energy expenditure, which is
mediated by the function of PGC-1β (9). PGC-1β and PGC-1α share extensive
sequence identity to each other (10,11),
and the primary structure of PGC-1β has several unique features of
primary structure such as LXXLL in its middle portion and the
absence of a proline-rich region at the C-terminus. Knockout
studies have also suggested functional differences between PGC-1α
and PGC-1β, such as lethality (12). We previously demonstrated that
Syvn1 is involved in the development of rheumatoid arthritis,
fibrosis, limb girdle muscular dystrophy, and liver cirrhosis
(13–17). We also described another unique
function of Syvn1; specifically, Syvn1 entraps and degrades tumor
suppressor p53 and NRF2 (17,18).
Nrf2 is a transcriptional factor that activates antioxidant and
cytoprotective genes that share a common a cis-acting
enhancer sequence, termed the antioxidant response element (ARE),
under conditions of oxidative stress. Several studies indicate that
Nrf2 is neuroprotective against neurotoxicity (19–21).
These results suggest that loss of Syvn1 expression might have
neuroprotective effects.
In the present study, we therefore investigated
whether Syvn1-deficient mice were resistant to cecal
ligation/perforation (CLP) treatment, which is a mouse model of
sepsis, and revealed that Syvn1 ablation mediates the activation of
tryptophan metabolism via KAT4 gene expression.
Materials and methods
Mice
All procedures involving animals were performed in
accordance with institutional and national guidelines for animal
experimentation, and were approved by the Institutional Animal Care
and Use Committee of Tokyo Medical University (#S-24021). Mice were
kept in specific pathogen free (SPF) under standard conditions
(20–26°C temperature; 40–65% humidity) with a 12-h light/12-h dark
cycle. F-1 Foods (5.1% fat, 21.3% protein) were purchased from
Funabashi farm (Chiba, Japan). All mice used in the study were of
the C57BL/6J background. Heterozygous Syvn1 mice and tamoxifen
(Tam)-inducible Syvn1 knockout mice were previously
described (9,13).
Sepsis model
Male C57/BL6 mice at 7–9 weeks of age were
anesthetized with 4–5% sevoflurane and a middle abdominal incision
was made along the ventral surface of the abdomen to expose the
cecum. Before perforation, feces were gently relocated towards the
distal cecum. The cecum was ligated at 5 mm from the distal end and
then punctured once with a 21-gauge needle, allowing the exposure
of feces (cecal ligation and puncture; CLP). A small droplet of
feces was then gently squeezed through both sides of the puncture.
The cecum was returned to the peritoneal cavity with careful
attention to ensure that feces did not contaminate the margins of
the abdominal and skin wound. The muscle and skin incision were
closed with 3-0 black silk. This model represents a high-grade
sepsis with 100% mortality in 72 h (22). Mice received a subcutaneous
injection of pre-warmed saline solution (37°C; 5 ml per 100 g body
weight). Mice receiving sham surgery underwent the same procedure
except that the cecum was neither ligated nor punctured (sham
group; S). At each experimental time point after performing the
procedure, mice were euthanized by cervical dislocation, and brain
and skeletal muscle tissue samples were obtained.
Measurement of metabolites
Brain tissues were obtained at 18 h after the CLP
procedure (n=2 for each group). Approximately 20 mg of frozen brain
tissues were immersed into 1,500 µl of 50% acetonitrile/Milli-Q
water containing internal standards (H3304-1002; Human Metabolome
Technologies, Inc., Tsuruoka, Japan) at 0°C in order to inactivate
enzymes. The tissue was homogenized three times at 1,500 rpm for
120s using a tissue homogenizer (BMS-M10N21; BMS Co., Ltd., Tokyo,
Japan) and then the homogenate was centrifuged at 2,300 × g
and 4°C for 5 min. Subsequently, 800 µl of the upper aqueous layer
was centrifugally filtered through a Millipore 5-kDa cut-off filter
at 9,100 × g and 4°C for 120 min to remove proteins. The
filtrate was centrifugally concentrated and resuspended in 50 µl of
Milli-Q water for CE-MS analysis. Metabolome measurements were
performed by a facility service at Human Metabolome Technology
Inc., Tsuruoka, Japan.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from the skeletal muscle and brain of the
mice was purified by using ISOGEN (Nippon Gene, Tokyo, Japan)
according to the manufacturer's instructions and reverse
transcribed by using ReverTra Ace with random primers (Toyobo,
Osaka, Japan). qPCR was performed by using LightCycler 480 Probes
Master (Roche Diagnostics, Mannheim, Germany) and the Step One Plus
Detection System (Applied Biosystems, Life Technologies, Tokyo,
Japan). The Relative standard curve method (23) was used in this study and expression
levels were determined relative to that of 18s rRNA. Primers and
probes used in this study are shown in Table I.
 | Table I.Primer sequences and length of
specific polymerase chain reaction products. |
Table I.
Primer sequences and length of
specific polymerase chain reaction products.
| Gene |
Directiona | Primer
sequence | Probeb |
|---|
| Syvn1 | F |
5′-CTGGGTATCCTGGACTTCCTC-3′ | 89 |
|
| R |
5′-AAGCACCATGGTCATCAGAA-3′ |
|
| KAT1 | F |
5′-CAGAGCAGCGCTATTGTTTG-3′ | 81 |
|
| R |
5′-GCAGACAGTCTAGGCCAGAAA-3′ |
|
| KAT3 | F |
5′-TTACACGTGTGCGACTCCTT-3′ | 27 |
|
| R |
5′-GCTTGATATCGATCCAAAACG-3′ |
|
| KAT4 | F |
5′-ATGGCTGCTGCCTTTCAC-3′ | 17 |
|
| R |
5′-GATCTGGAGGTCCCATTTCA-3′ |
|
| 18sRNA | F |
5′-GCAATTATTCCCCATGAACG-3′ | 48 |
|
| R |
5′-GGGACTTAATCAACGCAAGC-3′ |
|
Plasmids, siRNA and antibodies
PGV-B2 (PicaGene Basic vector 2; Toyo Ink, Tokyo,
Japan) vector and pcDNA3 (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) vector were purchased. pcDNA3 hemagglutinin
antigen (HA) was constructed by inserting the HA sequence into
pcDNA3 (Invitrogen; Thermo Fisher Scientific, Inc.). The coding
sequence of human full-length NRF2 was PCR-amplified (primers,
5′-CAGTGTGCTGGAATTATGATGGACTTGGAGCTGCC-3′ and
5′-GATATCTGCAGAATTGTTTTTCTTAACATCTGGCTTCTTAC-3′) from HeLa cDNA and
full-length NRF2 was inserted into the pcDNA3 HA plasmid
(Invitrogen; Thermo Fisher Scientific, Inc.) for transient
transfection assays. The promoter of the KAT4 gene was
PCR-amplified (primers, 5′-AAAGCTAGCAAGCTTCATACTGTAAGC-3′ and
5′-AAACTCGAGAGAGCCGAGATCTGGGGAAG-3′) from the genome of C57BL/6J
mice and the fragment (−2553 to +30) was subcloned into PGV-B2
(PicaGene Basic vector 2; Toyo Ink). Plasmid sequences were
confirmed by sequencing. SYVN1 plasmids and siRNA against Syvn1
were previously described (9,13).
The following antibodies were used: Anti-tubulin (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany), and anti-HA (3F10; Roche
Diagnostics, Indianapolis, IN, USA), anti-KAT1 and KAT4 (Abcam,
Cambridge, UK), anti-KAT2 and KAT3 (Santa Cruz Biotechnology, Inc.,
Dallas, Texas, USA). The anti-Syvn1 rabbit polyclonal antibody that
was used was previously reported (18).
Cell culture and transient
transfection
A total of 293 cells and C2C12 cells were cultured
in Dulbecco's modified Eagle's medium as previously described
(9). Transient transfection was
performed with Lipofectamine 2000 according to the manufacturer's
protocol (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were
lysed with cell lysis buffer (Promega Corporation, Madison, WI,
USA) 24 h after transfection, and luciferase activity was measured.
To ensure equal amounts of DNA, empty plasmids were added to each
transfection.
Luciferase assay
The assay was performed as previously described
(24–26). Briefly, 293 cells were transiently
transfected with 50 ng KAT4-luc reporter plasmid, 0.1 ng pRL-CMV,
and 10, 50, 100 ng pcDNA3 HA-NRF2. For Syvn1 knockdown, 50 ng
KAT4-luc reporter plasmid, 0.1 ng pRL-CMV, and 10, 20 nM Syvn1
siRNA were transfected. For SYVN1 overexpression, 50 ng KAT4-luc
reporter plasmid, 0.1 ng pRL-CMV, and 10, 50, or 100 ng SYVN1
expression vector were transfected. 293 cells were transiently
transfected with 50 ng KAT4-luc reporter plasmid, 0.1 ng pRL-CMV,
and 50 ng pcDNA3 HA-NRF2 and/or 25 ng pcDNA3 HA PGC-1β. 293 cells
were transiently transfected with 50 ng KAT4-luc reporter plasmid,
0.1 ng pRL-CMV, and 50 ng pcDNA3 HA-NRF2 and/or 25 ng pcDNA3 HA
PGC-1α. After 24 h, cells were lysed with cell lysis buffer, which
was followed by measurement of luciferase activity. Each experiment
was performed at least three times.
RNA interference assay
siRNAs for Syvn1 were previously described (9). Transfection with siRNAs (20 µM) was
performed by using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Total
RNA from C2C12 cells was purified 3 d after transfection using
ISOGEN (Nippon Gene) according to the manufacturer's instructions,
and reverse transcribed using ReverTra Ace with random primers
(Toyobo).
Statistical analysis
All data are expressed as the means ± standard
deviation. Differences between two groups were examined by using
the Student's t-test and P<0.05 was considered to indicate a
statistically significant difference. One-way analysis of variance
with Tukey-Kramer post hoc analysis was used to determine
correlations in datasets containing multiple groups in luciferase
assays. All results were derived from at least three independent
experiments.
Results
Metabolome analysis of Syvn1-deficient
septic mice
To investigate the role of Syvn1 in sepsis, we
performed the metabolome analysis using brain tissue of WT and
heterozygous Syvn1-deficient mice because the homozygous mice die
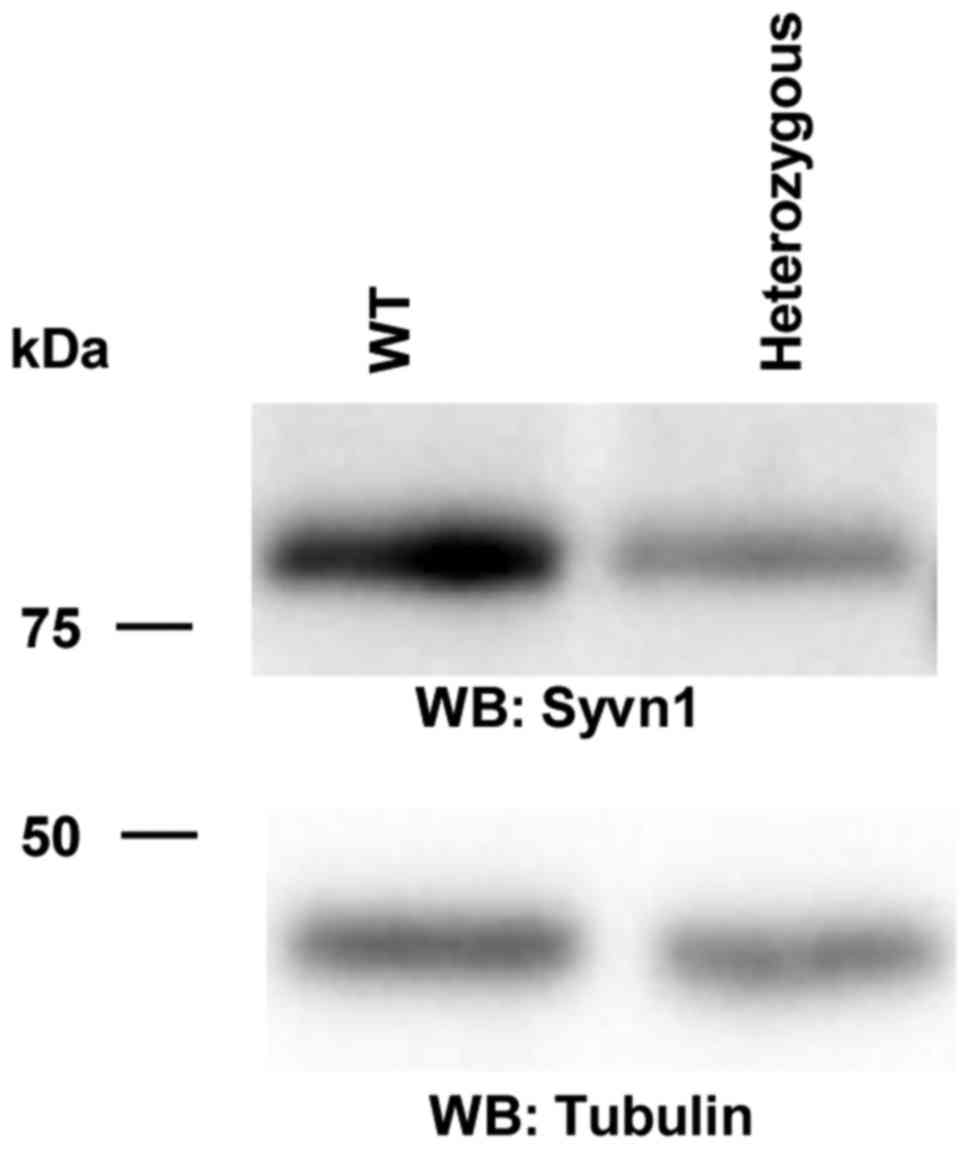
in utero. We first examined the expression of Syvn1 in the brain.
Western blotting showed that the expression of Syvn1 was decreased
in the heterozygous mice (Fig. 2).
As shown in Table II, KYN was not
detected in sham mice [WT (sham) and heterozygous Syvn1 mice
(sham)] and was increased in CLP-induced septic mice at 18 h [WT
(CLP)]. Interestingly, the quantity of KYN decreased in CLP-induced
septic Syvn1-deficient mice [heterozygous Syvn1 mice (CLP)]. This
result suggests that decreased expression of Syvn1 might induce KYN
metabolism.
 | Table II.Metabolome analysis. |
Table II.
Metabolome analysis.
|
| Case |
|---|
|
|
|
|---|
|
| WT (sham) | Heterozygous Syvn1
(sham) | WT (CLP) | Heterozygous Syvn1
(CLP) |
|---|
|
|
|
|
|
|
|---|
| Treatment | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 |
|---|
| Kynurenine
(nmol/g) | ND | ND | ND | ND |
1.1×10−04 |
3.1×10−04 | ND | ND |
Expression of KAT genes in Syvn1-KO
mice
The conversion of KYN to KYNA is catalyzed by KATs,
which have been detected in the brain and skeletal muscle.
Therefore, we examined the expression level of the KAT1-4
genes in the brain and the skeletal muscle tissue of tamoxifen
(Tam)-inducible Syvn1 knockout (KO) mice (the post-neonatal
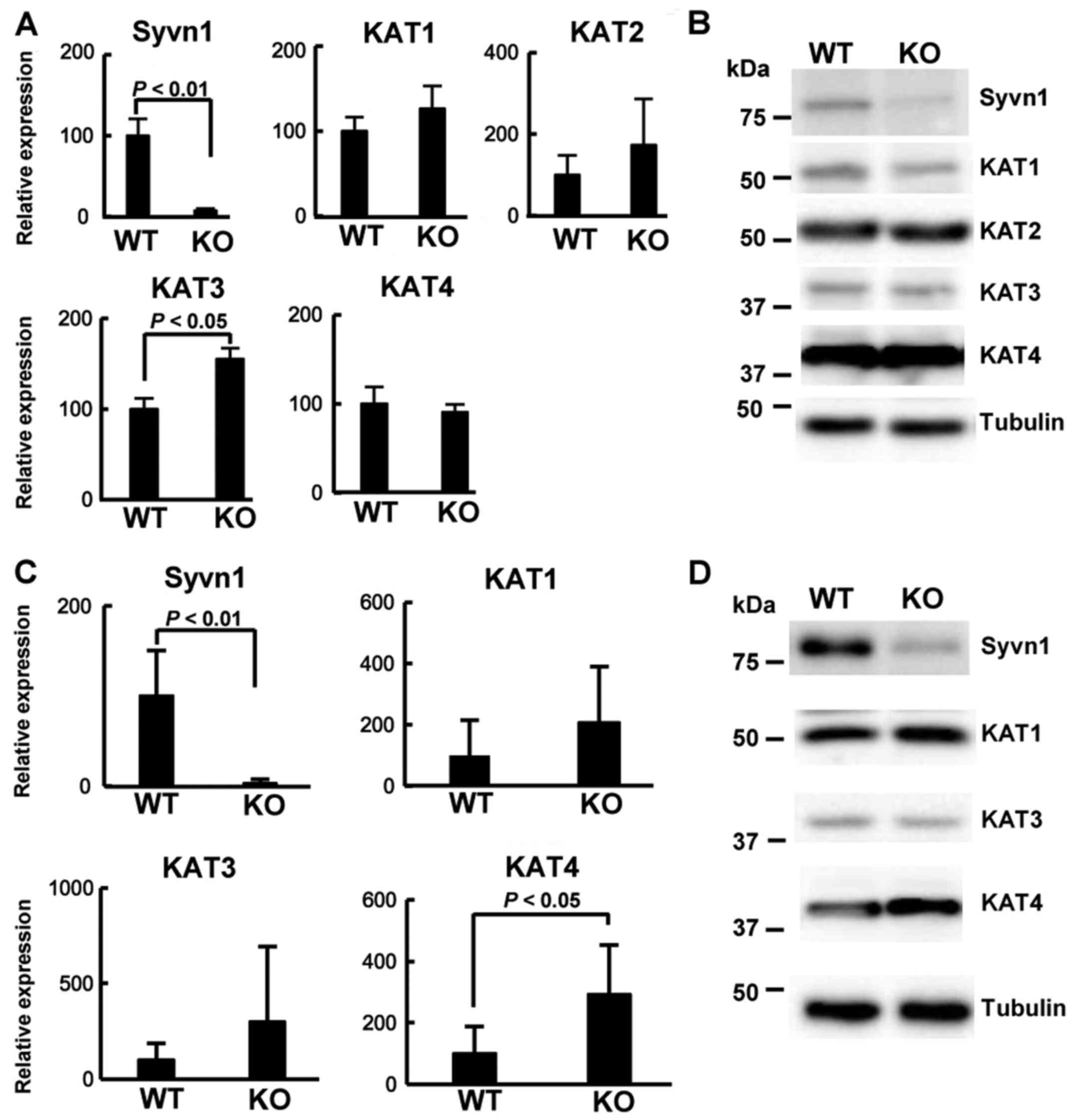
knockout mice) (9). Real-time PCR
assay showed that the expression of Syvn1 was very low in
the brain tissue of KO mice and that the expression of KAT3
was significantly increased in KO mice (Fig. 3A). To examine the protein level, we
performed western blotting using brain extracts from KO mice and WT
mice. As shown in Fig. 3B, the
expression of Syvn1 was very low. However, the expression level of
KAT1-4 was similar between WT mice and KO mice. We next
investigated the mRNA levels by real-time PCR assay. As shown in
Fig. 3C, the expression of
Syvn1 was very low in the skeletal muscle tissue of KO mice.
The expression of KAT4 was significantly increased in KO
mice compared to that in WT mice (P<0.05), and the expression of
KAT1 and KAT3 was similar between WT mice and KO
mice. KAT2 was not detected in the skeletal muscle tissue.
Western blotting using the skeletal muscle extracts revealed that
the expression of Syvn1 was very low and that the expression of
KAT4 was clearly increased in the skeletal muscle tissue of KO mice
(Fig. 3D). These results suggest
that KAT4 could be an important gene and Syvn1 might
regulate the expression of KAT4.
Syvn1 represses KAT4 expression
To examine the role of Syvn1 in KAT4
expression, we performed luciferase assay using the KAT4
promoter (−2553/+30) reporter constructs. We used 293 cells, which
have high transfection efficiency, because we have used 293 cells
as model cells for a long time to analyze transcriptional
regulation (9,25,27).
As shown in Fig. 4A, treatment
with a siRNA against Syvn1 (siSyvn1) induced the luciferase
reporter activity, 1.2-fold (10 µM) and 1.4-fold (20 µM), compared
to that with control siRNA (siControl). Overexpression of Syvn1
repressed the reporter activity in a dose-dependent manner
(Fig. 4B). To further examine
whether Syvn1 regulates the KAT4 expression, we performed
knock-down assays in C2C12 cells. C2C12 cells were treated with
control siRNA (siControl) or siRNA for Syvn1 (siSyvn1) for 3 d and
total RNA was purified. Then, we performed RT-PCR and real-time PCR
assays to measure the expression of KAT1-4. With siSyvn1
treatment, the expression of Syvn1 decreased to 40% that of control
levels. The expression of KAT4 was significantly increased
in cells treated with siSyvn1 compared to that in cells treated
with control siRNA (siControl). In addition, the expression of
KAT3 and KAT4 was not different between siSyvn1 and
siControl conditions (Fig.
4C).
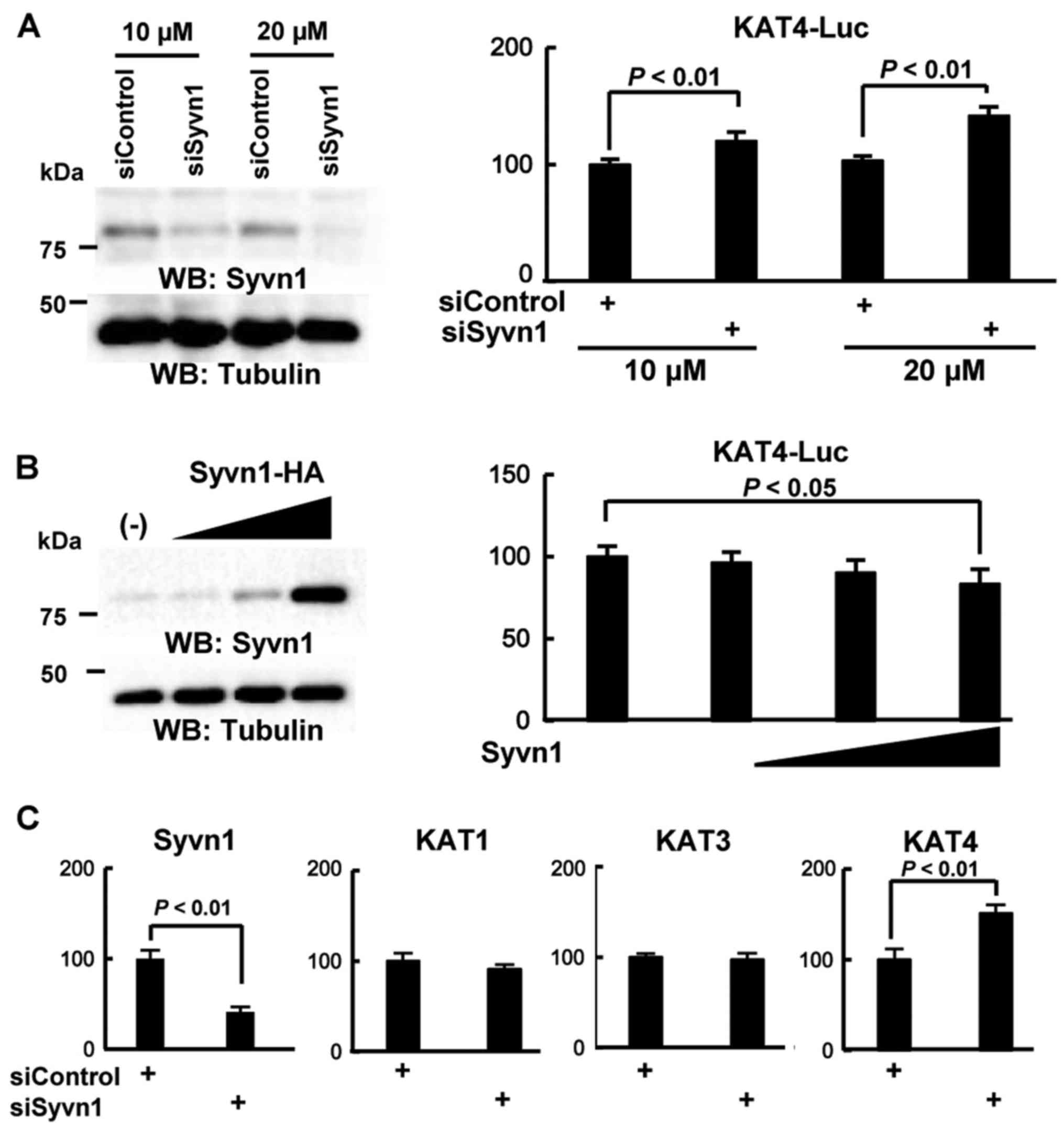 | Figure 4.Syvn1 represses KAT4 expression based
on KAT4 promoter-driven luciferase reporter assays. (A)
Effect of Syvn1 knockdown by siRNA. 293 cells were transiently
transfected with a reporter plasmid containing the KAT4 promoter,
pRL-CMV, control siRNA (siControl) or siRNA for Syvn1
(siSyvn1). Data were analyzed by a Student's t-test and expressed
as mean ± SD. Western blotting was performed with anti-Syvn1 and
anti-tubulin antibodies. (B) Effect of Syvn1 overexpression. 293
cells were transiently transfected with a reporter plasmid
containing the KAT4 promoter, pRL-CMV, and the HA-tagged Syvn1
(Syvn1-HA)-expression vector (10, 50, and 100 ng). Analysis of
variance with Tukey-Kramer post hoc analysis and expressed as mean
± SD. Western blotting was performed with anti-Syvn1 and
anti-tubulin antibodies. (C) Effect of Syvn1 on KAT4 expression.
C2C12 cells were transiently transfected with siControl or siSyvn1.
After 3 days, total RNA were purified and qPCR was performed.
Individual measurements were standardized using 18S RNA, and the
average for siControl was set to 100. Data were analyzed by
performing a Student's t-test and expressed as mean ± SD. The
experiment was performed three times. KAT, kynurenine
aminotransferase; Syvn1, synoviolin; HA, hemagglutinin antigen. |
NRF2 and PGC-1β activate the KAT4
promoter
We previously demonstrated that Syvn1 regulates the
transcriptional factor, NRF2, and the transcriptional coactivator,
PGC-1β, which activates NRF2- and PPAR-mediated transcription
(12). Therefore, we analyzed the
transcriptional factor binding sites of the above factors in the
KAT4 promoter (−2553/+30) using TF BIND (http://tfbind.hgc.jp/). As shown in Fig. 5A, one PPAR responsive element (PRE)
and 17 NRF2 binding sites were detected in the KAT4
promoter. To examine the role of NRF2 and PGC-1β, we performed
luciferase assays using the KAT4 promoter (−2553/+30). NRF2
significantly activated the luciferase reporter activity in a
dose-dependent manner (Fig. 5B),
whereas PPAR-α did not induce the reporter activity (data not
shown). As shown in Fig. 5C, NRF2
and PGC-1β synergistically activated the reporter activity. PGC-1α
also induced reporter activity, and NRF2 and PGC-1α additively
activated the reporter activity (Fig.
5D). In addition, NRF2 rescued the repressive effect induced by
overexpression of Syvn1 in a dose-dependent manner (Fig. 5E). These results suggest that the
NRF2-PGC-1β pathway induces the expression of KAT4.
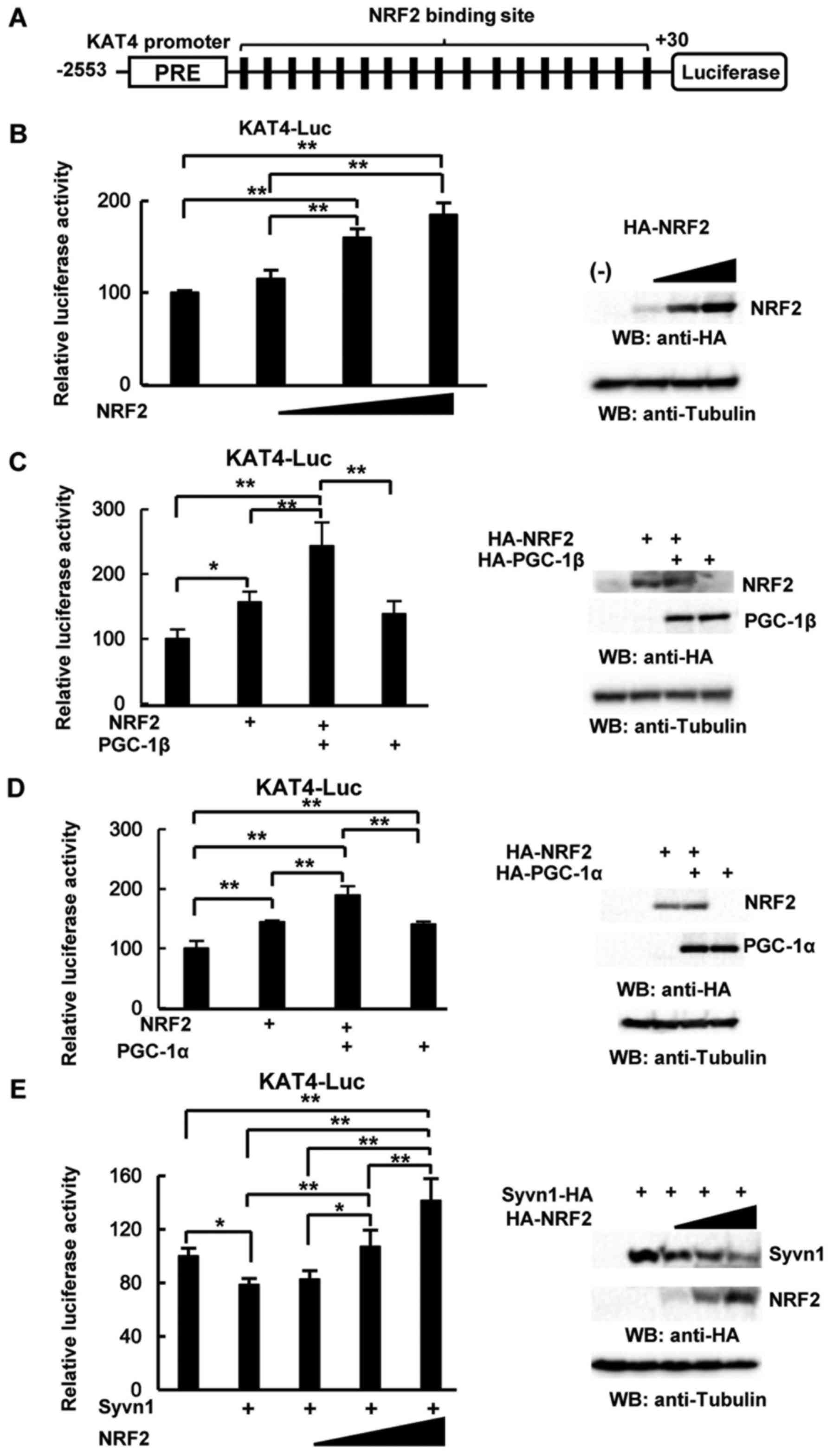 | Figure 5.The NRF2/PGC-1β pathway activates the
KAT4 promoter. (A) Schematic representation of the
KAT4 promoter (−2553/+30). PRE: PPAR responsive element,
black box: NRF2 binding site (−2123/-2114, −2029/-2020,
−1981/-1972, −1957/-1948, −1742/-1733, −1732/-1723, −1459/-1450,
−1386/-1377, −1378/-1369, −1308/-1299, −1106/-1097, −759/-750,
−359/-350, −250/-241, −105/-96, −46/-37, and +2/+11). (B) Effect of
NRF2 overexpression on KAT4 promoter-driven luciferase
reporter activity. 293 cells were transiently transfected with a
reporter plasmid containing the KAT4 promoter, pRL-CMV, and the
HA-NRF2-expression vector (10, 50, and 100 ng). Western blotting
was performed with an anti-HA and anti-tubulin antibodies. (C)
Effect of NRF2 and PGC-1β overexpression on the KAT4
promoter-driven luciferase reporter activity. 293 cells were
transiently transfected with a reporter plasmid containing the KAT4
promoter, pRL-CMV, 50 ng of the HA-NRF2-expression vector, and 25
ng of the HA-PGC-1β-expression vector. Western blotting was
performed with an anti-HA and anti-tubulin antibodies. (D) Effect
of NRF2 and PGC-1α overexpression on KAT4 promoter-driven
luciferase reporter activity. A total of 293 cells were transiently
transfected with a reporter plasmid containing the KAT4 promoter,
pRL-CMV, 50 ng of the HA-NRF2-expression vector, and 25 ng of the
HA-PGC-1α-expression vector. Western blotting was performed with an
anti-HA and anti-tubulin antibodies. (E) NRF2 overexpression
abrogates the effect of Syvn1 overexpression on KAT4
promoter-driven luciferase reporter activity. 293 cells were
transiently transfected with a reporter plasmid containing the KAT4
promoter, pRL-CMV, 100 ng of the Syvn1-HA-expression vector, and
the HA-NRF2-expression vector (10, 50, and 100 ng). Western
blotting was performed with an anti-HA and anti-tubulin antibodies.
(B-D) Data were analyzed by performing a Tukey-Kramer post hoc
analysis and expressed as mean ± SD (*P<0.05, **P<0.01). The
experiment was performed three times. KAT, kynurenine
aminotransferase; Syvn1, synoviolin; HA, hemagglutinin antigen;
PGC, proliferator-activated receptor (PPAR)-PPAR coactivator. |
Discussion
In the present study, we investigated the role of
Syvn1 in a mouse model of sepsis. We showed that the level of KYN
was elevated in the brain tissue of septic WT mice. However, KYN
was not detected in septic Syvn1-deficient mice. In addition,
expression of KAT4, which encodes one of the enzymes that
converts KYN into KYNA, was elevated in the skeletal muscle tissue
of Syvn1-KO mice. Moreover, Syvn1 knockdown induced KAT4
promoter-driven reporter activity, whereas overexpression of Syvn1
repressed this effect. The KAT4 promoter was also activated
by the NRF2-PGC-1β pathway. NRF2 and PGC-1β are target proteins of
Syvn1-induced degradation (9,17).
Taken together, these results suggest that Syvn1 deficiency might
induce KYN metabolism via the NRF2-PGC-1β-KAT4 pathway.
KATs are critical enzymes that catalyze the
conversion of KYN to KYNA. Recent studies indicates that exercise
induces the expression of KATs expression in the skeletal muscle
(7,8,28).
Analysis of skeletal muscle-specific PGC-1α-transgenic mice
revealed that an increased expression of KATs in skeletal muscle
shifts the KYN metabolism towards enhanced synthesis of KYNA,
resulting in a decrease in KYN levels, and thereby protecting the
tissue from stress-induced damage. This novel function of PGC-1α in
the regulation of the KYN metabolism suggests communication between
the skeletal muscle and brain. In the present study, we showed that
KAT4 expression was significantly increased in the skeletal
muscle tissue of Syvn1-deficient mice and that the NRF2-PGC-1β
pathway activates the KAT4 promoter. We previously
demonstrated that Syvn1 interacts with NRF2 and PGC-1β, and induces
the degradation of these factors through ubiquitination. Therefore,
in Syvn1-deficient mice, accumulated NRF2 and PGC-1β could activate
KAT4 expression in the skeletal muscle to enhance the
synthesis of KYNA, resulting in decreased KYN in the brain
tissue.
Syvn1 is involved in both acute and chronic
inflammation. We previously demonstrated that Syvn1 overexpression
in transgenic mice led to advanced arthropathy through the
suppression of apoptosis in synoviocytes and that heterozygous
Syvn1-mice were resistant to arthritis and fibrosis (13,15).
In addition, we recently showed that Syvn1 is involved in the
development of obesity, limb girdle muscular dystrophy, and liver
cirrhosis. These studies indicate that Syvn1 is a key factor for
diseases associated with chronic inflammation. In the present
study, we examined the role of Syvn1 in a CLP-induced septic mouse
model, which is an example of acute inflammation. Proinflammatory
cytokines such as tumor necrosis factor (TNF)-α and interleukin
(IL)-1β are induced in the mouse sepsis model and septic patients
(5,29,30).
Previous studies demonstrated that Syvn1 is a key target for
inflammatory cytokines such as TNF-α, IL-1, and IL-17 (5,31,32),
and that the expression of Syvn1 is transcriptionally regulated by
Ets transcription factors, GABPα, GABPβ, and ILF-3 (33,34),
which are the downstream of inflammatory cytokines signaling. These
results prompted us to speculate that Syvn1 expression might be
induced by inflammatory cytokines during sepsis and induce the
degradation of NRF2 and PGC-1β in the skeletal muscle tissue.
Further studies are warranted to unveil the detailed role of Syvn1
in sepsis.
KYN is a key immune mediator. During inflammation,
increases in cytokines such as interferon (IFN)-γ, TNF-α, and IL-1β
activate indoleamine 2,3-dioxygenase (IDO), and tryptophan is
metabolized into the toxic metabolite KYN via IDO. Several studies
indicate that the development of mood symptoms with inflammation is
associated with reduced circulating tryptophan levels and
concomitant increases in serum levels of KYN (35,36).
In addition, recent studies show that KYN and proinflammatory
cytokines such as TNF-α and IL-1β are induced in mouse sepsis
models and human septic patients (5,29,30).
Therefore, the KYN pathway is one of targets for the treatment of
SAE. The IDO inhibitor, 1-methyl-D, was shown to attenuate
neuroinflammation through the repression of proinflammatory
cytokines and KYN, and protect against sepsis-induced cognitive
impairment in a mouse model (5).
In the present study, we showed that the induction of KYN by CLP is
not observed in Syvn1-deficient mice. We previously developed a
Syvn1 inhibitor, LS-102, and demonstrated that the Syvn1 inhibitor
attenuated arthritis, fibrosis, obesity, limb girdle muscular
dystrophy, and liver cirrhosis (13–17).
Although further studies are needed to investigate the effect of
this Syvn1 inhibitor on SAE, Syvn1 might represent a novel target
for the treatment of SAE.
Taken together, in the present study, we provide
evidence that Syvn1 regulates the NRF2-PGC-1β-KAT4 pathway in a
mouse model of sepsis. Further analysis of Syvn1 will be helpful to
understand its physiological and clinical significance.
Acknowledgements
The authors thank Mr. Shyouichiro Shibata (Institute
of Medical Science, Tokyo Medical University) for technical
assistance. We also thank all members of Professor Toshihiro
Nakajima's laboratory (Institute of Medical Science, Tokyo Medical
University).
Funding
This study was funded in part by grants from the
Naito Foundation, Natural Science Scholarship Daiichi-Sankyo
Foundation of Life Science, Mitsubishi, Tanabe Pharma Corporation,
Bureau of Social Welfare and Public Health, Academic contribution
of Pfizer, Eisai, Santen Pharmaceutical, Abbvie, Astellas, Takeda
Science Foundation, AstraZeneca (R&D Grant 2013), and ONO
Medical Research Foundation. This study was supported in part by
funds provided through a MEXT-Supported program of the Strategic
Research Foundation at Private Universities (grant no. S1411011,
2014-2018) from the Ministry of Education, Culture, Sports, Science
and Technology of Japan. This study was also supported in part by
the Japan Society for the Promotion of Science KAKENHI (grant nos.
23659176, 26670479, 26461478 and 16H05157) and Industry-university
cooperation (BioMimetics Sympathies Inc.).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YI, HF, HU and TN conceived the project and designed
the experiments. YI, MC, NT, YO, NU, FN and HU performed the sepsis
model and analyzed the metabolome data. YI, HF, SA, MY and TN
performed experiments and analyzed data. YI, HF and TN wrote the
manuscript. YI, HF, SA, MC, NT, MY, YO, NU, FN, HU and TN discussed
the results and commented on the manuscript.
Ethics approval and consent to
participate
All procedures involving animals were performed in
accordance with institutional and national guidelines for animal
experimentation, and were approved by the Institutional Animal Care
and Use Committee of Tokyo Medical University (approval no.
S-28044).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Syvn1
|
synoviolin
|
|
SAE
|
sepsis-associated encephalopathy
|
|
CLP
|
cecal ligation/perforation
|
|
KYN
|
kynurenine
|
|
KYNA
|
kynurenic acid
|
|
KATs
|
kynurenine aminotransferases
|
|
PGC-1β
|
Peroxisome proliferator-activated
receptor coactivator 1β
|
|
QA
|
quinolinic acid
|
|
3-HAA
|
3-hydroxykynurenine
|
|
SPF
|
specific pathogen-free
|
References
|
1
|
Sprung CL, Peduzzi PN, Shatney CH, Schein
RM, Wilson MF, Sheagren JN and Hinshaw LB: Impact of encephalopathy
on mortality in the sepsis syndrome. The Veterans Administration
Systemic Sepsis Cooperative Study Group. Crit Care Med. 18:801–806.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eidelman LA, Putterman D, Putterman C and
Sprung CL: The spectrum of septic encephalopathy. Definitions,
etiologies, and mortalities. JAMA. 275:470–473. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zwilling D, Huang SY, Sathyasaikumar KV,
Notarangelo FM, Guidetti P, Wu HQ, Lee J, Truong J,
Andrews-Zwilling Y, Hsieh EW, et al: Kynurenine 3-monooxygenase
inhibition in blood ameliorates neurodegeneration. Cell.
145:863–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stone TW and Darlington LG: The kynurenine
pathway as a therapeutic target in cognitive and neurodegenerative
disorders. Br J Pharmacol. 169:1211–1227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao R, Kan MQ, Wang SG, Yang RH and Zhang
SG: Disrupted tryptophan metabolism induced cognitive impairment in
a mouse model of sepsis-associated encephalopathy. Inflammation.
39:550–560. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moroni F, Carpenedo R, Cozzi A, Meli E,
Chiarugi A and Pellegrini-Giampietro DE: Studies on the
neuroprotective action of kynurenine mono-oxygenase inhibitors in
post-ischemic brain damage. Adv Exp Med Biol. 527:127–136. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Agudelo LZ, Femenia T, Orhan F,
Porsmyr-Palmertz M, Goiny M, Martinez-Redondo V, Correia JC, Izadi
M, Bhat M, Schuppe-Koistinen I, et al: Skeletal muscle PGC-1α1
modulates kynurenine metabolism and mediates resilience to
stress-induced depression. Cell. 159:33–45. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ruas JL, White JP, Rao RR, Kleiner S,
Brannan KT, Harrison BC, Greene NP, Wu J, Estall JL, Irving BA, et
al: A PGC-1α isoform induced by resistance training regulates
skeletal muscle hypertrophy. Cell. 151:1319–1331. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fujita H, Yagishita N, Aratani S,
Saito-Fujita T, Morota S, Yamano Y, Hansson MJ, Inazu M, Kokuba H,
Sudo K, et al: The E3 ligase synoviolin controls body weight and
mitochondrial biogenesis through negative regulation of PGC-1β.
EMBO J. 34:1042–1055. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kressler D, Schreiber SN, Knutti D and
Kralli A: The PGC-1-related protein PERC is a selective coactivator
of estrogen receptor alpha. J Biol Chem. 277:13918–13925. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin J, Puigserver P, Donovan J, Tarr P and
Spiegelman BM: Peroxisome proliferator-activated receptor gamma
coactivator 1beta (PGC-1beta), A novel PGC-1-related transcription
coactivator associated with host cell factor. J Biol Chem.
277:1645–1648. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Scarpulla RC: Nuclear control of
respiratory gene expression in mammalian cells. J Cell Biochem.
97:673–683. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amano T, Yamasaki S, Yagishita N,
Tsuchimochi K, Shin H, Kawahara K, Aratani S, Fujita H, Zhang L,
Ikeda R, et al: Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel
pathogenic factor for arthropathy. Genes Dev. 17:2436–2449. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bianchini E, Fanin M, Mamchaoui K, Betto R
and Sandonà D: Unveiling the degradative route of the V247M
α-sarcoglycan mutant responsible for LGMD-2D. Hum Mol Genet.
23:3746–3758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hasegawa D, Fujii R, Yagishita N,
Matsumoto N, Aratani S, Izumi T, Azakami K, Nakazawa M, Fujita H,
Sato T, et al: E3 ubiquitin ligase synoviolin is involved in liver
fibrogenesis. PLoS One. 5:e135902010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Shen Y, Ding Y, Liu Y, Su D and
Liang X: Hrd1 participates in the regulation of collagen I
synthesis in renal fibrosis. Mol Cell Biochem. 386:35–44. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu T, Zhao F, Gao B, Tan C, Yagishita N,
Nakajima T, Wong PK, Chapman E, Fang D and Zhang DD: Hrd1
suppresses Nrf2-mediated cellular protection during liver
cirrhosis. Genes Dev. 28:708–722. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamasaki S, Yagishita N, Sasaki T,
Nakazawa M, Kato Y, Yamadera T, Bae E, Toriyama S, Ikeda R, Zhang
L, et al: Cytoplasmic destruction of p53 by the endoplasmic
reticulum-resident ubiquitin ligase 'Synoviolin'. EMBO J.
26:113–122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Buendia I, Michalska P, Navarro E, Gameiro
I, Egea J and León R: Nrf2-ARE pathway: An emerging target against
oxidative stress and neuroinflammation in neurodegenerative
diseases. Pharmacol Ther. 157:84–104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen PC, Vargas MR, Pani AK, Smeyne RJ,
Johnson DA, Kan YW and Johnson JA: Nrf2-mediated neuroprotection in
the MPTP mouse model of Parkinson's disease: Critical role for the
astrocyte. Proc Natl Acad Sci USA. 106:2933–2938. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Innamorato NG, Rojo AI, García-Yagüe AJ,
Yamamoto M, de Ceballos ML and Cuadrado A: The transcription factor
Nrf2 is a therapeutic target against brain inflammation. J Immunol.
181:680–689. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rittirsch D, Huber-Lang MS, Flierl MA and
Ward PA: Immunodesign of experimental sepsis by cecal ligation and
puncture. Nat Protoc. 4:31–36. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak K: ABI Prism 7700 Sequence Detection
System, User Bulletin 2PE Applied Biosystems. Foster City, CA:
1997
|
|
24
|
Chakravarti D, LaMorte VJ, Nelson MC,
Nakajima T, Schulman IG, Juguilon H, Montminy M and Evans RM: Role
of CBP/P300 in nuclear receptor signalling. Nature. 383:99–103.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fujita H, Fujii R, Aratani S, Amano T,
Fukamizu A and Nakajima T: Antithetic effects of MBD2a on gene
regulation. Mol Cell Biol. 23:2645–2657. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakajima T, Fukamizu A, Takahashi J, Gage
FH, Fisher T, Blenis J and Montminy MR: The signal-dependent
coactivator CBP is a nuclear target for pp90RSK. Cell. 86:465–474.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fujita H, Ohshima T, Oishi T, Aratani S,
Fujii R, Fukamizu A and Nakajima T: Relevance of nuclear
localization and functions of RNA helicase A. Int J Mol Med.
15:555–560. 2005.PubMed/NCBI
|
|
28
|
Schlittler M, Goiny M, Agudelo LZ,
Venckunas T, Brazaitis M, Skurvydas A, Kamandulis S, Ruas JL,
Erhardt S, Westerblad H and Andersson DC: Endurance exercise
increases skeletal muscle kynurenine aminotransferases and plasma
kynurenic acid in humans. Am J Physiol Cell Physiol. 310:C836–C840.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schäfer ST, Franken L, Adamzik M, Schumak
B, Scherag A, Engler A, Schönborn N, Walden J, Koch S, Baba HA, et
al: Mitochondrial DNA: An endogenous trigger for immune paralysis.
Anesthesiology. 124:923–933. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Darcy CJ, Davis JS, Woodberry T, McNeil
YR, Stephens DP, Yeo TW and Anstey NM: An observational cohort
study of the kynurenine to tryptophan ratio in sepsis: Association
with impaired immune and microvascular function. PLoS One.
6:e211852011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Toh ML, Gonzales G, Koenders MI, Tournadre
A, Boyle D, Lubberts E, Zhou Y, Firestein GS, van den Berg WB and
Miossec P: Role of interleukin 17 in arthritis chronicity through
survival of synoviocytes via regulation of synoviolin expression.
PLoS One. 5:e134162010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Toh ML, Marotte H, Blond JL, Jhumka U,
Eljaafari A, Mougin B and Miossec P: Overexpression of synoviolin
in peripheral blood and synoviocytes from rheumatoid arthritis
patients and continued elevation in nonresponders to infliximab
treatment. Arthritis Rheum. 54:2109–2118. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Izumi T, Fujii R, Izumi T, Nakazawa M,
Yagishita N, Tsuchimochi K, Yamano Y, Sato T, Fujita H, Aratani S,
et al: Activation of synoviolin promoter in rheumatoid synovial
cells by a novel transcription complex of interleukin enhancer
binding factor 3 and GA binding protein alpha. Arthritis Rheum.
60:63–72. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tsuchimochi K, Yagishita N, Yamasaki S,
Amano T, Kato Y, Kawahara K, Aratani S, Fujita H, Ji F, Sugiura A,
et al: Identification of a crucial site for synoviolin expression.
Mol Cell Biol. 25:7344–7356. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Heisler JM and O'Connor JC: Indoleamine
2,3-dioxygenase-dependent neurotoxic kynurenine metabolism mediates
inflammation-induced deficit in recognition memory. Brain Behav
Immun. 50:115–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Maes M, Leonard BE, Myint AM, Kubera M and
Verkerk R: The new ‘5-HT’ hypothesis of depression: Cell-mediated
immune activation induces indoleamine 2,3-dioxygenase, which leads
to lower plasma tryptophan and an increased synthesis of
detrimental tryptophan catabolites (TRYCATs), both of which
contribute to the onset of depression. Prog Neuropsychopharmacol
Biol Psychiatry. 35:702–721. 2011. View Article : Google Scholar : PubMed/NCBI
|