Introduction
In recent years, obesity has been revealed to be
closely associated with metabolic abnormalities, which represents a
risk factor for the development of atherosclerosis (AS),
cardiovascular disease, cancer and other diseases (1). Metabolically healthy but obese (MHO)
is an obesity subgroup, which is characterized by obesity and high
insulin sensitivity, and accounts for 20–30% of patients with
obesity worldwide (2,3). Similarly, MHO may cause various
vascular diseases, including AS, cerebral infarction and large
artery embolism, which are induced by dysfunction of the vascular
endothelium (4,5).
AS is a cardiovascular disease, which exhibits a
relatively high incidence rate and may subsequently induce arterial
thrombosis in acute coronary syndromes, strokes and various other
diseases, which may pose a threat to human mortality (6). According to a previous study, the
pathogenesis of AS is highly complex (7). It has been widely established that
endothelial dysfunction is an important factor in the early stage
of AS (8). Endothelial dysfunction
results in functional cell alterations, and may be characterized by
the suppressed release of nitric oxide (NO) and NO bioavailability,
in addition to the enhanced expression of adhesion molecules and
chemokines (9). Interaction
between of these alterations and smooth muscle cells located in
blood vessels in turn alters vascular function and structure, which
ultimately leads to AS (8,9). Therefore, attenuation of endothelial
dysfunction may reduce the risk of the development of AS.
Krüppel-like factors (KLFs) are a class of zinc
finger DNA-binding transcription proteins, which are involved in
numerous pathophysiological processes, including cell
differentiation, apoptosis and tumor formation (10–12).
KLFs are closely associated with cardiovascular diseases, including
hypertension, AS and coronary heart disease (10–12).
KLF15 is a member of the zinc finger protein family (13). It has been previously demonstrated
that KLF15 is expressed in the heart, liver, kidney and numerous
other organs (14,15). Furthermore, KLF15 is involved in
the pathological processes of nephropathy, abnormal glucose
metabolism and myocardial injury (16,17).
However, the function of KLF15 in vascular endothelial dysfunction
remains unclear. Nuclear factor (NF)-κB is a transcription factor
that is highly expressed in mammals and highly conserved among
mammalian species (18).
Initially, NF-κB was considered to be a homologous/heterogeneous
dimer composed of Katanin p60 ATPase-containing subunit A1 and
transcription factor p65 (p65) subunits (18); however, subsequent studies have
revealed that there is an NF-κB protein family, which consists of
several polypeptides with a high degree of homology (18,19).
Abnormal activation of NF-κB may cause rheumatoid arthritis, AS,
inflammation and tumor formation (20). Furthermore, previous studies have
demonstrated that nuclear factor erythroid 2-related factor 2
(Nrf2) signaling may inhibit the activation of NF-κB during
inflammation (21,22). In addition, NF-κB and Nrf2 always
interact during oxidative stress and numerous inflammatory
responses (23).
In the present study, the function of KLF15 in
TNF-α-induced vascular endothelial dysfunction was investigated, in
addition to whether its underlying molecular mechanisms are
involved in the regulation of the NF-κB and Nrf2 signaling
pathways.
Materials and methods
Cell culture and treatment
The human umbilical vein fusion cell line (Eahy926)
was obtained from Shanghai Fuhengbio Biotechnology Co., Ltd.
(Shanghai, China). Cells were maintained in RPMI-1640 (Beijing Hua
Yueyang Biotechnology Co., Ltd., Beijing, China) supplemented with
10% fetal bovine serum (Jiangsu Enmoasai Biological Technology Co.,
Ltd., Changzhou, China) and 1% penicillin-streptomycin (Shanghai
Yuanmu Biotechnology Co., Ltd., Shanghai, China), in a 37°C
humidified incubator (MG80; Shanghai LNB Instruments Co., Ltd,
Shanghai China) with 5% CO2. Following culture, Eahy926
cells were treated with PBS (Control) or different concentrations
of TNF-α (1, 5, 10 and 20 ng/ml) in 37°C incubator for 48 h.
Cell transfection
Human pTA2-KLF15 (targeting sequence:
5′-ACAGAGACGTTGTGCTGCTTT-3′) and negative control pTA2 vectors were
synthesized by Genewiz Biotechnology Co., Ltd. (Suzhou, China).
Eahy926 cells were incubated in 6-well plates (2.5×104
cell/well) for 12 h at 37°C and, once 40–60% confluence had been
reached, the cells were transfected with the aforementioned vectors
(30 nM) using Lipofectamine™ 2000 transfection reagent
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). After 48 h of
transfection, cells were retained for subsequent experiments.
Transfection efficiency was evaluated by RT-qPCR and western blot
analysis.
MTT analysis
The viability of Eahy926 cells was investigated
using an MTT kit (Gefan, Shanghai, China), according to the
manufacturer's protocol. Firstly, Eahy926 cells were incubated in
96-well plates (2×103 cell/well) for 24 h and subsequently treated
with 0.1% PBS (control group), pTA2 vector (NC group), pTA2-KLF15
vector (KLF15 group), 10 ng/ml TNF-α (TNF-α group), pTA2 vector and
10 ng/ml TNF-α (TNF-α + NC group), and pTA2-KLF15 vector and 10
ng/ml TNF-α (TNF-α + KLF15 group). Following this, 15 µl MTT
solution was added to each well and the plates were subsequently
incubated at 37°C for 6 h. Following this, 0.2% dimethyl sulfoxide
was added to the cells and subsequently agitated using an for 15
sec at room temperature. Finally, the optical density (OD) values
were determined at 450 nm using a microplate reader (cat. no.
HBS-1096A; Nanjing Detie Experimental Equipment Co., Ltd., Nanjing,
China).
NO analysis
The quantity of NO released by Eahy926 cells was
determined using a NO detection kit (Beyotime Institute of
Biotechnology, Shanghai, China), according to the manufacturer's
protocol. Firstly, cells were digested using trypsin (Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China),
resuspended in RPMI-1640 medium, and the supernatant was
subsequently added to 96-well plates. Following this, 5 µl NADPH,
10 µl flavine adenine dinucleotide (FAD) and 5 µl nitrate reductase
was added to each well and subsequently incubated at room
temperature for 40 min. Following this, LDH was added to the plates
in a dropwise manner at 37°C for 18 min. Subsequently, cells were
fixed using Griess reagent I/II at room temperature for 10 min.
Finally, OD values were determined at 540 nm using a microplate
reader.
Cell adhesion analysis
The adhesive ability of Eahy926 cells was
investigated using a cell adhesion detection kit (BestBio Company,
Shanghai, China), according to the manufacturer's protocol.
Firstly, coating liquid (BestBio Company) was added into 96-well
plates in a drop-wise manner. Following this, plates were
transferred to a 4°C refrigerator (cat. no. BCD-216SDN; Qingdao
Haier, Co., Ltd., Qingdao, China) for 24 h. Cells were then
digested using trypsin, resuspended in RPMI-1640 medium and
subsequently incubated in 96-well plates (2×103 cells/well) in a
37°C incubator for 30 min. Following this, 20 µl staining solution
B (BestBio Company) was added to cells for 2 h. Finally, OD values
were determined at 450 nm using a microplate reader, and cells were
photographed under an inverted fluorescent microscope (×200; cat.
no. GMSP-5; Guangmi, Shanghai, China). The adhesion rate was
calculated using the following formula: Adhesion rate
(%)=[(ODtest-ODblank)/(ODcontrol-ODblank)]x100.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated and lysed using an RNA
extraction kit (BioTeke Corporation, Beijing, China). cDNA was
subsequently synthesized using a RevertAid™ cDNA
Synthesis kit (Thermo Fisher Scientific, Inc.). The temperature
protocol used for RT was as follows: 42°C for 60 min and 70°C for 5
min. Following this, qPCR was performed using a DreamTaq Green PCR
master Mix kit (Thermo Fisher Scientific, Inc.), which included the
following reagents: 25 µl Dream Taq Green PCR master Mix, 1 µl
forward primer, 1 µl reverse primer, 19 µl nuclease-free distilled
water, 4 µl cDNA, total volume 50 µl. The thermocycling conditions
used for qPCR were as follows: Initial degeneration at 94°C for 5
min; followed by 30 cycles of denaturation at 94°C for 30 sec and
annealing at 65°C for 30 sec; and a final extension at 72°C for 30
sec. Primer sequences used for qPCR are presented in Table I. β-actin was used as an internal
control. The 2−ΔΔCq method was used to determine gene
expression (24).
 | Table I.Sequences of primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Sequences of primers used for reverse
transcription-quantitative polymerase chain reaction.
| Primer name | Sequence,
5′→3′ | Product size,
bp |
|---|
| eNOS forward |
GCTAGCCAAAGTCACCATCG | 230 |
| eNOS reverse |
TGGAAAACAGGAGTGAGGCT |
|
| KLF15 forward |
TAGTCAACATCCAGGGGCAG | 212 |
| KLF15 reverse |
GGAAACTTCTGGCCCACAAG |
|
| MCP-1 forward |
AGCCACCTTCATTCCCCAAG | 192 |
| MCP-1 reverse |
GGGTCAGCACAGATCTCCTT |
|
| ICAM-1 forward |
CAGTCACCTATGGCAACGAC | 238 |
| ICAM-1 reverse |
GCAGCTTACAGTGACAGAGC |
|
| TGF-β1 forward |
TACAGCAACAATTCCTGGCG | 200 |
| TGF-β1 reverse |
GTGAACCCGTTGATGTCCA |
|
| p65 forward |
GAGGAGCACAGATACCACCA | 220 |
| p65 reverse |
AGCCTCATAGAAGCCATCCC |
|
| Nrf2 forward |
TGAGCCCAGTATCAGCAACA | 171 |
| Nrf2 reverse |
AGTGAAATGCCGGAGTCAGA |
|
| β-actin
forward |
CACCATGTACCCAGGCATTG | 180 |
| β-actin
reverse |
TCGTACTCCTGCTTGCTGAT |
|
Western blot analysis
Total proteins were isolated and lysed with
radioimmunoprecipitation assay buffer (cat. no. 20101ES60; Shanghai
Shengsheng Biotechnology Co., Ltd., Beijing, China), and the total
protein concentration was determined using a Bradford protein
detection kit (Yeasen, Beijing, China). Firstly, proteins (30
µg/lane) were separated via 12% SDS-PAGE analysis and then
transferred to a polyvinylidene difluoride membrane. Subsequently,
membranes were blocked with 5% non-fat milk at 37°C for 1 h. The
membrane was then incubated with anti-KLF15 (cat. no. ab2647;
1:1,000; Abcam, Cambridge, UK), anti-endothelial nitric oxide
synthase (eNOS; cat. no. AF950; 1:700; R&D Systems, Inc.,
Minneapolis, MN, USA), anti-monocyte chemoattractant protein-1
(MCP-1; cat. no. MAB679, 1:1,000; R&D Systems, Inc.),
anti-intercellular adhesion molecule-1 (ICAM-1; cat. no. BBA3;
1:800; R&D Systems, Inc.), anti-transforming growth factor-β1
(TGF-β1; cat. no. MAB1835; 1:1,200; R&D Systems, Inc.),
anti-phosphorylated (p)-p65 (cat. no. MAB7226; 1:1,000; R&D
Systems, Inc.), anti-Nrf2 (cat. no. MAB3925; 1:1,000; R&D
Systems, Inc.) and anti-β-actin (cat. no. MAB8969; 1:2,000; R&D
Systems, Inc.) at 4°C overnight. Following hybridization, the
primary antibodies were reclaimed and stored in a 4°C refrigerator,
and membranes were subsequently washed with TBS containing 0.1%
Tween-20. Following washing, membranes were incubated with the
following corresponding secondary antibodies at 37°C for 60 min:
Horseradish peroxidase (HRP)-tagged goat anti-mouse IgG H&L
(cat. no. ab6789; 1:7,000; Abcam), HRP-tagged rabbit anti-mouse IgG
H&L (cat. no. ab6728; 1:8,000; Abcam) and HRP-tagged rabbit
anti-goat IgG H&L (cat. no. ab97100; 1:8,000; Abcam). Finally,
the blots were visualized by enhanced chemiluminescent reagent (EMD
Millipore, Billerica, MA, USA). Images were captured with a Fuji
LAS-3000 imaging system (Fuji Photo Film Co., Ltd., Tokyo, Japan)
and analyzed with ImageJ 1.48u software (National Institutes of
Health, Bethesda, MD, USA).
Statistical analysis
All data were expressed as the mean ± standard
deviation using Microsoft Excel 2007 software (Microsoft
Corporation, Redmond, WA, USA). A Student's t-test was used to
compare the differences between two groups. Differences between
multiple groups were determined using one-way analysis of variance
followed by Tukey's post-hoc test. GraphPad Prism 7 (GraphPad
Software, Inc., La Jolla, CA, USA) was used to draw the graphs.
Experiments were performed in triplicate. P<0.05 was considered
to indicate a statistically significant difference.
Results
TNF-α induces dysfunction in Eahy926
cells
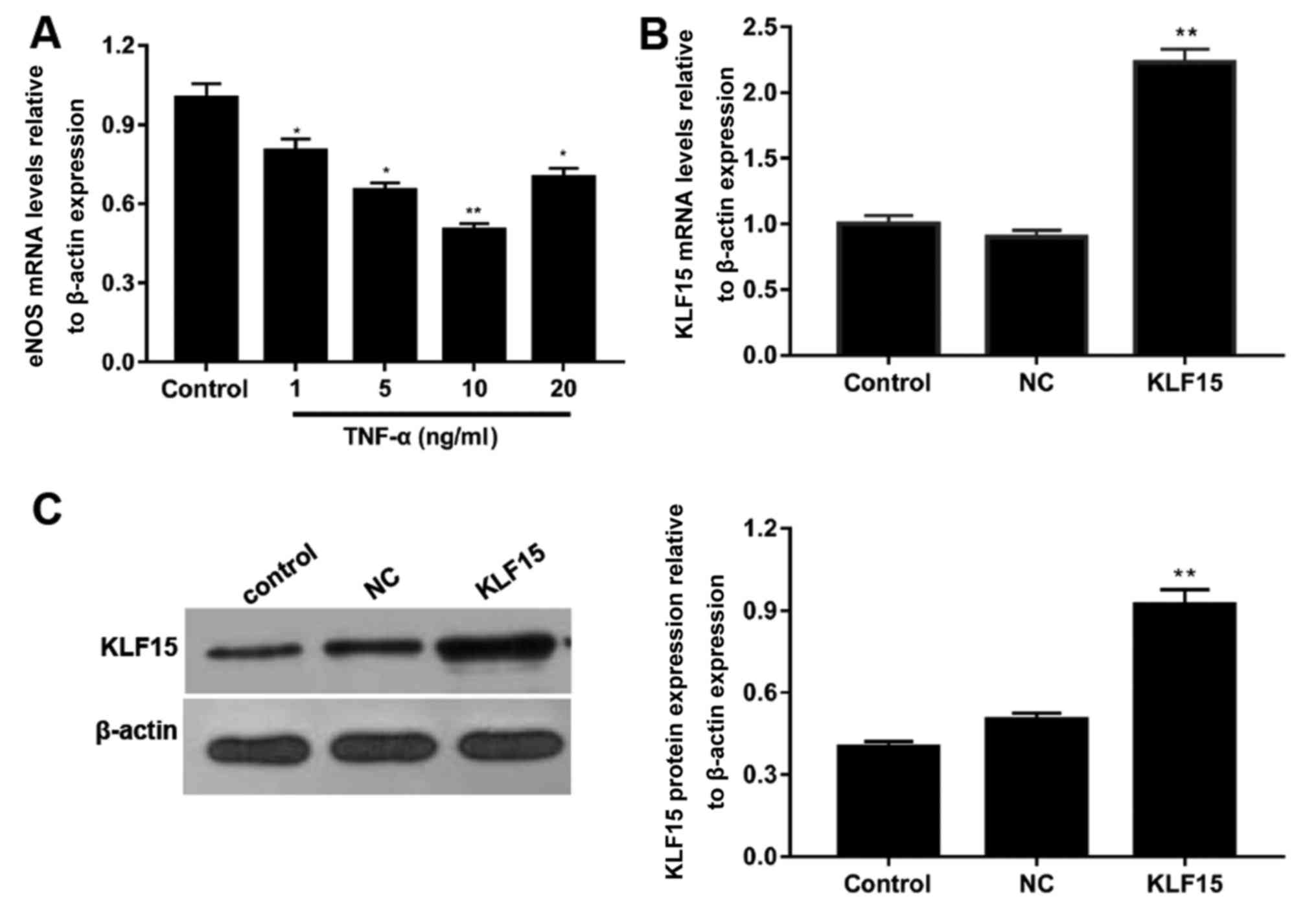
To determine the optimal concentration of TNF-α
required to induce dysfunction in Eahy926 cells, cells were treated
with various concentrations of TNF-α and the mRNA expression levels
of eNOS were subsequently determined using RT-qPCR. Following
treatment of cells with 1, 5, 10 and 20 ng/ml TNF-α, mRNA
expression levels of eNOS were decreased by ~20, 35, 50 and 30%
compared with the control group, respectively (Fig. 1A). Furthermore, the expression of
eNOS was most significantly suppressed in cells treated with 10
ng/ml TNF-α (Fig. 1A;
P<0.05).
To investigate the transfection efficiency of the
pTA2-KLF15 vector, RT-qPCR and western blot analyses were performed
to detect the mRNA and protein expression levels of KLF15,
respectively. Following the transfection of pTA2-KLF15 vector into
cells, the expression levels of KLF15 were revealed to be
significantly enhanced in the KLF15 group compared with the NC and
control groups (Fig. 1B and C;
P<0.01). Furthermore, the expression levels of KLF15 in NC and
control groups were revealed to be similar (Fig. 1B and C).
Overexpression of KLF15 attenuates the
viability of TNF-α induced Eahy926 cells
MTT assays were performed to investigate the
viability of Eahy926 cells. The results demonstrated that treatment
with TNF-α significantly suppressed the viability of cells compared
with non-treated cells. (Fig. 2A;
P<0.05 and P<0.01). Furthermore, when KLF15 was overexpressed
in Eahy926 cells, the viability of Eahy926 cells did not exhibit a
significant difference compared with the NC group; however, cell
viability was significantly enhanced following overexpression of
KLF15 in TNF-α-induced Eahy926 cells compared with the TNF-α + NC
group. Cells overexpressing KLF15 at the 48 h time interval
exhibited significantly suppressed cell viability, compared with
the 12 h time interval (Fig. 2A;
P<0.05).
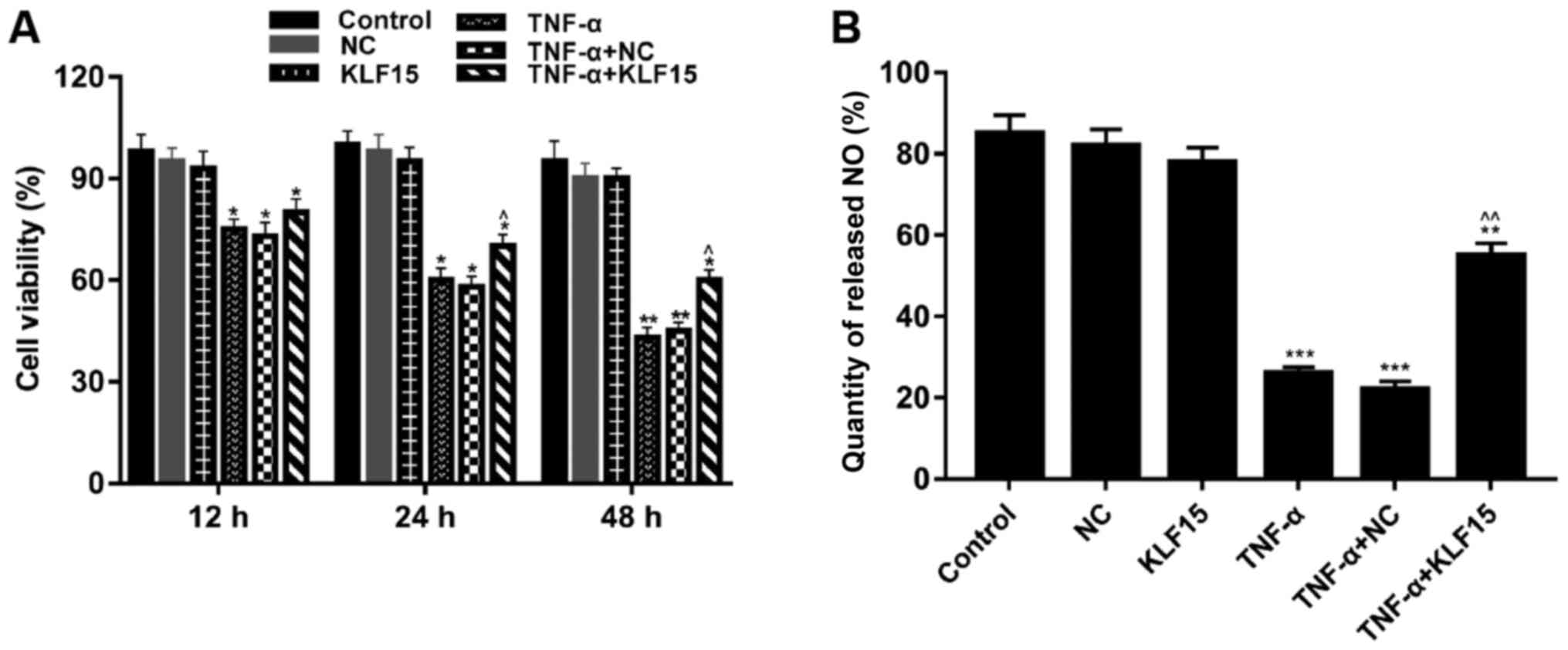 | Figure 2.Overexpression of KLF15 attenuates
the viability of Eahy926 cells treated with TNF-α. (A) Eahy926
cells were either treated with0.1% PBS (control), transfected with
pTA2 vector, transfected with pTA2-KLF15 vector, treated with 10
ng/ml TNF-α, transfected with pTA2 vector and treated with 10 ng/ml
TNF-α or transfected with pTA2-KLF15 vector and treated with 10
ng/ml TNF-α. The viability of Eahy926 cells was investigated using
an MTT kit. (B) An NO detection kit was used to investigate the
quantity of NO released by Eahy926 cells. *P<0.05, **P<0.01
and ***P<0.001 vs. respective NC. ^P<0.05,
^^P<0.01 and vs. respective TNF-α + NC. NC, negative
control pTA2 vector; KLF15, Krüppel-like factor 15; TNF-α, tumor
necrosis factor-α; NO, nitric oxide. |
To investigate the quantity of NO released by
Eahy926 cells, an NO detection kit was used. The results
demonstrated that treatment with TNF-α significantly decreased the
quantity of NO released by Eahy926 cells compared with the NC group
(Fig. 2B; P<0.001).
Furthermore, the results revealed that Eahy926 cells overexpressing
KLF15 did not exhibit a significant difference regarding the
quantity of released NO compared with the NC group (Fig. 2B). However, the quantity of
released NO was significantly enhanced following overexpression of
KLF15 in TNF-α-induced Eahy926 cells compared with the TNF-α + NC
group (Fig. 2B; P<0.01).
Overexpression of KLF15 suppresses
adhesion of TNF-α induced Eahy926 cells
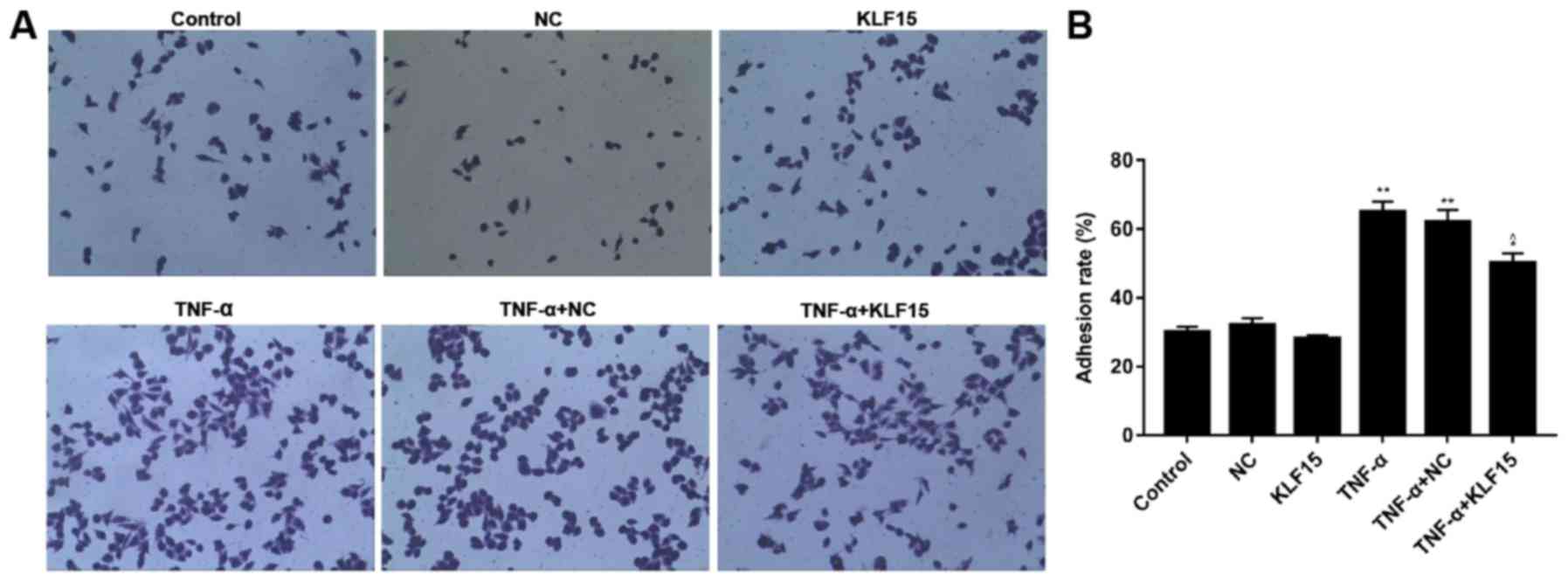
The rate of adhesion in Eahy926 cells was
investigated using a cell adhesion detection kit. The results
revealed that the rate of adhesion in Eahy926 cells overexpressing
KLF15 did not exhibit a marked difference compared with the NC
group, whereas TNF-α-induced Eahy926 cells exhibited a
significantly increased adhesion rate compared with the NC group
(Fig. 3; P<0.01). Furthermore,
overexpression of KLF15 in TNF-α-induced Eahy926 cells was revealed
to significantly suppress the rate of adhesion compared with the
TNF-α + NC group (Fig. 3;
P<0.05).
Overexpression of KLF15 regulates the
expression of factors associated with endothelial cells in Eahy926
cells
In order to determine the mRNA and protein
expression levels of factors associated with endothelial cells in
Eahy926 cells, RT-qPCR and western blot analyses were performed,
respectively. The results demonstrated that treatment with TNF-α
significantly upregulated them RNA and protein expression levels of
MCP-1, ICAM-1 and TGF-β1 in Eahy926 cells; whereas, treatment with
TNF-α significantly downregulated the mRNA and protein expression
levels of eNOS in Eahy926 cells (Fig.
4; P<0.05 and P<0.01). Furthermore, the results revealed
that overexpression of KLF15 in Eahy926 cells significantly
decreased the levels of MCP-1 mRNA and protein; however, the mRNA
and protein expression levels of eNOS, ICAM-1 and TGF-β1 in cells
overexpressing KLF15 did not exhibit significant differences
compared with the NC group. Furthermore, the mRNA and protein
expression levels of MCP-1, ICAM-1 and TGF-β1 were significantly
downregulated, and eNOS mRNA and protein expression levels were
significantly upregulated, following overexpression of KLF15 in
TNF-α-induced Eahy926 cells compared with the TNF-α + NC group
(Fig. 4; P<0.05, P<0.01 and
P<0.001).
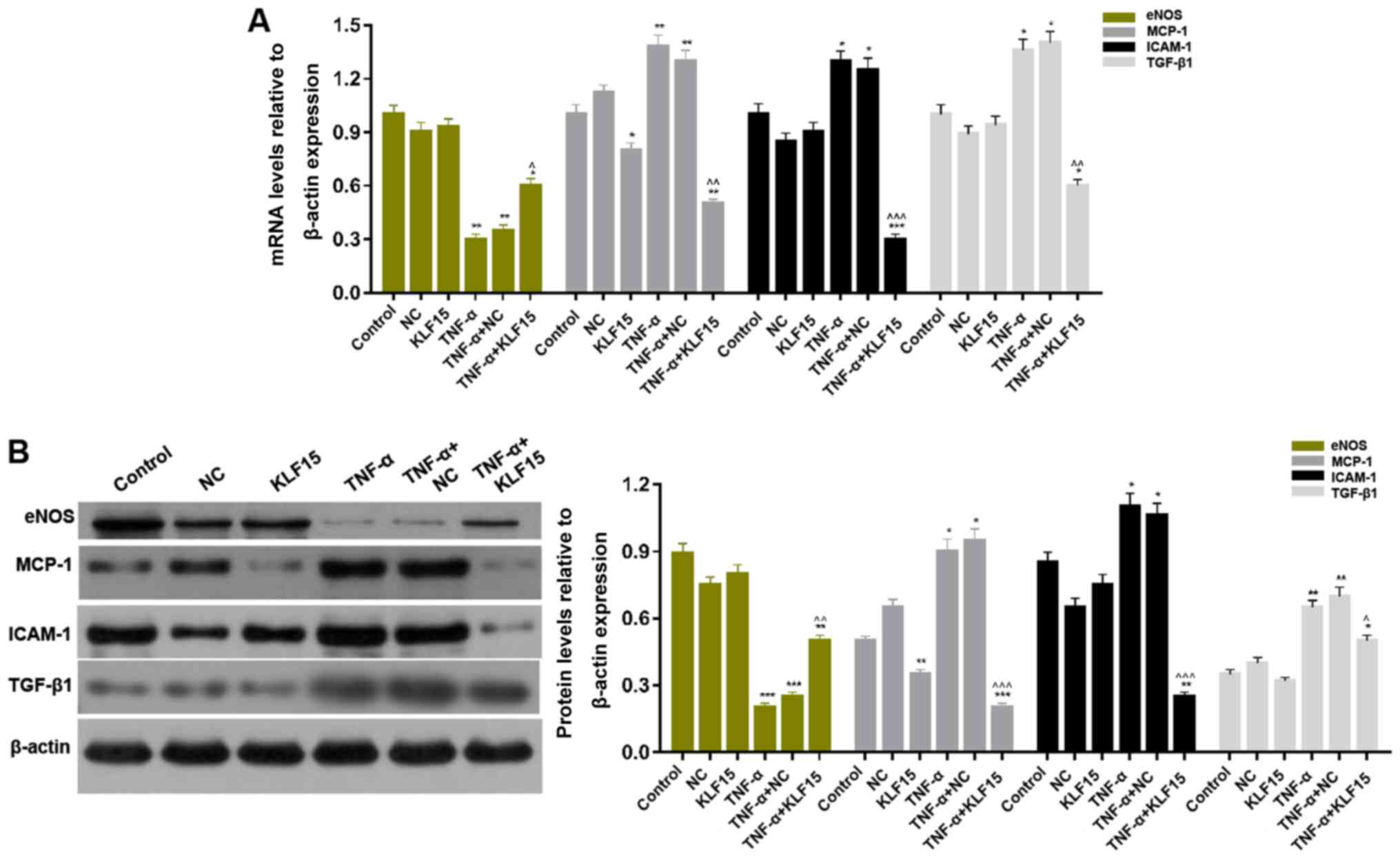 | Figure 4.Overexpression of KLF15 regulates
endothelial cell associated factors in Eahy926 cells. (A) mRNA
expression levels of eNOS, MCP-1, ICAM-1 and TGF-β1 were determined
via reverse transcription-quantitative polymerase chain reaction.
(B) The protein expression levels of eNOS, MCP-1, ICAM-1 and TGF-β1
were investigated via western blotting. β-actin was used as an
internal control. *P<0.05, **P<0.01 and ***P<0.001 vs.
respective NC. ^P<0.05, ^^P<0.01 and
^^^P<0.001 vs. respective TNF-α + NC. NC, negative
control pTA2 vector; KLF15, Krüppel-like factor 15; TNF-α, tumor
necrosis factor-α; eNOS, endothelial nitric oxide synthase; MCP-1,
monocyte chemoattractant protein-1; ICAM-1, intercellular adhesion
molecule-1; TGF-β1, transforming growth factor-β1. |
Overexpression of KLF15 regulates the
NF-κB and Nrf2 signaling pathways in Eahy926 cells
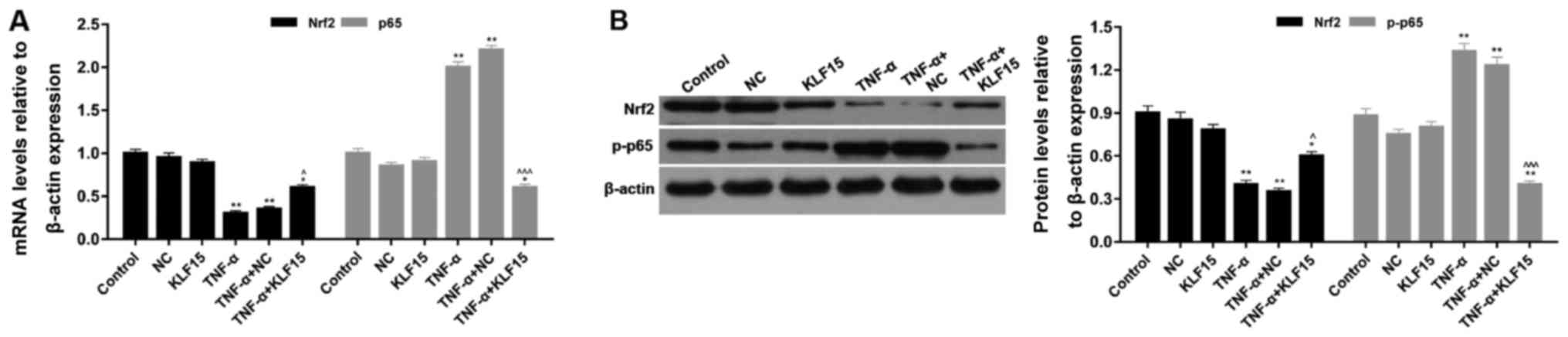
The mRNA and protein expression levels of
p65/phosphorylated (p)-p65 and Nrf2 were investigated using RT-qPCR
and western blot analyses, respectively. The results of the RT-qPCR
analyses demonstrated that treatment with TNF-α significantly
suppressed Nrf2 mRNA expression and significantly enhanced p65 mRNA
expression compared with the NC group (Fig. 5; P<0.01). Eahy926 cells
overexpressing KLF15 did not exhibit any significant differences
regarding the mRNA expression levels of p65 and Nrf2 compared with
the NC group in the absence of TNF-α (Fig. 5). However, when Eahy926 cells
overexpressed KLF15 were treated with TNF-α, the mRNA expression
levels of Nrf2 were significantly increased, and p65 mRNA
expression levels were significantly decreased, compared with the
TNF-α + NC group (Fig. 5;
P<0.05 and P<0.001). Furthermore, the results of the western
blot analyses revealed that the expression trends of p-p65 and Nrf2
protein were similar to the expression trends of p65 and Nrf2 mRNA
(Fig. 5; P<0.05, P<0.01 and
P<0.001). These results suggested that overexpression of KLF15
regulated NF-κB and Nrf2 signaling pathways in Eahy926 cells.
Discussion
NO is the predominant factor released by endothelial
cells, and it functions as a vasodilator that is able to attenuate
endothelial dysfunction (25).
Furthermore, NO functions as a intercellular signal and its
synthesis is regulated by three factors: eNOS gene expression
levels, eNOS activity and NO bioavailability (26). Thus, the function of endothelial
cells may be investigated via determination of the quantity of
released NO and the expression levels of eNOS. Furthermore, TNF-α
is frequently utilized to induce various injuries in scientific
research (27,28), and induces endothelial dysfunction
in diabetic mouse hearts (29).
The results of the present study demonstrated that eNOS mRNA
expression levels in Eahy926 cells treated with 1, 5 and 10 ng/ml
TNF-α decreased in a dose-dependent manner. However, when Eahy926
cells were treated with 20 ng/ml TNF-α, eNOS mRNA expression levels
were upregulated compared with cells treated with 10 ng/ml TNF-α.
It was hypothesized that self-repair mechanisms performed by
injured endothelial cells may be associated with increased
expression levels of eNOS. Therefore, treatment with 10 ng/ml TNF-α
was chosen to induce Eahy926 cells for the construction of the
vascular endothelial dysfunction model for subsequent experiments
in the present study.
KLFs are involved in numerous pathological processes
associated with cardiovascular disease and are associated with the
regulation of various signal transduction pathways (30). KLF2 has been demonstrated to induce
the expression of numerous bioactive factors that have a protective
role in the cardiovascular system (31). In addition, it has been revealed
that overexpression of KLF11 attenuates TNF-α-induced damage to
human umbilical vein endothelial cells (HUVECs) (32). Therefore, it was hypothesized that
KLF15 may have a role in the TNF-α-induced dysfunction of vascular
endothelial cells. The results demonstrated that overexpression of
KLF15 attenuated the viability of Eahy926 cells otherwise inhibited
by TNF-α, enhanced the quantity of released NO and enhanced eNOS
expression. These results suggested that overexpression of KLF15
attenuated TNF-α-induced dysfunction in Eahy926 cells.
Vascular endothelial injury may increase the
adhesion rate of endothelial cells or platelets, and induce the
formation of vasoactive molecules, including ICAM-1; chemotactic
factors, including MCP-1; and growth factors, including TGF-β
(33,34). Previous studies have revealed that
KLF2 is able to regulate the secretion and release of inflammatory
factors and adhesion factors, including ICAM-1, thereby inhibiting
the adhesion of leukocytes, platelets and vascular endothelial
cells (33,35). Furthermore, HUVECs overexpressing
KLF11 have been demonstrated to exhibit suppressed expression
levels of MCP-1 and ICAM-1 (32).
Considering the results of the aforementioned studies, the present
study aimed to investigate whether overexpression of KLF15 affected
adhesive ability and the expression levels of MCP-1, ICAM-1 and
TGF-β1 in Eahy926 cells. The results revealed that overexpression
of KLF15 decreased the rate of adhesion and attenuated the
expression levels of MCP-1, ICAM-1 and TGF-β1 in Eahy926 cells
induced by TNF-α.
An increasing number of studies have suggested that
activation of the NF-κB signaling pathway has an important role in
endothelial dysfunction (36,37).
The activation of NF-κB is closely associated with the upregulation
of MCP-1 and ICAM-1 expression levels, which represent biomarkers
of endothelial dysfunction (38,39).
KLF11 is able to regulate the expression of downstream inflammatory
and adhesion factors by binding top65 (32). Furthermore, previous studies have
demonstrated that KLF2 is able to activate the expression of Nrf2,
which subsequently stimulates the production of antioxidants in
endothelial cells (40). Thus, we
hypothesized that overexpression of KLF15 may have a protective
effect on TNF-α-induced dysfunction in Eahy926 cells, which may
regulate NF-κB and Nrf2 signaling pathways. The results of the
present study revealed that Eahy926 cells overexpressed KLF15
exhibited activated Nrf2 signaling and inhibited NF-κB signaling,
which attenuated TNF-α-induced dysfunction in endothelial
cells.
In conclusion, the results of the present study
revealed that TNF-α was able to induce vascular endothelial
dysfunction in Eahy926 cells at an optimum concentration of 10
ng/ml. Furthermore, overexpression of KLF15 in Eahy926 cells was
demonstrated to have a protective effect on the TNF-α-induced
dysfunction of endothelial cells via activation of Nrf2 signaling
and inhibition of NF-κB signaling. Therefore, KLF15 may represent a
novel therapeutic target for the treatment of patients with
atherosclerosis.
Acknowledgements
Not applicable.
Funding
The present study was funded by Qingdao City South
District Science and Technology Program (grant no. 2538), the Youth
Fund of The Affiliated Hospital of Qingdao University (grant no.
2616) and the Qingdao University ‘Clinical Medicine +X’ Engineering
Fund (grant no. 2017M24).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
BL, LLX, HFW and SX made substantial contributions
to the conception and design of the study. XMY and MJG implemented
the experiments. XMY, WL and XZS were responsible for analyzing and
interpreting the data. BL and MJG drafted the manuscript. All
authors were responsible for giving approval of the final version
of the manuscript to be published.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Salzer L, Tenenbaum-Gavish K and Hod M:
Metabolic disorder of pregnancy (understanding pathophysiology of
diabetes and preeclampsia). Best Pract Res Clin Obstet Gynaecol.
29:328–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ding W, Cheng H, Chen F, Yan Y, Zhang M,
Zhao X, Hou D and Mi J: Adipokines are associated with hypertension
in Metabolically Healthy Obese (MHO) children and adolescents: A
prospective population-based cohort study. J Epidemiol. 28:19–26.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karelis AD, Brochu M and Rabasa-Lhoret R:
Can we identify metabolically healthy but obese individuals (MHO)?
Diabetes Metab. 30:569–572. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim TJ, Shin HY, Chang Y, Kang M, Jee J,
Choi YH, Ahn HS, Ahn SH, Son HJ and Ryu S: Metabolically healthy
obesity and the risk for subclinical atherosclerosis.
Atherosclerosis. 262:191–197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moon S, Oh CM, Choi MK, Park YK, Chun S,
Choi M, Yu JM and Yoo HJ: The influence of physical activity on
risk of cardiovascular disease in people who are obese but
metabolically healthy. PLoS One. 12:e01851272017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Caffarelli C, Montagnani A, Nuti R and
Gonnelli S: Bisphosphonates, atherosclerosis and vascular
calcification: Update and systematic review of clinical studies.
Clin Interv Aging. 12:1819–1828. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moore KJ and Tabas I: Macrophages in the
pathogenesis of atherosclerosis. Cell. 145:341–355. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mitra R, O'Neil GL, Harding IC, Cheng MJ,
Mensah SA and Ebong EE: Glycocalyx in atherosclerosis-relevant
endothelium function and as a therapeutic target. Curr Atheroscler
Rep. 19:632017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bonetti PO, Lerman LO and Lerman A:
Endothelial dysfunction: A marker of atherosclerotic risk.
Arterioscler Thromb Vasc Biol. 23:168–175. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bieker JJ: Krüppel-like factors: Three
fingers in many pies. J Biol Chem. 276:34355–34358. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chiambaretta F, De Graeve F, Turet G,
Marceau G, Gain P, Dastugue B, Rigal D and Sapin V: Cell and tissue
specific expression of human Krüppel-like transcription factors in
human ocular surface. Mol Vis. 10:901–909. 2004.PubMed/NCBI
|
|
12
|
Wang N, Liu ZH, Ding F, Wang XQ, Zhou CN
and Wu M: Down-regulation of gut-enriched Kruppel-like factor
expression in esophageal cancer. World J Gastroenterol. 8:966–970.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Y and Cai L: Does krüppel like factor
15 play an important role in the left ventricular hypertrophy of
patients with type 2 diabetes? EBioMedicine. 20:17–18. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shou F, Xu F, Li G, Zhao Z, Mao Y, Yang F,
Wang H and Guo H: RASSF1A promoter methylation is associated with
increased risk of thyroid cancer: A meta-analysis. Onco Targets
Ther. 10:247–257. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mori T, Sakaue H, Iguchi H, Gomi H, Okada
Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, et
al: Role of Krüppel-like factor 15 (KLF15) in transcriptional
regulation of adipogenesis. J Biol Chem. 280:12867–12875. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Leenders JJ, Wijnen WJ, van der Made I,
Hiller M, Swinnen M, Vandendriessche T, Chuah M, Pinto YM and
Creemers EE: Repression of cardiac hypertrophy by KLF15: Underlying
mechanisms and therapeutic implications. PLoS One. 7:e367542012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Patel SK, Wai B, Lang CC, Levin D, Palmer
CNA, Parry HM, Velkoska E, Harrap SB, Srivastava PM and Burrell LM:
Genetic variation in kruppel like factor 15 is associated with left
ventricular hypertrophy in patients with type 2 diabetes: Discovery
and replication cohorts. EBioMedicine. 18:171–178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang C and Lin X: Analysis of epidermal
growth factor-induced NF-κB signaling. Methods Mol Biol.
1280:75–102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Crofford LJ, Tan B, McCarthy CJ and Hla T:
Involvement of nuclear factor kappa B in the regulation of
cyclooxygenase-2 expression by interleukin-1 in rheumatoid
synoviocytes. Arthritis Rheum. 40:226–236. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Aggarwal BB: Nuclear factor-kappaB: The
enemy within. Cancer Cell. 6:203–208. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cuadrado A, Martín-Moldes Z, Ye J and
Lastres-Becker I: Transcription factors NRF2 and NF-κB are
coordinated effectors of the Rho family, GTP-binding protein RAC1
during inflammation. J Biol Chem. 289:15244–15258. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim JK, Lee JE, Jung EH, Jung JY, Jung DH,
Ku SK, Cho IJ and Kim SC: Hemistepsin A ameliorates acute
inflammation in macrophages via inhibition of nuclear factor-κB and
activation of nuclear factor erythroid 2-related factor 2. Food
Chem Toxicol. 111:176–188. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wardyn JD, Ponsford AH and Sanderson CM:
Dissecting molecular cross-talk between Nrf2 and NF-κB response
pathways. Biochem Soc Trans. 43:621–626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Myers PR, Guerra R Jr and Harrison DG:
Release of NO and EDRF from cultured bovine aortic endothelial
cells. Am J Physiol. 256:H1030–H1037. 1989.PubMed/NCBI
|
|
26
|
Nathan C and Xie QW: Nitric oxide
synthases: Roles, tolls, and controls. Cell. 78:915–918. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pang Y, Zheng B, Fan LW, Rhodes PG and Cai
Z: IGF-1 protects oligodendrocyte progenitors against
TNFalpha-induced damage by activation of PI3K/Akt and interruption
of the mitochondrial apoptotic pathway. Glia. 55:1099–1107. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang J, Yang X, Wang H, Zhao B, Wu X, Su
L, Xie S, Wang Y, Li J, Liu J, et al: PKCζ as a promising
therapeutic target for TNFα-induced inflammatory disorders in
chronic cutaneous wounds. Int J Mol Med. 40:1335–1346. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee J, Lee S, Zhang H, Hill MA, Zhang C
and Park Y: Interaction of IL-6 and TNF-α contributes to
endothelial dysfunction in type 2 diabetic mouse hearts. PLoS One.
12:e01871892017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang E, Nayak L and Jain MK: Krüppel-like
factors in endothelial cell biology. Curr Opin Hematol. 24:224–229.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Parmar KM, Larman HB, Dai G, Zhang Y, Wang
ET, Moorthy SN, Kratz JR, Lin Z, Jain MK, Gimbrone MA Jr and
García-Cardeña G: Integration of flow-dependent endothelial
phenotypes by Kruppel-like factor 2. J Clin Invest. 116:49–58.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fan Y, Guo Y, Zhang J, Subramaniam M, Song
CZ, Urrutia R and Chen YE: Krüppel-like factor-11, a transcription
factor involved in diabetes mellitus, suppresses endothelial cell
activation via the nuclear factor-κB signaling pathway.
Arterioscler Thromb Vasc Biol. 32:2981–2988. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han JM, Li H, Cho MH, Baek SH, Lee CH,
Park HY and Jeong TS: Soy-leaf extract exerts atheroprotective
effects via modulation of krüppel-like factor 2 and adhesion
molecules. Int J Mol Sci. 18:pii: E373. 2017. View Article : Google Scholar
|
|
34
|
Jain MK, Sangwung P and Hamik A:
Regulation of an inflammatory disease: Krüppel-like factors and
atherosclerosis. Arterioscler Thromb Vasc Biol. 34:499–508. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
SenBanerjee S, Lin Z, Atkins GB, Greif DM,
Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, et
al: KLF2 is a novel transcriptional regulator of endothelial
proinflammatory activation. J Exp Med. 199:1305–1315. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bodiga VL, Kudle MR and Bodiga S:
Silencing of PKC-α, TRPC1 or NF-κB expression attenuates
cisplatin-induced ICAM-1 expression and endothelial dysfunction.
Biochem Pharmacol. 98:78–91. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang X, Guo D, Lin C, Shi Z, Qian R, Fu W,
Liu J, Li X and Fan L: hCLOCK causes Rho-kinase-mediated
endothelial dysfunction and NF-κB-mediated inflammatory responses.
Oxid Med Cell Longev. 2015:6718392015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiao W: Advances in NF-kappaB signaling
transduction and transcription. Cell Mol Immunol. 1:425–435.
2004.PubMed/NCBI
|
|
39
|
Zhou Z, Connell MC and MacEwan DJ:
TNFR1-induced NF-kappaB, but not ERK, p38MAPK or JNK activation,
mediates TNF-induced ICAM-1 and VCAM-1 expression on endothelial
cells. Cell Signal. 19:1238–1248. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fledderus JO, Boon RA, Volger OL, Hurttila
H, Ylä-Herttuala S, Pannekoek H, Levonen AL and Horrevoets AJ: KLF2
primes the antioxidant transcription factor Nrf2 for activation in
endothelial cells. Arterioscler Thromb Vasc Biol. 28:1339–1346.
2008. View Article : Google Scholar : PubMed/NCBI
|