Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common neoplasm and the third most frequent cause of
cancer-associated mortality (1).
The majority of patients with HCC suffering from hepatitis B virus
(HBV) infection have cirrhosis secondary to chronic
necroinflammation (2). HBV, an
oncogenic virus, likely causes HCC via direct (integration of its
DNA into the host genome) and indirect (necroinflammation and
regeneration injury) pathways (3).
The aberrant expression of genes, which involves numerous types of
RNA, serves an important role in the occurrence and development of
HCC. MicroRNAs (miRNAs) and mRNA have been reported to be involved
in the pathogenesis of HCC. In addition, miRNAs have clinical value
in the diagnosis of HCC, since they are present in the blood and
thus may be used as diagnostic markers, and in turn as potential
targets for specific systemic treatments (4).
Mature human miRNA is a class of single-stranded,
small non-coding RNAs that are ~22 nucleotides in length (5). miRNAs serve a key regulatory role in
gene expression at the posttranscriptional level. miRNAs act by
binding to imperfectly complementary sites within the
3′-untranslated regions of their target mRNAs, inhibiting
translation or initiating degradation of the target mRNA through
the miRNA-associated RNA-induced silencing complex (miRISC).
Recruitment of the miRISC may thus modulate the expression of
targeted protein-coding genes (6).
A single mRNA transcript may have a number of miRNA response
elements for different miRNAs and, conversely, one miRNA may target
as many as 100 different mRNAs in a gene regulatory network
(7,8). miRNAs are involved in a series of
crucial biological processes (9).
The most notable alterations in miRNA expression are observed in
cancer. Certain miRNAs may function as oncogenes or tumor
suppressor genes by targeting their corresponding mRNAs, and
certain dysregulated miRNAs are able to promote tumorigenesis and
cancer progression (10,11). As has been widely indicated, the
expression of miRNAs and their corresponding target genes is often
inversely modulated in different backgrounds (12). Mounting evidence has suggested the
importance of miRNAs in the modulation of gene expression, cellular
proliferation, cellular mobility, cellular differentiation,
apoptosis and tumorigenesis (13).
In recent years, miRNA and mRNA expression profile
studies have identified a large number of genes with differential
expression in HCC (14–25). Numerous differentially expressed
miRNAs and mRNAs have been identified to be involved in the
occurrence and development of HCC, and thus may be potential
prognostic and diagnostic markers (17,26).
Studying the interaction of miRNAs and mRNAs has become an
important part of cancer research.
The microarray is a powerful tool for conveniently
and quickly analyzing miRNAs relevant to cancer. More importantly,
it also simultaneously profiles the mRNA targets of the miRNAs,
thereby providing insights into the interaction between the
cancer-associated miRNAs and their target mRNAs (27). Collectively, the predicted target
genes, network diagrams, and Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analyses may be
useful for determining the mechanism of HCC tumorigenesis.
In the present study, the expression profiles of
miRNAs and mRNAs in HBV-associated HCC were identified by
polymerase chain reaction (PCR) microarray, and the interactions
between the differentially expressed miRNAs and their targets were
analyzed using bioinformatics methods.
Patients and methods
Patients and clinical specimens
A total of five HCC tissues and paired adjacent
non-tumor tissues (NTs) were collected as surgical specimens
between June 2012 and December 2013 at Beijing Youan Hospital,
Capital Medical University (Beijing, China). All tissue specimens
were immediately preserved in RNAfixer reagent (Bioteke, Beijing,
China) and stored at −80°C until use. The NTs were taken 5 cm from
the edge of the cancer and contained no obvious tumor cells, as
evaluated by an experienced pathologist. The five HCC patients were
diagnosed with HBV infection; serum positive for hepatitis B
surface antigen identified by ELISA method (ELISA kit Cat. no.
185982, Roche Diagnostics, Basel, Switzerland) was defined as HBV
infection. Tumors were staged according to the
tumor-node-metastasis staging system (28) and Barcelona Clinic Liver Cancer
system (29) (Table I). No radiotherapy, chemotherapy or
targeted therapy was administered prior to the isolation of tissue
specimens. The protocol was approved by the appropriate ethics
committees in Beijing Youan Hospital and was conducted in
compliance with the Declaration of Helsinki. Written consent was
obtained from all participants.
 | Table I.Clinical details of the five patients
with hepatocellular carcinoma. |
Table I.
Clinical details of the five patients
with hepatocellular carcinoma.
| Case ID | Sex | Age, years | HBV-DNA load,
IU/ml | Liver
cirrhosis |
Differentiation | Stage, TNM | Stage, BCLC |
|---|
| Case 1 | M | 44 | <100 | None | Moderate | T1N0M0 | A |
| Case 2 | M | 63 |
4.01×106 | Yes | Moderate | T1N0M0 | A |
| Case 3 | M | 44 |
9.85×102 | Yes | Poor | T3aN0M0 | B |
| Case 4 | F | 45 |
1.98×103 | Yes | Moderate | T1N0M0 | A |
| Case 5 | M | 59 |
1.60×106 | Yes | Poor | T1N0M0 | A |
Total RNA extraction and quality
control
Total tissue RNA was extracted from the HCC tissues
and paired adjacent NTs using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
following the manufacturer's protocol. Contaminating DNA was
removed from the RNA preparations using DNase I. The RNA was
purified using an RNeasy® MinElute™ Cleanup
kit (Qiagen GmbH, Hilden, Germany). Subsequently, the RNA
concentration of the samples was determined via the absorbance at
260 nm using a NanoDrop ND-1000 instrument (Thermo Fisher
Scientific, Inc., Wilmington, DE, USA). The integrity of the RNA
was assessed by electrophoresis on a denaturing agarose gel.
cDNA synthesis and quantitative
(q)PCR
cDNA was synthesized using a miRNA First-Strand cDNA
Synthesis kit (Arraystar, Inc., MD, USA) and operated according
manufacturer's protocol. Briefly, total RNA was ligated with the
3′-ligation adapter by RNA ligase. The cRNA template was obtained
by a reverse transcriptase reaction at 42°C for 60 min, then
inactivated at 85°C for 5 min. The reaction system included 10 µl
ligated product, 1 µl Moloney murine leukemia virus reverse
transcriptase, 8.5 µl RT Reaction Master Mix (contain dNTP) and 0.5
µl RNase inhibitor. qPCR was performed using miRStar™ Hu
man Cancer Focus miRNA & Target mRNA PCR Array (Arraystar,
Inc., Rockville, MD, USA) on an ABI PRISM7900 Real-time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). This array
contained 184 critical tumor-associated miRNAs and 178 well-defined
mRNA targets of these miRNAs. The PCR reaction system included
diluted cDNA template, PCR primer mix and SYBR® Green
Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The PCR cycling conditions were as follows: 95°C for 10 min,
followed by 40 cycles of 95°C for 10 sec and 60°C for 1 min.
Following qPCR, the amplification products were optically measured
referred to the instruction manual and the resulting melting curves
were analyzed.
Data analysis and statistical
analysis
The initial data analysis was performed using the
software (version SDS 2.4) supplied with the qPCR instrument to
obtain raw Cq values. Normalization and further data analysis was
performed using GenEx qPCR analysis software (version 6.1,
www.exiqon.com/mirna-pcranalysis). qPCR reactions
results were calculated using the 2−∆∆Cq method
(30). The resulting data were
analyzed by two-tailed Student's t-test. Fold change (FC) ≥2 and
P<0.05 was considered to indicate a statistically significant
difference.
miRNA target prediction, GO and KEGG
pathway analyses
miRNA target prediction was performed using miRWalk
software (version 7.0, http://mirwalk.umm.uni-heidelberg.de). A total of five
algorithms were used for miRNA target gene prediction, namely
miRWalk (version 7.0, mirwalk.umm.uni-heidelberg.de), miRanda (www.microrna.org/microrna/home.do),
miRDB (Last modified: May 03, 2016, http://mirdb.org/mirdb/), RNA22 (version 2, cm.jefferson.edu/rna22/Interactive/l)
and Targetscan (release 7.1, www.targetscan.org). Only when one gene was confirmed
by all five algorithms was the gene identified as a target gene, in
order to increase the accuracy of the target gene prediction. GO
functional enrichment and KEGG pathway enrichment analyses for
differentially expressed genes were performed using the cluster
profiler package of R software (version 3.3.3, http://www.r-project.org) based on the GO database
(www.geneontology.org) and the Database
for Annotation, Visualization, and Integrated Discovery (DAVID;
version 6.7, http://david.ncifcrf.gov). Fisher's exact test was
used to identify whether there was more overlap between the
differentially expressed gene list and the GO annotation list
compared with that which would be expected by chance.
Results
Identification of differentially
expressed miRNAs and mRNAs
In our study, 32 differentially expressed miRNAs
(four upregulated miRNAs and 28 downregulated miRNAs) and 16
differentially expressed mRNAs (11 upregulated mRNAs and five
downregulated mRNAs) were identified [fold change (FC)≥2 and
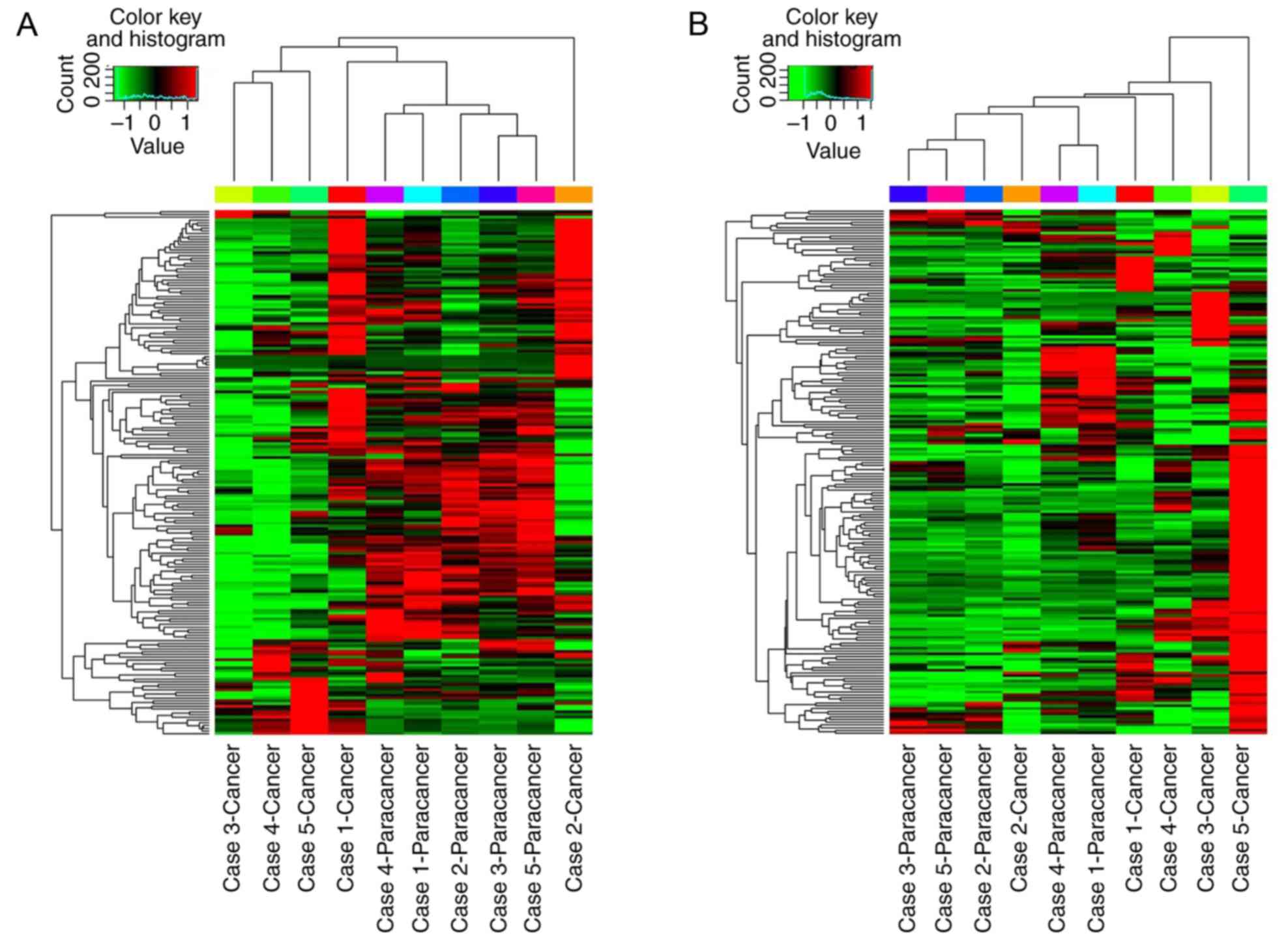
P≤0.05]. The global miRNA and mRNA expression patterns were
evaluated using a hierarchical clustering plot (Fig. 1). The results demonstrated that
each differentially expressed miRNA was able to regulate one or
more mRNAs. Similarly, one mRNA may be regulated by one or more
miRNAs. The differentially expressed miRNAs and mRNAs with FC≥2 and
P≤0.05 are presented in Tables II
and III. In the HCC tissues,
hsa-miR-96-5p and hsa-miR-18b-5p were the most significantly
upregulated miRNAs, while hsa-miR-451a and hsa-miR-199a-5p were the
most significantly downregulated miRNAs, compared with their levels
in the NTs. In turn, the upregulated miRNAs were indicated to
negatively regulate their target mRNAs; for instance, upregulated
hsa-miR-96-5p and hsa-miR-18b-5p suppressed the mRNA expression of
forkhead box O1 (FOXO1) and MET transcriptional regulator MACC1
(MACC1), respectively. The downregulated miRNAs were indicated to
positively regulate their target mRNAs; downregulated
hsa-miR-199a-5p facilitated an increase in the mRNA expression of
cyclin dependent kinase 4 (CDK4) and insulin like growth factor 2
(IGF2).
 | Table II.Concurrent analysis of differential
miRNA and relevant mRNA expression. |
Table II.
Concurrent analysis of differential
miRNA and relevant mRNA expression.
| miRNA ID | Fold change | T-test P-value | mRNA ID | Fold change | T-test P-value |
|---|
| hsa-miR-96-5p | 3.23 | 0.015466 | FOXO1 | −3.06 | 0.003590 |
| hsa-miR-18b-5p | 3.30 | 0.028242 | MACC1 | −9.90 | 0.038804 |
| hsa-miR-18a-5p | 2.79 | 0.047523 | MACC1 | −9.90 | 0.038804 |
| hsa-miR-182-5p | 2.14 | 0.017416 | ZEB2 | −3.32 | 0.022872 |
| hsa-miR-142-5p | −2.95 | 0.006185 | RECK | 2.23 | 0.046041 |
|
|
|
| MET | 2.30 | 0.007944 |
| hsa-let-7a-5p | −2.26 | 0.000101 | DMTF1 | 2.14 | 0.012181 |
|
|
|
| CD34 | 3.06 | 0.026660 |
| hsa-miR-342-3p | −2.20 | 0.005877 | E2F1 | 3.70 | 0.013213 |
| hsa-miR-30e-3p | −2.07 | 0.009148 | CDK4 | 3.57 | 0.027208 |
|
|
|
| PTK2 | 3.13 | 0.008627 |
| hsa-let-7b-5p | −2.53 | 0.000082 | DMTF1 | 2.14 | 0.012181 |
|
|
|
| CD34 | 3.06 | 0.026660 |
| hsa-miR-145-5p | −3.92 | 0.008139 | IGF2 | 83.52 | 0.025926 |
| hsa-miR-101-3p | −3.05 | 0.001338 | EZH2 | 7.73 | 0.006231 |
|
|
|
| MET | 2.30 | 0.007944 |
|
hsa-miR-200b-3p | −3.53 | 0.043589 | RECK | 2.23 | 0.046041 |
|
hsa-miR-130a-3p | −2.97 | 0.005732 | MET | 2.30 | 0.007944 |
|
|
|
| CDK4 | 3.57 | 0.027208 |
| hsa-miR-422a | −2.64 | 0.000209 | SMO | 2.53 | 0.046150 |
|
hsa-miR-130b-3p | −2.39 | 0.002171 | MET | 2.30 | 0.007944 |
|
|
|
| CDK4 | 3.57 | 0.027208 |
| hsa-miR-150-5p | −2.57 | 0.046146 | CDK4 | 3.57 | 0.027208 |
|
hsa-miR-199a-5p | −9.21 | 0.000657 | CDK4 | 3.57 | 0.027208 |
|
|
|
| IGF2 | 83.52 | 0.025926 |
| hsa-let-7c-5p | −2.45 | 0.000184 | DMTF1 | 2.14 | 0.012181 |
|
|
|
| CD34 | 3.06 | 0.026660 |
| hsa-let-7e-5p | −2.11 | 0.003287 | DMTF1 | 2.14 | 0.012181 |
|
|
|
| CD34 | 3.06 | 0.026660 |
| hsa-miR-143-3p | −2.68 | 0.025475 | DMTF1 | 2.14 | 0.012181 |
|
|
|
| SMO | 2.53 | 0.046150 |
| hsa-miR-26a-5p | −2.11 | 0.000546 | CCNE1 | 8.13 | 0.027245 |
|
hsa-miR-181a-5p | −2.23 | 0.000035 | RECK | 2.23 | 0.046041 |
|
|
|
| MET | 2.30 | 0.007944 |
|
|
|
| IGF2 | 83.52 | 0.025926 |
|
|
|
| SMO | 2.53 | 0.046150 |
| hsa-miR-122-5p | −2.68 | 0.015077 | MET | 2.30 | 0.007944 |
| hsa-miR-505-3p | −2.54 | 0.002885 | PTK2 | 3.13 | 0.008627 |
|
|
|
| SMO | 2.53 | 0.046150 |
|
|
|
| MET | 2.30 | 0.007944 |
| hsa-miR-26b-5p | −2.05 | 0.001286 | CCNE1 | 8.13 | 0.027245 |
|
hsa-miR-125b-5p | −3.22 | 0.000950 | SMO | 2.53 | 0.046150 |
| hsa-miR-223-3p | −2.32 | 0.003535 | E2F1 | 3.70 | 0.013213 |
|
|
|
| IGF2 | 83.52 | 0.025926 |
| hsa-miR-100-5p | −3.01 | 0.034014 | – | – | – |
| hsa-miR-188-5p | −2.48 | 0.003011 | – | – | – |
| hsa-miR-22-3p | −2.63 | 0.000404 | – | – | – |
| hsa-miR-451a | −10.55 | 0.000566 | – | – | – |
| hsa-miR-99a-5p | −3.23 | 0.007100 | – | – | – |
 | Table III.Concurrent analysis of differential
mRNA and relevant miRNA expression. |
Table III.
Concurrent analysis of differential
mRNA and relevant miRNA expression.
| mRNA ID | Fold change | T-test P-value | miRNA ID | Fold change | T-test P-value |
|---|
| CCNE1 | 8.13 | 0.027245 | hsa-miR-26a-5p | −2.11 | 0.000546 |
|
|
|
| hsa-miR-26b-5p | −2.05 | 0.001286 |
| IGF2 | 83.52 | 0.025926 | hsa-miR-145-5p | −3.92 | 0.008139 |
|
|
|
|
hsa-miR-181a-5p | −2.23 | 0.000035 |
|
|
|
|
hsa-miR-199a-5p | −9.21 | 0.000657 |
|
|
|
| hsa-miR-223-3p | −2.32 | 0.003535 |
| CDK4 | 3.57 | 0.027208 |
hsa-miR-130a-3p | −2.97 | 0.005732 |
|
|
|
|
hsa-miR-130b-3p | −2.39 | 0.002171 |
|
|
|
| hsa-miR-150-5p | −2.57 | 0.046146 |
|
|
|
|
hsa-miR-199a-5p | −9.21 | 0.000657 |
|
|
|
| hsa-miR-30e-3p | −2.07 | 0.009148 |
| E2F1 | 3.70 | 0.013213 | hsa-miR-223-3p | −2.32 | 0.003535 |
|
|
|
| hsa-miR-342-3p | −2.20 | 0.005877 |
| PTK2 | 3.13 | 0.008627 | hsa-miR-30e-3p | −2.07 | 0.009148 |
|
|
|
| hsa-miR-505-3p | −2.54 | 0.002885 |
| CD34 | 3.06 | 0.026660 | hsa-let-7a-5p | −2.26 | 0.000101 |
|
|
|
| hsa-let-7b-5p | −2.53 | 0.000082 |
|
|
|
| hsa-let-7c-5p | −2.45 | 0.000184 |
|
|
|
| hsa-let-7e-5p | −2.11 | 0.003287 |
| EZH2 | 7.73 | 0.006231 | hsa-miR-101-3p | −3.05 | 0.001338 |
| SMO | 2.53 | 0.046150 |
hsa-miR-125b-5p | −3.22 | 0.000950 |
|
|
|
| hsa-miR-143-3p | −2.68 | 0.025475 |
|
|
|
|
hsa-miR-181a-5p | −2.23 | 0.000035 |
|
|
|
| hsa-miR-422a | −2.64 | 0.000209 |
|
|
|
| hsa-miR-505-3p | −2.54 | 0.002885 |
| MET | 2.30 | 0.007944 | hsa-miR-101-3p | −3.05 | 0.001338 |
|
|
|
| hsa-miR-122-5p | −2.68 | 0.015077 |
|
|
|
|
hsa-miR-130a-3p | −2.97 | 0.005732 |
|
|
|
|
hsa-miR-130b-3p | −2.39 | 0.002171 |
|
|
|
| hsa-miR-142-5p | −2.95 | 0.006185 |
|
|
|
|
hsa-miR-181a-5p | −2.23 | 0.000035 |
|
|
|
| hsa-miR-505-3p | −2.54 | 0.002885 |
| DMTF1 | 2.14 | 0.012181 | hsa-let-7b-5p | −2.53 | 0.000082 |
|
|
|
| hsa-let-7c-5p | −2.45 | 0.000184 |
|
|
|
| hsa-let-7e-5p | −2.11 | 0.003287 |
|
|
|
| hsa-miR-143-3p | −2.68 | 0.025475 |
| RECK | 2.23 | 0.046041 | hsa-miR-142-5p | −2.95 | 0.006185 |
|
|
|
|
hsa-miR-181a-5p | −2.23 | 0.000035 |
|
|
|
|
hsa-miR-200b-3p | −3.53 | 0.043589 |
| CORO1A | −14.43 | 0.014401 |
hsa-miR-517a-3p | 1.60 | 0.357626 |
| MACC1 | −9.90 | 0.038804 | hsa-miR-18a-5p | 2.79 | 0.047523 |
|
|
|
| hsa-miR-18b-5p | 3.30 | 0.028242 |
| CCL4 | −4.72 | 0.002124 | hsa-miR-183-5p | 1.86 | 0.037847 |
| ZEB2 | −3.32 | 0.022872 | hsa-miR-182-5p | 2.14 | 0.017416 |
| FOXO1 | −3.06 | 0.003590 | hsa-miR-96-5p | 3.23 | 0.015466 |
IGF2 and cyclin E1 (CCNE1) were the most
significantly upregulated mRNAs, while coronin 1A (CORO1A) and
MACC1 were the most significantly downregulated mRNAs. The
high-level expression of IGF2 and CCNE1 was due to the
downregulation of hsa-miR-145-5p, hsa-miR-181a-5p, hsa-miR-199a-5p
and hsa-miR-223-3p, and hsa-miR-26a-5p and hsa-miR-26b-5p,
respectively. The low-level expression of CORO1A and MACC1 was due
to the upregulation of hsa-miR-517a-3p, and hsa-miR-18a-5p and
hsa-miR-18b-5p, respectively.
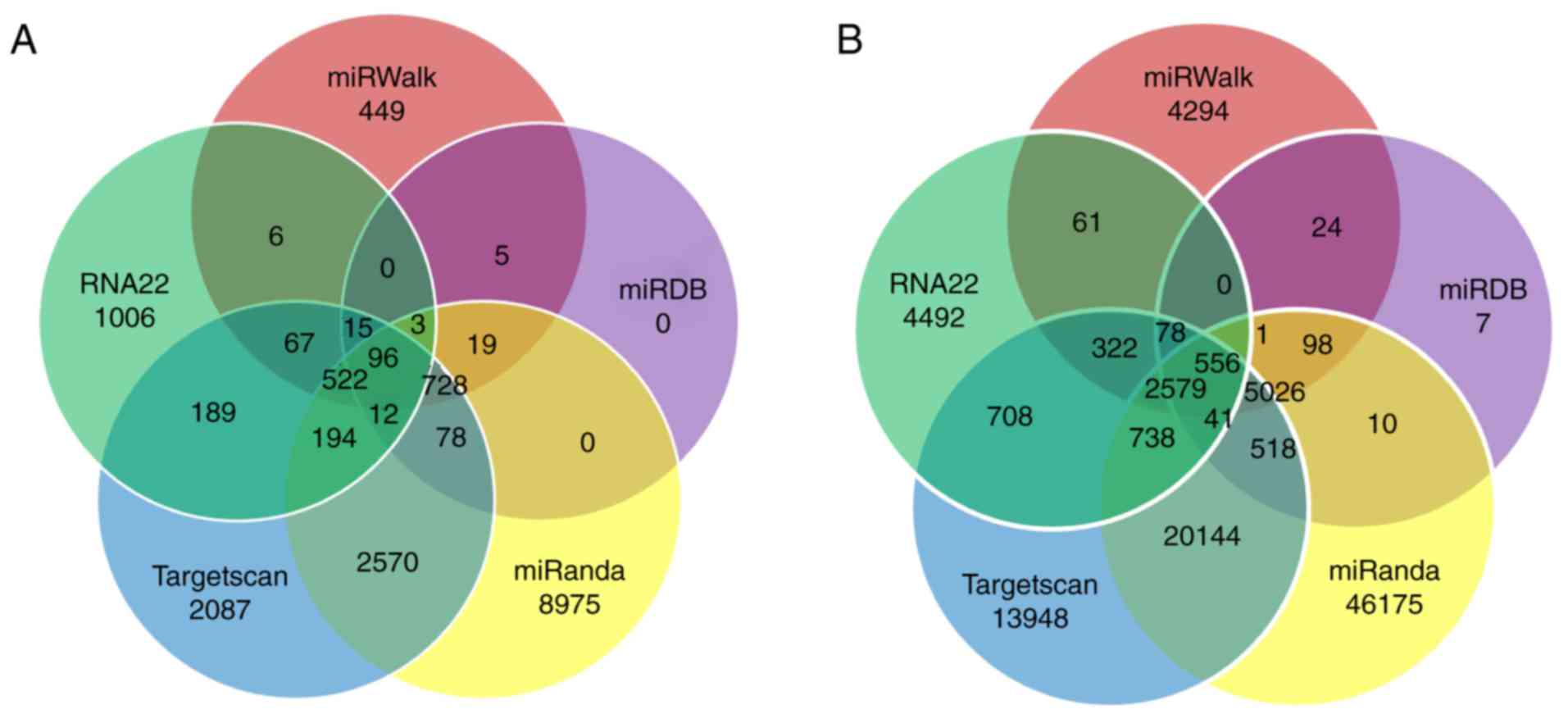
miRNA target prediction
The miRNA target prediction results demonstrated
that a single miRNA may regulate one or more target mRNAs.
Collectively, the four upregulated miRNAs targeted 96 targeted
genes, and the 21 downregulated miRNAs targeted 556 genes. The
differentially expressed miRNA-targeted genes that overlapped
between the five databases (miRanda, miRDB, miRWalk, RNA22 and
Targetscan) are presented in Fig.
2.
GO and KEGG pathway analyses of
differentially expressed miRNAs and mRNAs
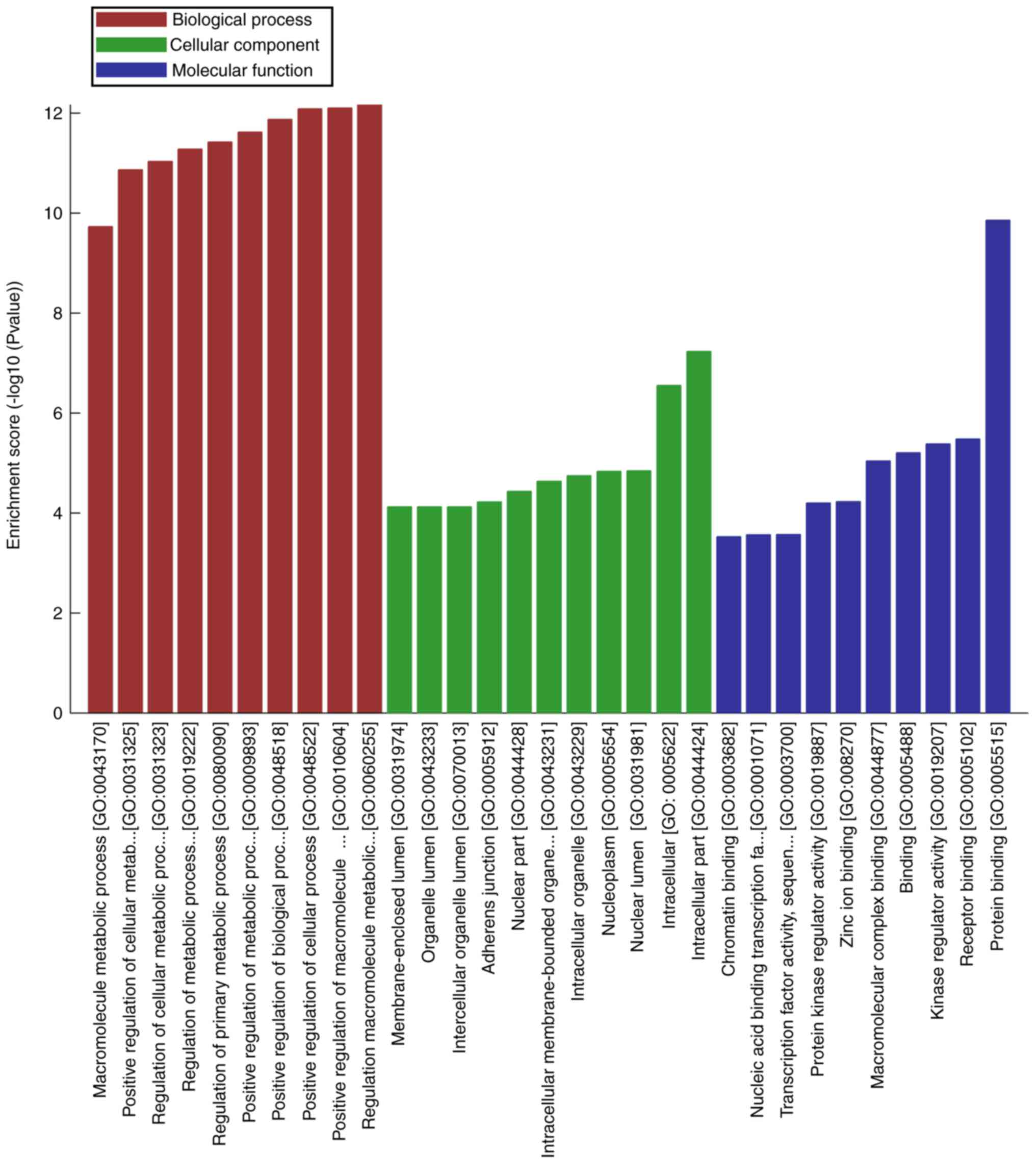
GO terms cover three domains: Biological process
(BP), cellular component (CC) and molecular function (MF). In the
present study, numerous GO terms were associated with oncogenesis,
including: ‘Positive regulation of cellular process’, ‘positive
regulation of biological process’ and ‘positive regulation of
metabolic process’ for BP; ‘intracellular part’, ‘intracellular’
and ‘nuclear lumen’ for CC; and ‘protein binding’, ‘receptor
binding’, ‘kinase regulator activity’, ‘protein kinase regulator
activity’, ‘transcription factor activity’ and ‘nucleic acid
binding transcription factor activity’ for MF. The top 10 most
significantly enriched terms for each category are presented in
Fig. 3.
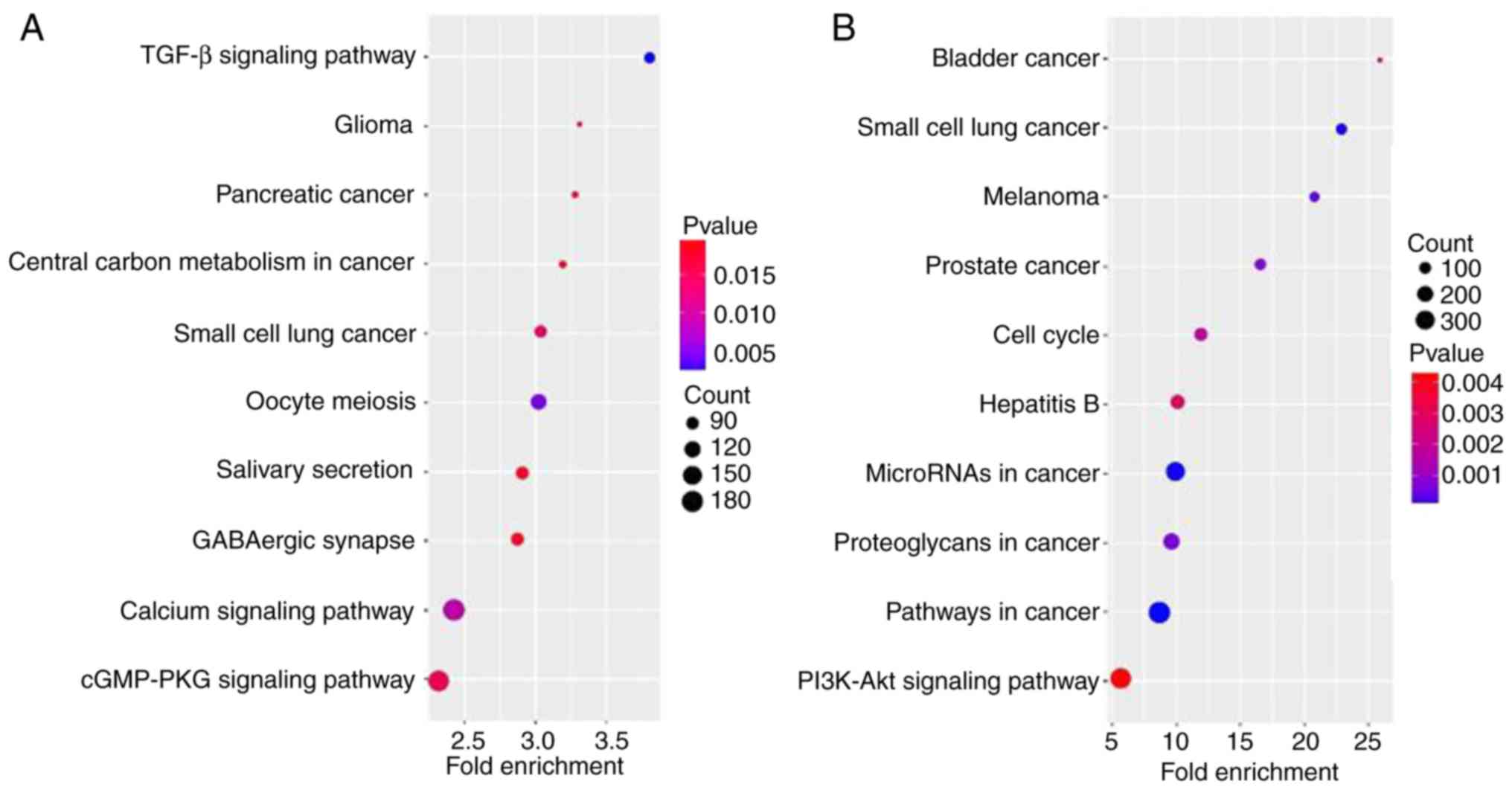
Pathway analysis is a type of functional analysis
that maps genes to KEGG pathways. As signal transduction may be
involved in HCC, the associated pathways were analyzed, according
to the functions and interactions of the differential genes. A
total of 10 pathways associated with the most dysregulated miRNAs
and mRNAs, and with the lowest P-values, were determined. The most
enriched pathways targeted by the dysregulated miRNAs and mRNAs
were associated with cancer, including ‘small cell lung cancer’,
‘pancreatic cancer’, ‘glioma’, ‘prostate cancer’, ‘melanoma’ and
‘bladder cancer’, in addition to oncogenesis pathways, including
‘TGF-beta signaling pathway’, ‘cGMP-PKG signaling pathway’,
‘calcium signaling pathway’, ‘PI3K-Akt signaling pathway’ and
‘pathways in cancer’ (Fig. 4).
This suggested that the dysregulated miRNAs and mRNAs identified in
the present study may regulate the oncogenesis of HCC through these
pathways.
Discussion
The present study identified four upregulated and 28
downregulated miRNAs, and 11 upregulated and five downregulated
mRNAs in HCC tissues compared with their expression levels in NTs.
A single differentially expressed miRNA may regulate one or more
mRNAs. Similarly, a single differentially expressed mRNA may be
regulated by one or more miRNAs. Upregulated miRNAs suppress target
mRNAs, while downregulated miRNAs are matched by an increase in
their target mRNAs. Furthermore, numerous GO terms and enriched
pathways associated with dysregulated miRNAs/mRNAs are associated
with oncogenesis.
In the present study, through coexpression analysis,
it was observed that upregulated hsa-miR-96-5p suppressed the mRNA
expression of FOXO1. Acting as an oncomiR, miR-96 has previously
been identified to be upregulated in HCC and is associated with
metastatic features, including the presence of microvascular
invasion, advanced tumor differentiation and shorter
recurrence-free survival (31).
Ectopic expression of miR-96-5p induces effects on insulin-like
growth factor binding protein-3, IGF2 and insulin-like growth
factor-1 receptor transcripts (32). Additionally, upregulated miR-96
promotes the proliferation of human HCC cells. On the contrary,
downregulated miR-96 led to reduced HCC cell proliferation and
migration (33). However, the
serum level of miR-96 in HCC patients is markedly higher compared
with that in patients with liver cirrhosis or chronic hepatitis B
infection and healthy controls, and may serve as a promising
biomarker in patients with HCC patients and chronic HBV infection
(34).
The upregulated expression of hsa-miR-18b-5p
suppressed the mRNA expression of MACC1 in the present study.
hsa-miR-18, which belongs to the oncomiR-1 or miR-17-92 cluster, is
regarded as an oncogene that is closely associated with the
occurrence and development of different types of cancer. miR-18
targets the expression of numerous genes, including heat shock
factor 2, c-Myc, E2F transcription factor 1 (E2F1), phosphatase and
tensin homolog (PTEN), Bim, tissue growth factor, SMAD family
member 3, transforming growth factor (TGF)-β, trinucleotide repeat
containing 6B, phosphoinositide 3-kinase (PI3K), cellular tumor
antigen p53 and angiogenesis inhibitor thrombospondin-1, to thus
regulate the associated signaling pathways (35). Overexpression of miRs in the
miR-17-92 cluster has been identified during the development of
cirrhosis, and also subsequently during the development of HCC, and
may be regulated by c-Myc and E2F1 (36). miR-18b expression in
poorly-differentiated HCC was reported to be significantly higher
compared with well-differentiated HCC (36). Additionally, overexpression of
miR-18b accelerates cell proliferation and the loss of cell
adhesion ability, and following surgical resection, patients with
HCC exhibiting high miR-18b expression have a significantly shorter
relapse-free period compared with those with low expression
(37). Overall, the concentration
of miR-18a in the plasma/serum of patients with cancer is increased
compared with that of controls, and miR-18 may be a potential
diagnostic biomarker for numerous types of cancer (35,38).
hsa-miR-451a was demonstrated to be downregulated in
HCC tissues in the present study. miR-451 has been demonstrated to
be downregulated in a number of human malignancies and is
correlated with tumor progression. miR-451 was observed to be
downregulated in HCC tissues, and significantly correlated with
advanced clinical stage, metastasis and poorer disease-free or
overall survival (39). miR-451
has been indicated to inhibit cell growth, induce
G0/G1 arrest and promote apoptosis in HCC
cells by regulating the epithelial-mesenchymal transition process.
The oncogene c-Myc is the direct and functional target of miR-451.
miR-451 downregulation-induced c-Myc overexpression leads to the
activation of extracellular signal regulated kinase (ERK)1/2
signaling (38). miR-451 may also
function as a potential suppressor of tumor angiogenesis by
targeting interleukin-6 receptor-signal transducer and activator of
transcription 3-vascular endothelial growth factor signaling in HCC
(40).
Abnormal expression of miR-199a has been observed in
various tumors, where it may influence the regulation of
proliferation, metabolism, invasion, metastasis, angiogenesis and
apoptosis of the tumor cells. The expression of miR-199a has been
identified to be downregulated in non-small cell lung cancer,
colorectal cancer (41), breast
cancer (42), bladder cancer
(43) and esophageal cancer
(44); however, miR-199a is
expressed at a high level in gastric cancer and positively
regulates gastric cancer cell proliferation, migration and invasion
(45). The different expression
patterns of miR-199a in various tumor tissues suggests that
miR-199a may serve as a tumor promoter or tumor suppressor.
However, miR-199a/b-3p is consistently downregulated in HCC, and
its reduction is significantly correlated with a poor survival rate
in patients with HCC (18,46). miR-199a/b-3p may target
tumor-promoting p21 (Rac1) activated kinase 4 (PAK4) to suppress
HCC growth by inhibiting the PAK4/Raf/MEK/ERK pathway in
vitro and in vivo (47). miR-199a-5p and let-7c cooperatively
and efficiently inhibit HCC cell migration and invasion by
targeting the metastasis promoter mitogen-activated protein kinase
kinase kinase kinase 3 (MAP4K3) and, consequently, MAP4K3-mediated
drug sensitization (48).
Decreased expression of miR-199a-5p contributes to increased cell
invasion through functional deregulation of discoidin domain
receptor tyrosine kinase 1 activity in HCC (46). miR-199a is significantly
downregulated in the tissues and sera of patients with HCC.
miR-199a may be used as a potential circulating biomarker for HCC
(49,50). In the present study, downregulated
miR-199a-5p elevated the expression of its targets CDK4 and
IGF2.
GO enrichment and KEGG pathway analyses are widely
used to determine the organization and functional annotation of
molecular components (51,52). The development of HCC is associated
with alterations in molecular functions and biological pathways
(53,54). In the present study, various GO
terms and KEGG pathways were indicated to be involved in HCC
tumorigenesis. A number of BP terms were significantly enriched for
cellular, biological and metabolic processes. Additionally,
numerous MF terms were enriched for protein binding, receptor
binding, protein kinase regulator activity, nucleic acid binding
and transcription factor activity. Thus, through GO enrichment
analysis, it was possible to determine the biological functions of
the differentially expressed genes.
The most enriched pathways were observed to be
associated with cancer (small cell lung cancer, pancreatic cancer,
glioma, prostate cancer, melanoma and bladder cancer) and oncogenic
pathways [TGF-β signaling pathway, cyclic guanosine monophosphate
(cGMP)-protein kinase cGMP-dependent 1 signaling pathway, calcium
signaling pathway, and PI3K-RAC-α serine/threonine-protein kinase
signaling pathway]. Certain differentially expressed miRNAs and
mRNAs have been reported to be associated with small cell lung
cancer, glioma and pancreatic cancer. This three-way association
ought to be the focus of future studies (18).
In conclusion, the present study identified four
upregulated and 28 downregulated miRNAs, and 11 upregulated and
five downregulated mRNAs by PCR microarray. Upregulated
hsa-miR-96-5p and hsa-miR-18b-5p suppressed FOXO1 and MACC1 mRNA
expression. Furthermore, downregulated hsa-miR-199a-5p increased
CDK4 and IGF2 mRNA expression. The high-level expression of IGF2
and CCNE1 mRNAs was a result of hsa-miR-145-5p, hsa-miR-181a-5p and
hsa-miR-199a-5p downregulation, and the low-level expression of
CORO1A and MACC1 mRNAs was a result of upregulated hsa-miR-517a-3p
and hsa-miR-18a-5p, and hsa-miR-18b-5p, respectively. Furthermore,
various GO terms and KEGG pathways associated with the dysregulated
miRNAs and mRNAs were likely involved in HCC tumorigenesis.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Sci-Tech
Support Plan (grant no. 2015BAI02B00) the and Beijing Natural
Science Foundation (grant no. 7174314).
Availability of data and materials
The datasets used or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XWC and SCC was involved in the design of the
experiment, drafted and revised the manuscript, collected and
processed the specimens, was responsible for the collection and
analysis of the data. ZLQ and CL were involved in the design of the
experiment, collected and processed the specimens and provided
final approval of the version to be published. All the authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of Beijing Youan Hospital, Capital Medical University
(Beijing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Herszényi L and Tulassay Z: Epidemiology
of gastrointestinal and liver tumors. Eur Rev Med Pharmacol Sci.
14:249–258. 2010.PubMed/NCBI
|
|
2
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Michielsen P and Ho E: Viral hepatitis B
and hepatocellular carcinoma. Acta Gastroenterol Belg. 74:4–8.
2011.PubMed/NCBI
|
|
4
|
Li G, Cai G, Li D and Yin W: MicroRNAs and
liver disease: Viral hepatitis, liver fibrosis and hepatocellular
carcinoma. Postgrad Med J. 90:106–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zamore PD and Haley B: Ribo-gnome: The big
world of small RNAs. Science. 309:1519–1524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Masaki T: MicroRNA and hepatocellular
carcinoma. Hepatol Res. 39:751–752. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nelson KM and Weiss GJ: MicroRNAs and
cancer: Past, present, and potential future. Mol Cancer Ther.
7:3655–3660. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Djuranovic S, Nahvi A and Green R:
miRNA-mediated gene silencing by translational repression followed
by mRNA deadenylation and decay. Science. 336:237–240. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell1. 16:281–297. 2004.
View Article : Google Scholar
|
|
14
|
Gu Z, Zhang C and Wang J: Gene regulation
is governed by a core network in hepatocellular carcinoma. BMC Syst
Biol. 6:322012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao B, Ning S, Li J, Liu H, Wei W, Wu F,
Tang Y, Feng Y, Li K and Zhang L: Integrated analysis of
differentially expressed mRNAs and miRNAs between hepatocellular
carcinoma and their matched adjacent normal liver tissues. Oncol
Rep. 34:325–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zeng L, Yu J, Huang T, Jia H, Dong Q, He
F, Yuan W, Qin L, Li Y and Xie L: Differential combinatorial
regulatory network analysis related to venous metastasis of
hepatocellular carcinoma. BMC Genomics. 13 Suppl 8:S142012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang D, Tan J, Xu Y, Tan X, Han M, Tu Y,
Zhu Z, Zen J, Dou C and Cai S: Identification of MicroRNAs and
target genes involvement in hepatocellular carcinoma with
microarray data. Hepatogastroenterology. 62:378–382.
2015.PubMed/NCBI
|
|
18
|
Wang W, Zhao LJ, Tan YX, Ren H and Qi ZT:
Identification of deregulated miRNAs and their targets in hepatitis
B virus-associated hepatocellular carcinoma. World J Gastroenterol.
18:5442–5453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han K, Li J, Zhao H, Liang P, Huang X,
Zheng L, Li Y, Yang T and Wang L: Identification of the typical
miRNAs and target genes in hepatocellular carcinoma. Mol Med Rep.
10:229–235. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He TL, Zheng KL, Li G, Song B and Zhang
YJ: Identification of typical miRNAs and target genes in
hepatocellular carcinoma by DNA microarray technique. Eur Rev Med
Pharmacol Sci. 18:108–116. 2014.PubMed/NCBI
|
|
21
|
Wen LM, Wu WZ and Peng XC: Identifying
significant pathways of hepatitis B virus-related hepatocellular
carcinoma based on crosstalk and network pathways. Genet Mol Res.
15:2016. View Article : Google Scholar :
|
|
22
|
Li J, Shi W, Gao Y, Yang B, Jing X, Shan
S, Wang Y and Du Z: Analysis of microRNA expression profiles in
human hepatitis B virus-related hepatocellular carcinoma. Clin Lab.
59:1009–1015. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thurnherr T, Mah WC, Lei Z, Jin Y, Rozen
SG and Lee CG: Differentially expressed miRNAs in hepatocellular
carcinoma target genes in the genetic information processing and
metabolism pathways. Sci Rep. 28:200652016. View Article : Google Scholar
|
|
24
|
Katayama Y, Maeda M, Miyaguchi K, Nemoto
S, Yasen M, Tanaka S, Mizushima H, Fukuoka Y, Arii S and Tanaka H:
Identification of pathogenesis-related microRNAs in hepatocellular
carcinoma by expression profiling. Oncol Lett. 4:817–823. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Guo X, Xiong L, Yu L, Li Z, Guo
Q, Li Z, Li B and Lin N: Comprehensive analysis of
microRNA-regulated protein interaction network reveals the tumor
suppressive role of microRNA-149 in human hepatocellular carcinoma
via targeting AKT-mTOR pathway. Mol Cancer. 13:2532014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He D, Liu ZP, Honda M, Kaneko S and Chen
L: Coexpression network analysis in chronic hepatitis B and C
hepatic lesions reveals distinct patterns of disease progression to
hepatocellular carcinoma. J Mol Cell Biol. 4:140–152. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang Y, Li D, Yang Y and Jiang G: An
integrated analysis of the effects of microRNA and mRNA on
esophageal squamous cell carcinoma. Mol Med Rep. 12:945–952. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Okuda K, Ohtsuki T, Obata H, Tomimatsu M,
Okazaki N, Hasegawa H, Nakajima Y and Ohnishi K: Natural history of
hepatocellular carcinoma and prognosis in relation to treatment.
Study of 850 patients. Cancer. 56:918–928. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bruix J and Sherman M: American
Association for the Study of Liver Diseases: Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leung WK, He M, Chan AW, Law PT and Wong
N: Wnt/β-catenin activates MiR-183/96/182 expression in
hepatocellular carcinoma that promotes cell invasion. Cancer Lett.
362:97–105. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Assal RA, El Tayebi HM, Hosny KA, Esmat G
and Abdelaziz AI: A pleiotropic effect of the single clustered
hepatic metasta miRs miR-96-5p and miR-182-5p on insulin-like
growth factor II, insulin-like growth factor-1 receptor and
insulin-like growth factor-binding protein-3 in hepatocellular
carcinoma. Mol Med Rep. 12:645–650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Baik SH, Lee J, Lee YS, Jang JY and Kim
CW: ANT2 shRNA downregulates miR-19a and miR-96 through the
PI3K/Akt pathway and suppresses tumor growth in hepatocellular
carcinoma cells. Exp Mol Med. 25:e2222016. View Article : Google Scholar
|
|
34
|
Chen Y, Dong X, Yu D and Wang X: Serum
miR-96 is a promising biomarker for hepatocellular carcinoma in
patients with chronic hepatitis B virus infection. Int J Clin Exp
Med. 8:18462–18468. 2015.PubMed/NCBI
|
|
35
|
Komatsu S, Ichikawa D, Takeshita H,
Morimura R, Hirajima S, Tsujiura M, Kawaguchi T, Miyamae M, Nagata
H, Konishi H, et al: Circulating miR-18a: A sensitive cancer
screening biomarker in human cancer. In Vivo. 28:293–297.
2014.PubMed/NCBI
|
|
36
|
Tan W, Li Y, Lim SG and Tan TM:
miR-106b-25/miR-17-92 clusters: Polycistrons with oncogenic roles
in hepatocellular carcinoma. World J Gastroenterol. 20:5962–5972.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Murakami Y, Tamori A, Itami S, Tanahashi
T, Toyoda H, Tanaka M, Wu W, Brojigin N, Kaneoka Y, Maeda A, et al:
The expression level of miR-18b in hepatocellular carcinoma is
associated with the grade of malignancy and prognosis. BMC Cancer.
13:992013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Al-Kafaji G, Al-Naieb ZT and Bakhiet M:
Increased oncogenic microRNA-18a expression in the peripheral blood
of patients with prostate cancer: A potential novel non-invasive
biomarker. Oncol Lett. 11:1201–1206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang JY, Zhang K, Chen DQ, Chen J, Feng
B, Song H, Chen Y, Zhu Z, Lu L, De W, et al: MicroRNA-451:
Epithelial-mesenchymal transition inhibitor and prognostic
biomarker of hepatocelluar carcinoma. Oncotarget. 6:18613–18630.
2015.PubMed/NCBI
|
|
40
|
Liu X, Zhang A, Xiang J, Lv Y and Zhang X:
miR-451 acts as a suppressor of angiogenesis in hepatocellular
carcinoma by targeting the IL-6R-STAT3 pathway. Oncol Rep.
36:1385–1392. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Han Y, Kuang Y, Xue X, Guo X, Li P, Wang
X, Guo X, Yuan B, Zhi Q and Zhao H: NLK, A novel target of
miR-199a-3p, functions as a tumor suppressor in colorectal cancer.
Biomed Pharmacother. 68:497–505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li SQ, Wang ZH, Mi XG, Liu L and Tan Y:
MiR-199a/b-3p suppresses migration and invasion of breast cancer
cells by downregulating PAK4/MEK/ERK signaling pathway. IUBMB Life.
67:768–777. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou M, Wang S, Hu L, Liu F, Zhang Q and
Zhang D: miR-199a-5p suppresses human bladder cancer cell
metastasis by targeting CCR7. BMC Urol. 16:642016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Byrnes KA, Phatak P, Mansour D, Xiao L,
Zou T, Rao JN, Turner DJ, Wang JY and Donahue JM: Overexpression of
miR-199a-5p decreases esophageal cancer cell proliferation through
repression of mitogen-activated protein kinase kinase kinase-11
(MAP3K11). Oncotarget. 7:8756–8770. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Song G, Zeng H, Li J, Xiao L, He Y, Tang Y
and Li Y: miR-199a regulates the tumor suppressor mitogen-activated
protein kinase kinase kinase 11 in gastric cancer. Biol Pharm Bull.
33:1822–1827. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shen Q, Cicinnati VR, Zhang X, Iacob S,
Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G and
Beckebaum S: Role of microRNA-199a-5p and discoidin domain receptor
1 in human hepatocellular carcinoma invasion. Mol Cancer.
9:2272010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong
Q, Qin L, Wu X, Zheng Y, Yang Y, et al: Identification of miRNomes
in human liver and hepatocellular carcinoma reveals miR-199a/b-3p
as therapeutic target for hepatocellular carcinoma. Cancer Cell.
19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu L, Lu L, Zheng A, Xie J, Xue Q, Wang
F, Wang X, Zhou H, Tong X, Li Y, et al: MiR-199a-5p and let-7c
cooperatively inhibit migration and invasion by targeting MAP4K3 in
hepatocellular carcinoma. Oncotarget. 8:13666–13677.
2017.PubMed/NCBI
|
|
49
|
Amr KS, Ezzat WM, Elhosary YA, Hegazy AE,
Fahim HH and Kamel RR: The potential role of miRNAs 21 and 199-a in
early diagnosis of hepatocellular carcinoma. Gene. 575:66–70. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kamel RR, Amr KS, Afify M, Elhosary YA,
Hegazy AE, Fahim HH and Ezzat WM: Relation between microRNAs and
Apoptosis in Hepatocellular Carcinoma. Maced J Med Sci. 4:31–37.
2016. View Article : Google Scholar
|
|
51
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lovering RC, Camon EB, Blake JA and Diehl
AD: Access to immunology through the Gene Ontology. Immunology.
125:154–160. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pinyol R, Nault JC, Quetglas IM,
Zucman-Rossi J and Llovet JM: Molecular profiling of liver tumors:
Classification and clinical translation for decision making. Semin
Liver Dis. 34:363–375. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shanbhogue AK, Prasad SR, Takahashi N,
Vikram R and Sahani DV: Recent advances in cytogenetics and
molecular biology of adult hepatocellular tumors: Implications for
imaging and management. Radiology. 258:673–693. 2011. View Article : Google Scholar : PubMed/NCBI
|