Introduction
Breast cancer (BC) ranks as the most common
malignancy in women worldwide and ranks as the second most common
cause of cancer-associated mortality (1,2). The
incidence of BC is increasing; the latest cancer statistics from
the USA estimated that the expected numbers of new cancer cases and
mortalities could reach 66,120 and 40,920, respectively, in 2018
(3). The human epidermal growth
factor receptor 2 (HER2), progesterone receptor (PR), and estrogen
receptor (ER) were established as the biomarkers of BC, and BC can
be classified into four molecular subtypes depending on the
expression of HER2, PR and ER: HER2(+), triple negative breast
cancer, Luminal A and Luminal B. Currently, advanced therapeutic
approaches have been applied in BC cases to improve the 5-year
survival rate based on the above classification, including
chemotherapy, surgical techniques and adjuvant radiotherapy
(4–8). Nevertheless, the 5-year survival rate
of BC patients with distant metastasis and tumor progression is
only 26%. Additionally, only 1.9% of patients under 50 with BC
received a BC diagnosis, but ~80% of BC patients over 50 received a
BC diagnosis. Therefore, an improved understanding of potential
treatment targets is imperative to improve the 5-year survival rate
and diagnosis of patients with BC (9,10).
MicroRNAs (miRNAs/miRs) are small, non-coding RNAs
of ~22 nucleotides. They regulate the expression of proteins by
silencing the transcripts of target genes or inhibiting the
translation of mRNA (11,12). Extensive studies have established
that miRNAs are crucial in the diagnosis, proliferation, prognosis,
invasion, apoptosis, migration and metastasis of cancer (13–17).
For example, Liang et al suggested that miRNA-10b was a
suppressor in BC growth, migration, proliferation and invasion
(18). Du et al reported
that miR-124 inhibited the proliferation and migration of BC by
targeting snail family transcriptional repressor 2 (19).
Located at 14q32.33 chromosome, miR-203a-3p may
possess a vital role in cancer. It has been reported that
miR-203a-3p can suppress hepatocellular carcinoma progression by
targeting homeobox D3 through the EGFR signaling pathway (20). However, only one study has examined
the role of miR-203a-3p in BC based on 109 BC cases and matched
normal breast (21). Therefore, it
is critical to establish the molecular mechanism of miR-203a-3p in
BC with a large number of samples. The present study estimated the
expression of precursor miR-203a and miR-203a-3p in BC tissue and
adjacent breast tissue by combing data from The Cancer Genome Atlas
(TCGA), Gene Expression Omnibus (GEO) and University of California
Santa Cruz (UCSC) Xena projects. In addition, the potential
molecular mechanisms of miR-203a-3p in BC were investigated through
gene ontology (GO) enrichment, Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathways analysis and protein-protein interaction
(PPI).
Materials and methods
Expression of miRNA in TCGA and UCSC
Xena projects
The TCGA data with level 3 miRNA-Seq profiles and
full annotation of clinical parameters were acquired from TCGA
(http://cancergenome.nih.gov/) (22). Additionally, the expression of
miR-203a-3p was downloaded from the UCSC Xena project (http://xena.ucsc.edu/) (23).
Selection of BC microarrays from GEO
data
The GEO (https://www.ncbi.nlm.nih.gov/geo/) (24) was used to download BC-associated
microarrays with the following prerequisites: (Breast OR mammary)
AND (carcinoma OR tumor OR tumor OR neoplas* OR adenocarcinoma OR
malignan* OR cancer). Microarrays were selected using the following
criteria: The microarrays should include BC tissue and adjacent
breast tissue, and the expression of miR-203a-3p in the two types
of tissue should be provided. A gene expression profile named
GSE50697 was screened to identify the differentially expressed
genes (DEGs).
Selection of prospective DEGs and
target genes of miR-203a-3p in BC
The prospective target genes of miR-203a-3p were
obtained from miRWalk2.0 databases (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
(25), which included 12 online
prediction tools: miRDB, miRNAMap, RNAhybrid, miRBridge, miRMap,
PICTAR2, PITA, MicroT4, TargetScan, miRWalk2.0, miRanda and RNA22.
Prospective target genes were selected if they appeared at least
four times in the above 12 online prediction tools to augment the
accuracy of the prediction. Gene Expression Profiling Interactive
Analysis (http://gepia.cancer-pku.cn/index.html) (26) was performed to acquire the DEGs
from TCGA with P<0.05 and log2 fold change >1. DEGs from GEO
were achieved by using GEO2R (ncbi.nlm.nih.gov/geo/geo2r/) to analyze GSE50697 with
P<0.05 and log2 fold change <-1.
Bioinformatics analyses
Venn diagrams were created to obtain the
intersection of prospective target genes, such as DEGs from GEO and
DEGs from TCGA, and to identify the potential target genes of
miR-203a-3p in BC (27).
Subsequently, GO and KEGG pathway analyses were used to confirm the
potential mechanism of miR-203a-3p in BC (28,29).
PPI analysis was also undertaken using the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) version 9.1
database (https://string-db.org/) (30–32)
to generate an association between the possible target genes and
hub genes were selected by counting the number of edges and
nodes.
Statistical analyses
Student's t-test was used to evaluate statistically
significant differences between two groups. Simultaneously, one way
analysis variance and Dunnett's test were carried out to estimate
statistically significant differences between multiple groups. The
receiver operating characteristic (ROC) curve was adopted to assess
the distinguishability of precursor miR-203a and miR-203a-3p
between BC tissue and adjacent breast tissue. The Kaplan-Meier
survival analysis was undertaken to evaluate the prognostic value
of precursor miR-203a in BC. The log-rank test was used to compare
high and low precursor miR-203a expression groups. STATA version
12.0 (StataCorp LP, College Station, TX, USA) was used to perform
the statistical analyses of the meta-analysis in the present study.
The standard mean difference (SMD) with a random effects model was
used to measure the expression of miR-203a-3p in BC tissue and
adjacent breast tissue. To identify the heterogeneity of the
studies, a heterogeneity test was performed and the level of
I2 calculated simultaneously. An influence analysis was
also conducted to ensure the source of heterogeneity. Concurrently,
a funnel plot asymmetry test was undertaken to assess the
publication bias, with P<0.05 indicating significant publication
bias. The distinguishability of miR-203a-3p in BC tissue and
adjacent breast tissue was estimated using a summarized ROC (sROC)
approach, with an area under the curve (AUC) >0.7 indicting an
ability to distinguish miR-203a-3p in BC. Spearman's correlation
analysis was used to verify the correlation between miR-203a-3p and
target genes based on TCGA data. r>0 and r<0 indicated a
positive and negative correlation, respectively,
Results
Clinical value of precursor miR-203a
and miR-203a-3p in BC, using TCGA and UCSC Xena data
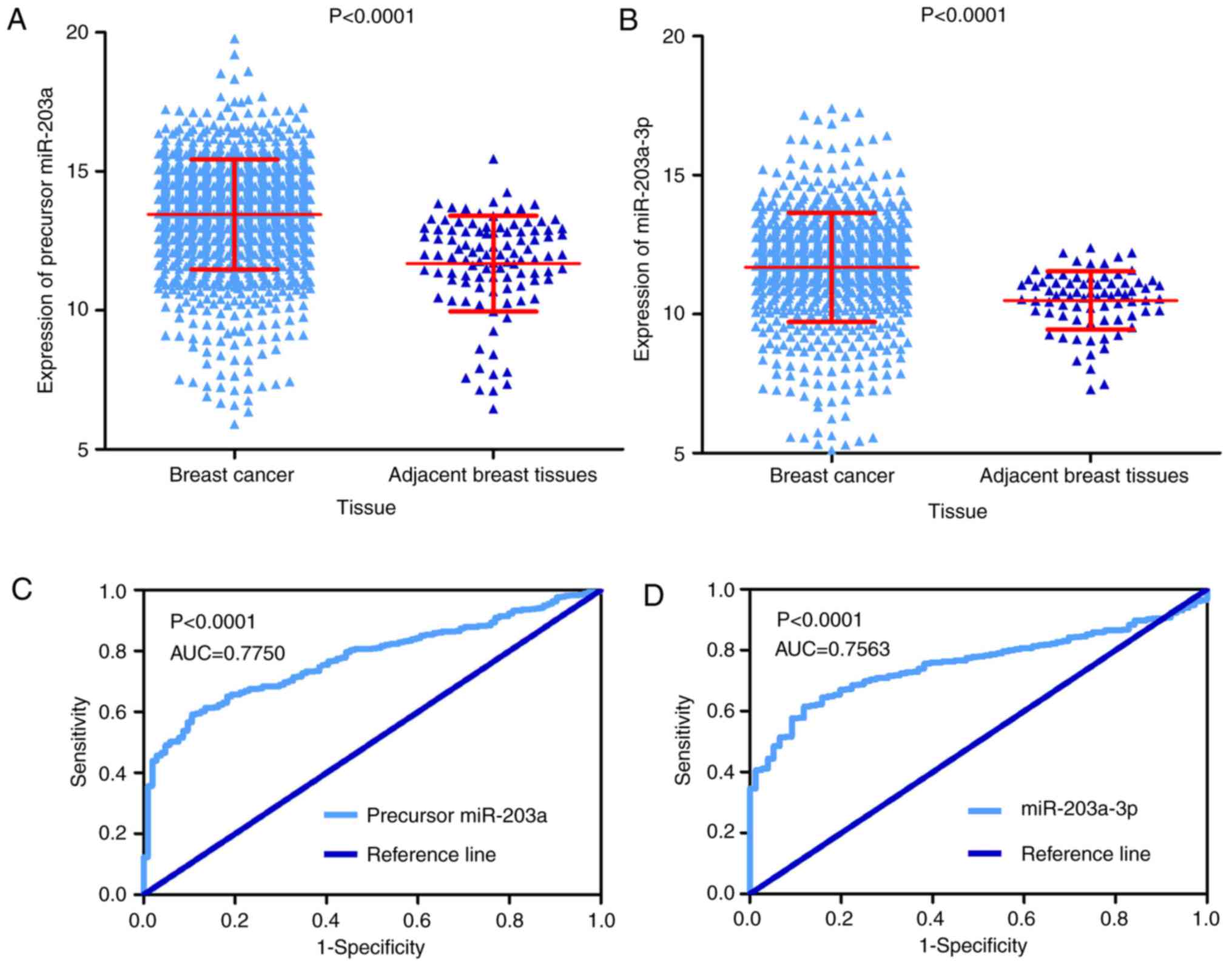
The expression of precursor miR-203a was markedly
elevated in 1,077 BC tissue cases compared to 104 adjacent breast
tissue cases according to TCGA project data (13.45±1.97 vs.
11.69±1.72, P<0.001; Fig. 1A).
Subsequently, the expression of miR-203a-3p was substantially
upregulated in 756 BC tissue cases compared to 76 adjacent breast
tissue cases in UCSC Xena project data (11.68±1.97 vs. 10.49±1.05;
P<0.001; Fig. 1B). Regarding
the distinguishability of precursor miR-203a and miR-203a-3p, the
AUC of ROC curve was 0.775 (P<0.0001; Fig. 1C) with a sensitivity of 59.24% and
a specificity of 89.42%, which implied that precursor miR-203a
could be used to distinguish between BC tissue and adjacent breast
tissue. The AUC of ROC in the UCSC Xena project was 0.756
(P<0.0001; Fig. 1D) with a
sensitivity of 61.51% and a specificity of 88.16 %, which indicated
that miR-203a-3p could be used to distinguish between BC tissue and
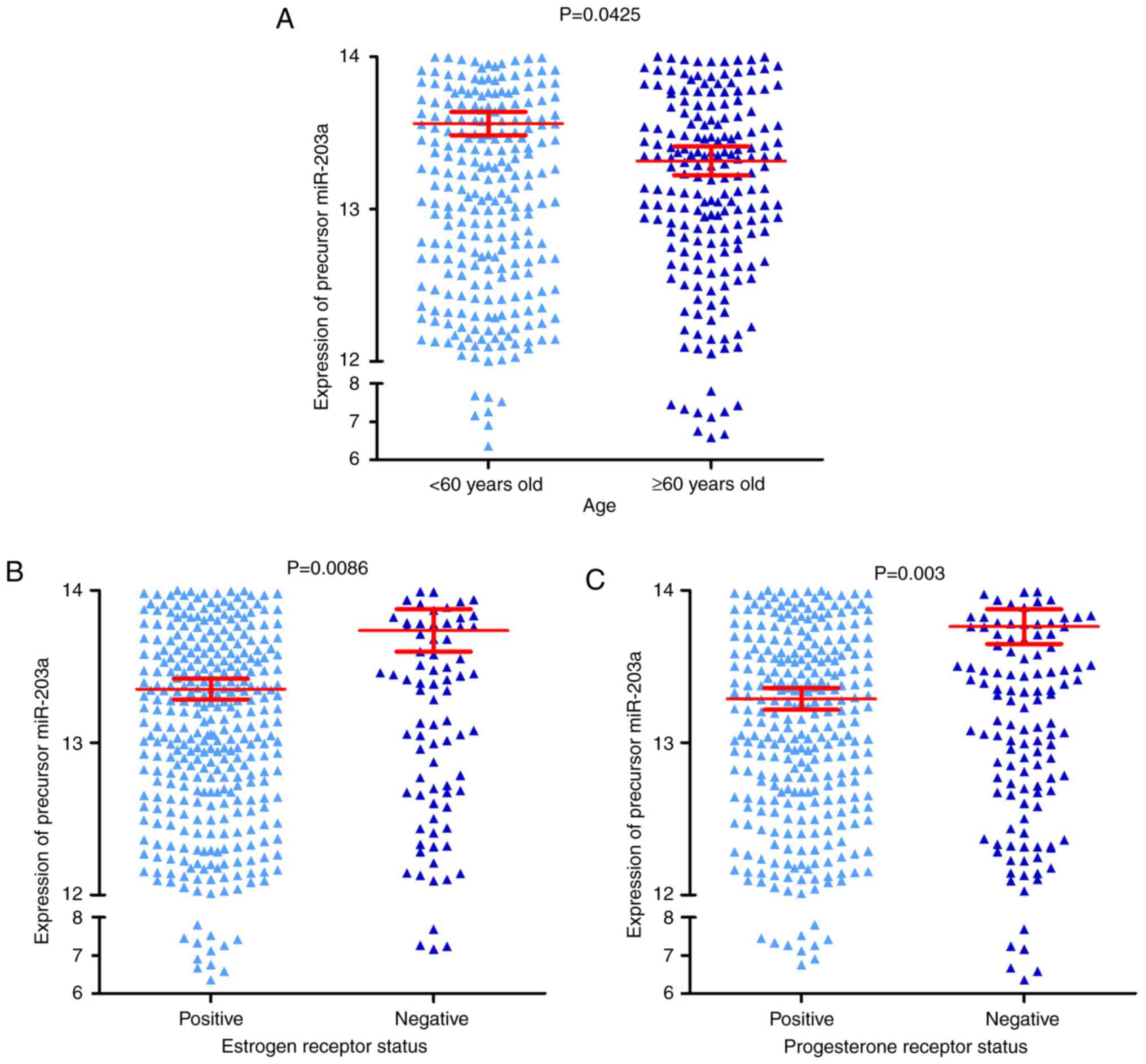
adjacent breast tissue. It was also identified that the expression
of precursor miR-203a was increased in three groups, including the
<60 years old group, the negative ER group and the negative PR
group, compared with their corresponding groups, the ≥60 years old
group, the positive ER group and the positive PR group (all
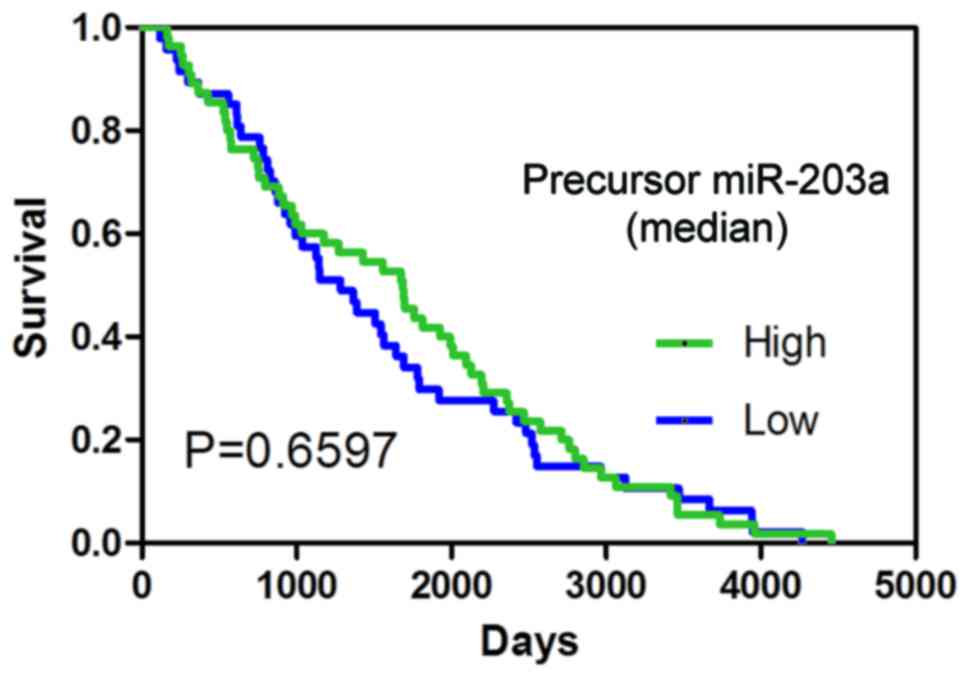
P<0.05; Fig. 2A-C, and Table I). The result of the survival
analysis indicated that precursor miR-203a possessed no prognostic
value in BC (Fig. 3).
 | Table I.Expression of precursor miR-203a in
groups divided from clinical parameters. |
Table I.
Expression of precursor miR-203a in
groups divided from clinical parameters.
|
|
| miR-203a
expression |
|---|
|
|
|
|
|---|
| Clinical
parameters | n | Mean ± standard
deviation | T or F | P-value |
|---|
| Tissue |
|
| 9.83a |
<0.001c |
|
Normal | 104 | 11.69±1.72 |
|
|
| Breast
cancer | 1,077 | 13.45±1.97 |
|
|
| Age (years) |
|
| −2.01a | 0.045c |
|
≤60 | 592 | 13.56±1.88 |
|
|
|
>60 | 485 | 13.32±2.08 |
|
|
| Sex |
|
| −1.10a | 0.295 |
|
Female | 1,065 | 13.46±1.98 |
|
|
|
Male | 12 | 12.91±1.71 |
|
|
| Vital status |
|
| −0.84a | 0.401 |
|
Alive | 975 | 13.43±1.98 |
|
|
|
Dead | 102 | 13.60±1.89 |
|
|
| Pathologic
stage |
|
|
F=1.253b |
|
| Stage
I | 181 | 13.44±1.65 |
| 0.289 |
| Stage
II | 609 | 13.46±2.11 |
|
|
| Stage
III | 244 | 13.50±1.89 |
|
|
| Stage
IV | 20 | 12.61±1.89 |
|
|
| T |
|
|
F=0.707b |
|
| T1 | 279 | 13.54±1.72 |
| 0.548 |
| T2 | 620 | 13.45±2.07 |
|
|
| T3 | 135 | 12.27±2.00 |
|
|
| T4 | 40 | 13.26±2.16 |
|
|
| N |
|
| −1.14a |
|
| No | 508 | 13.37±2.00 |
| 0.254 |
|
Yes | 549 | 13.51±1.96 |
|
|
| M |
|
| 1.81a |
|
| No | 893 | 13.44±1.94 |
| 0.085 |
|
Yes | 21 | 12.69±1.87 |
|
|
| Estrogen receptor
status |
|
| −2.63a |
|
|
Positive | 795 | 13.35±1.91 |
| 0.009c |
|
Negative | 232 | 13.74±2.13 |
|
|
| Progesterone
receptor status |
|
| −3.52a |
|
|
Positive | 689 | 13.29±1.88 |
|
<0.001c |
|
Negative | 335 | 13.76±2.09 |
|
|
| HER2 status |
|
| 0.60a |
|
|
Positive | 164 | 13.55±0.15 |
| 0.549 |
|
Negative | 564 | 13.45±0.08 |
|
|
Clinical value of miR-203a-3p in BC,
using GEO data
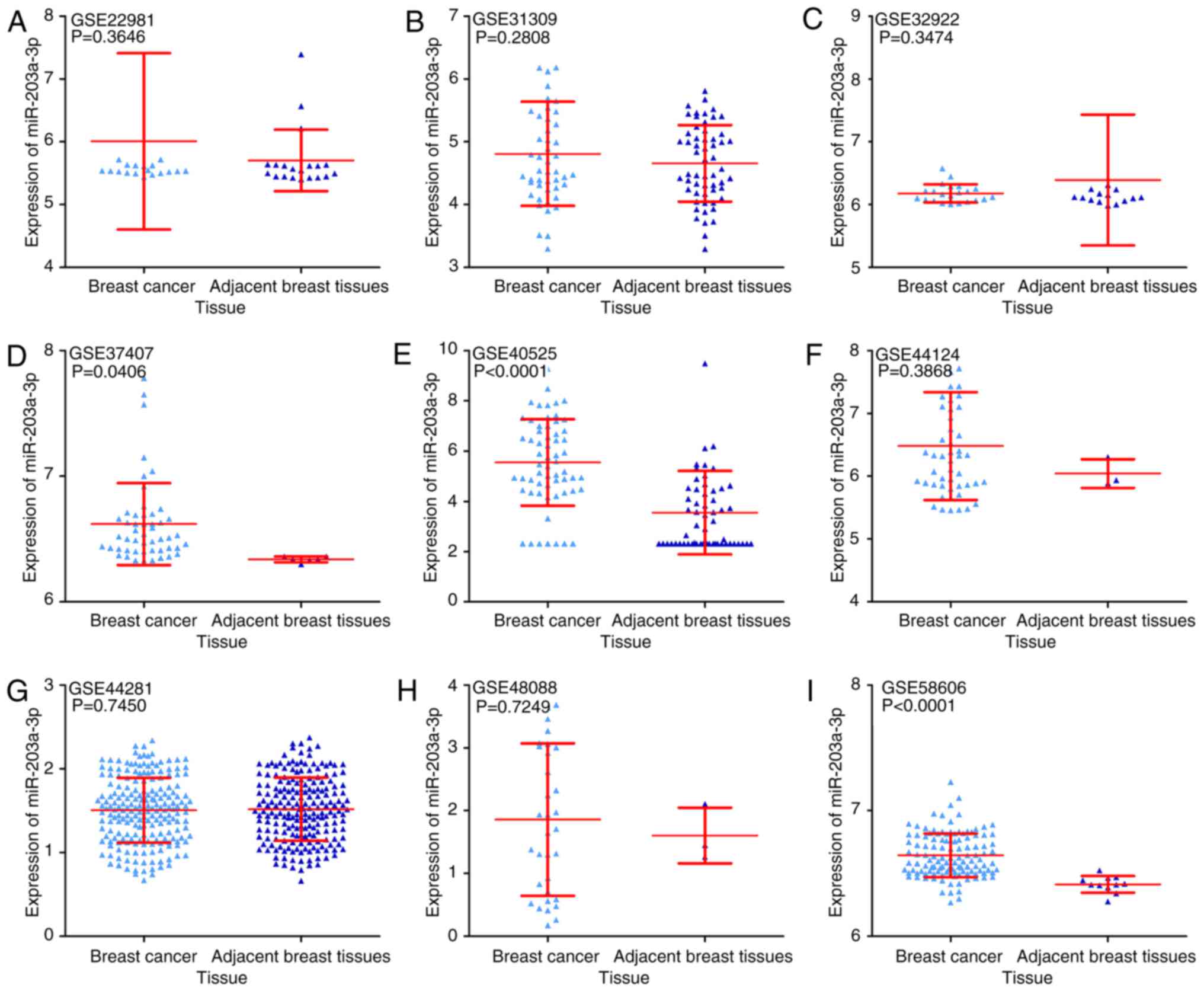
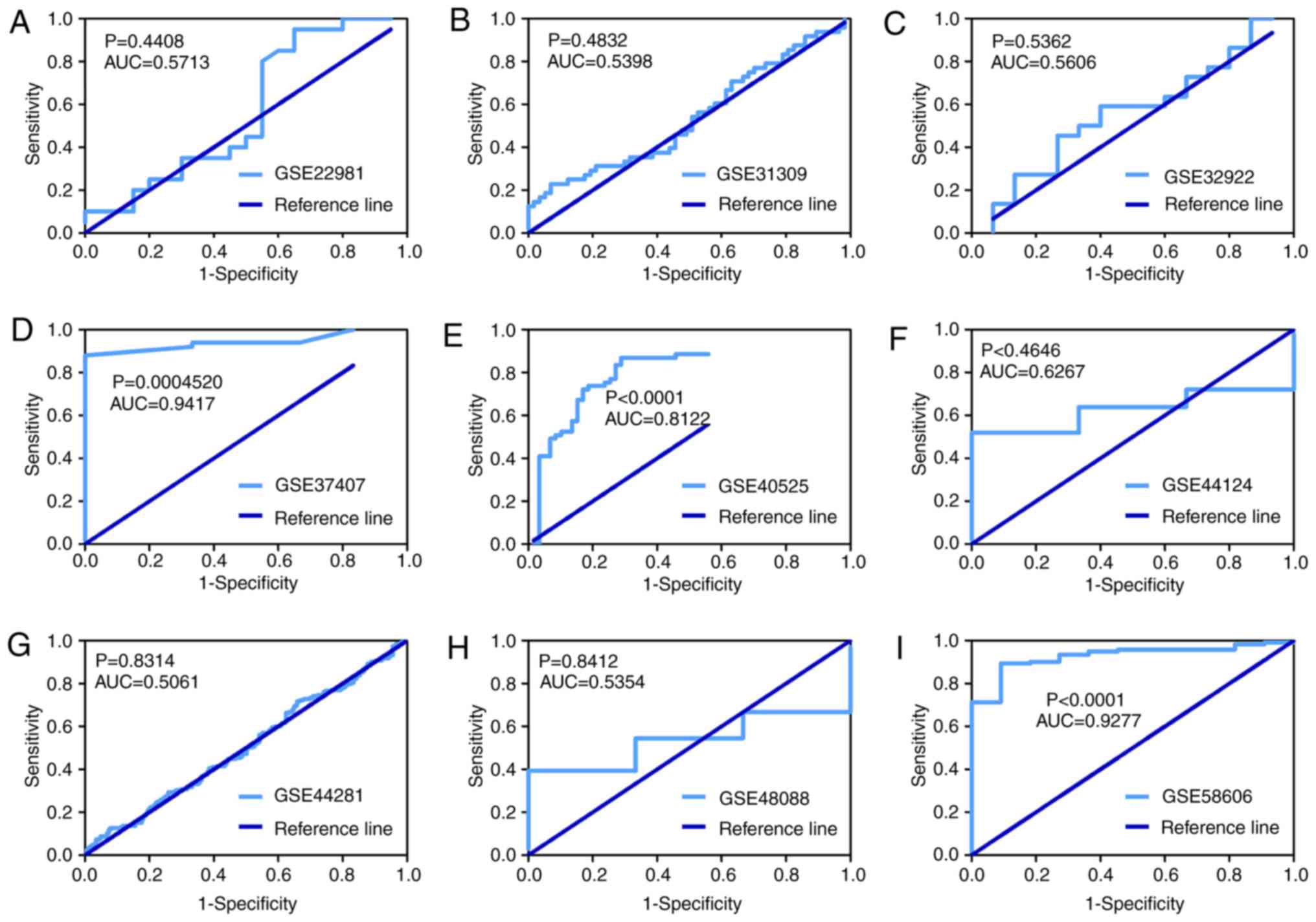
Finally, nine GEO microarrays with 611 BC tissue
samples and 379 adjacent breast tissue samples were selected for
further analysis (Fig. 4). It was
identified that the expression of miR-203a-3p was significantly
upregulated in BC tissue compared with adjacent breast tissue in 3
GEO microarrays (GSE37407, GSE40525 and GSE58606, all P<0.05;
Fig. 5). The ROC curve of these
three microarrays also implied that miR-203a-3p could be used to
distinguish between BC tissue and adjacent breast tissue (Fig. 6).
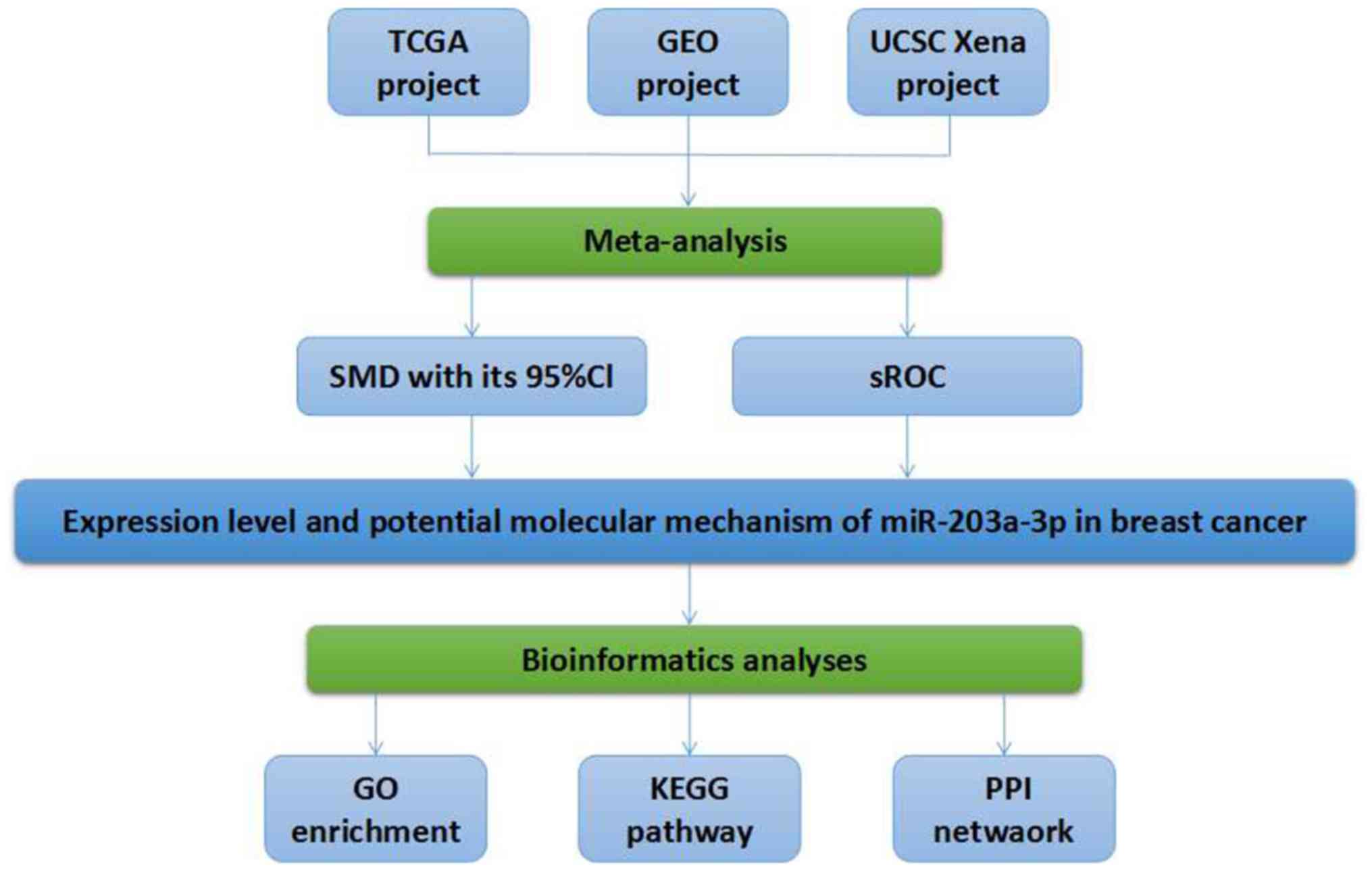 | Figure 4.Flow chart of the present study.
TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; UCSC,
University of California Santa Cruz; SMD, standard mean difference;
CI, confidence interval; sROC, summarized receiver operating
characteristic; miR, microRNA; GO, gene ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; PPI, protein-protein
interaction. |
Meta-analysis
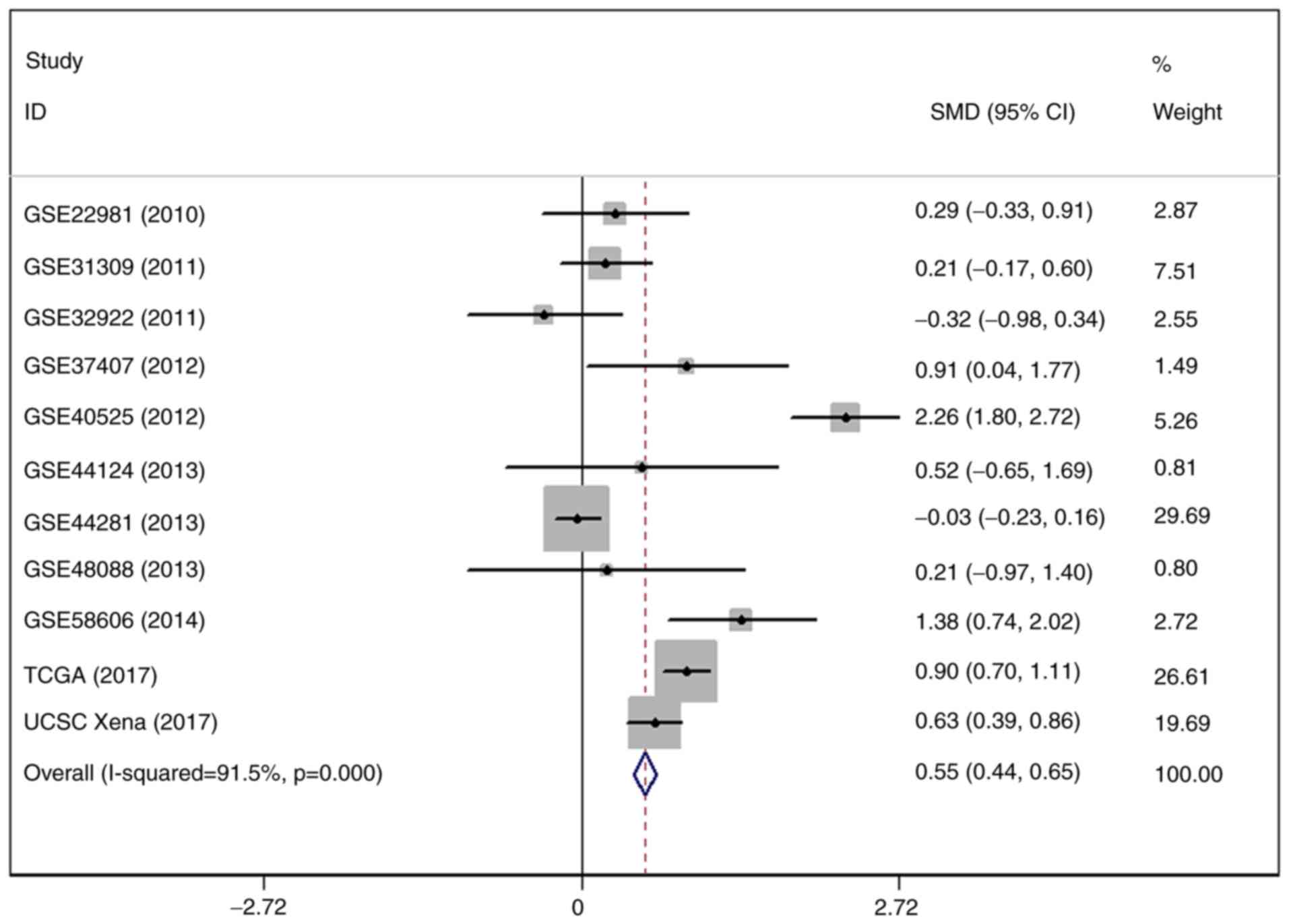
The result of SMD revealed that the expression of
miR-203a-3p was markedly increased in 2,444 BC tissue cases
compared with 559 adjacent breast tissue cases. The heterogeneity
test indicated that there was significant heterogeneity in the
included studies (I2=91.5%; P=0.000; 95% CI, 0.44–0.65;
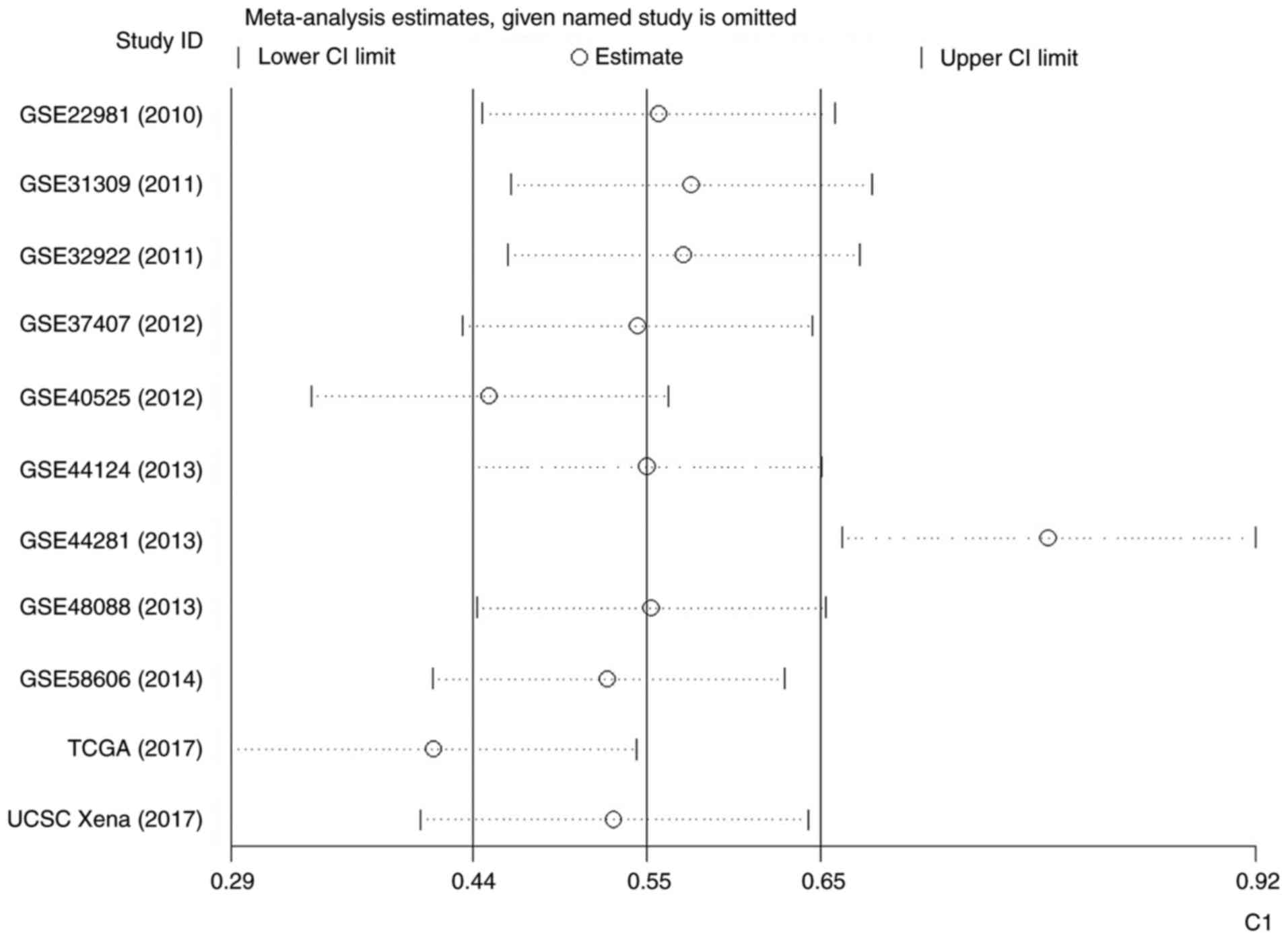
Fig. 7). Therefore, an influence
analysis was conducted to seek the source of heterogeneity and it
was identified that GSE44281 was significantly different from the
other 10 studies (Fig. 8).
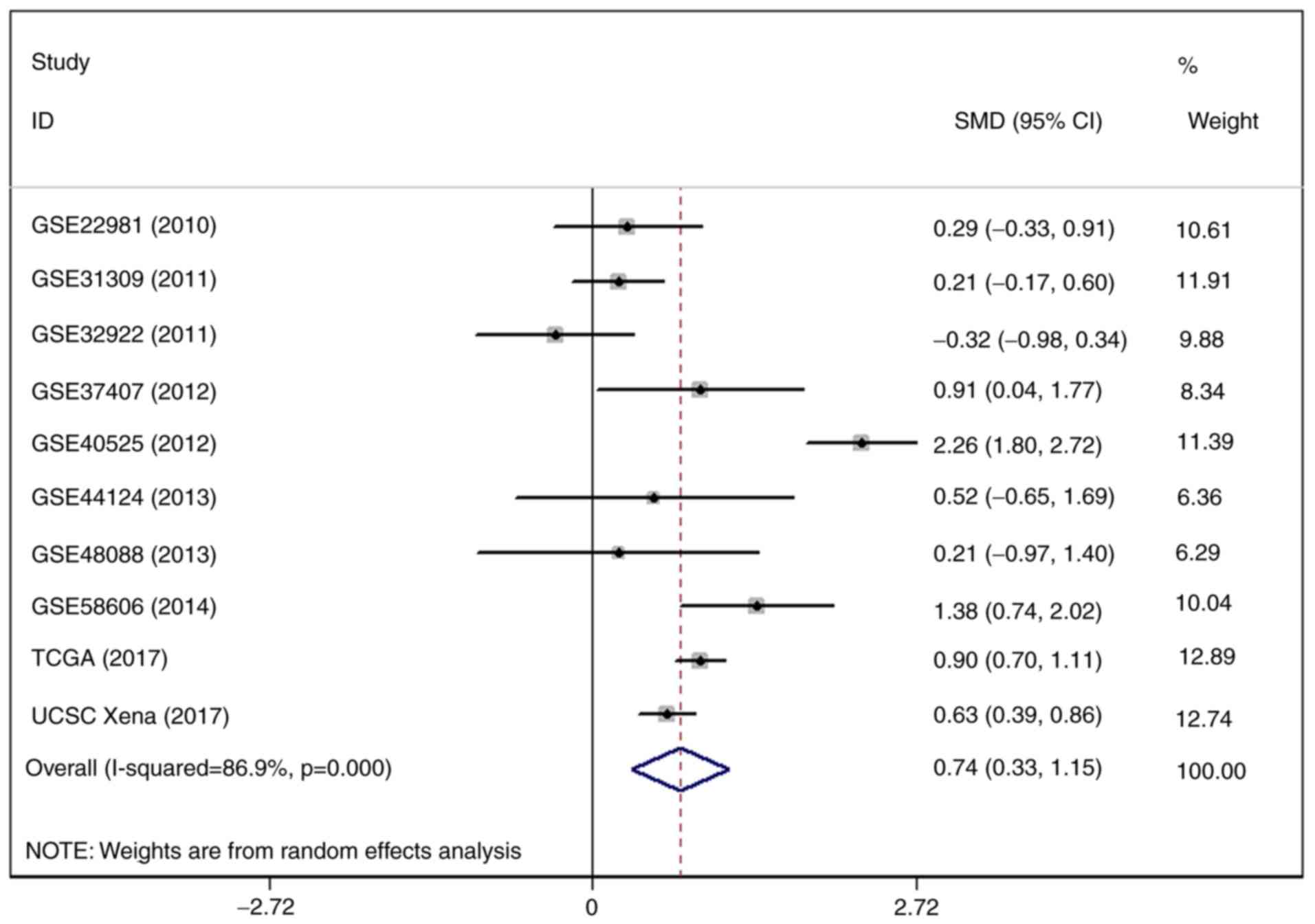
Following the omission of GSE44281, the level of I2 was
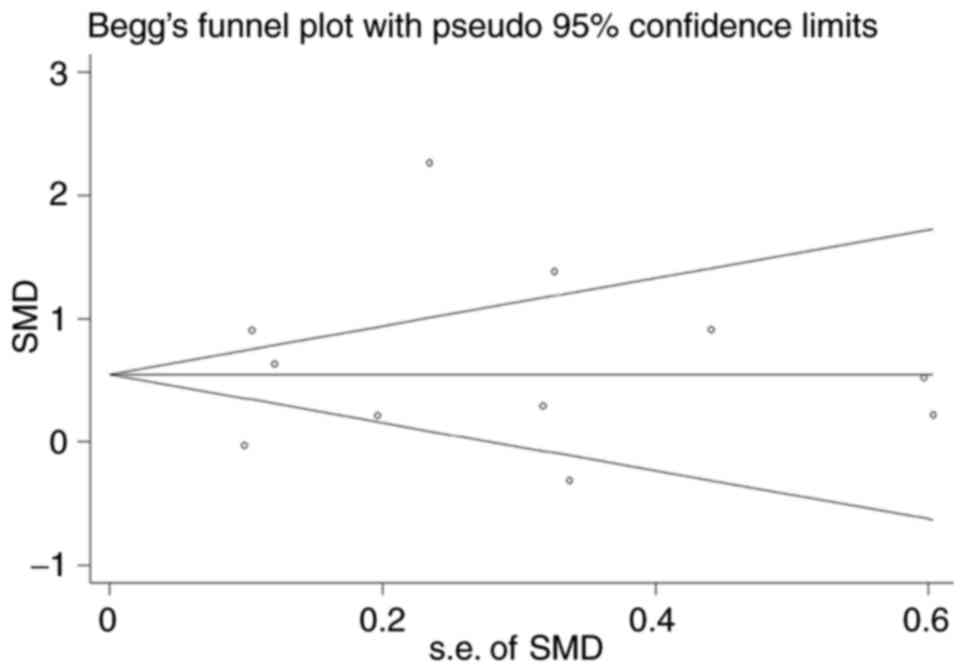
decreased, but still reached 86.9% (Fig. 9). The outcome of a funnel plot
asymmetry test indicated that no publication bias was identified in
the included studies (Fig. 10).
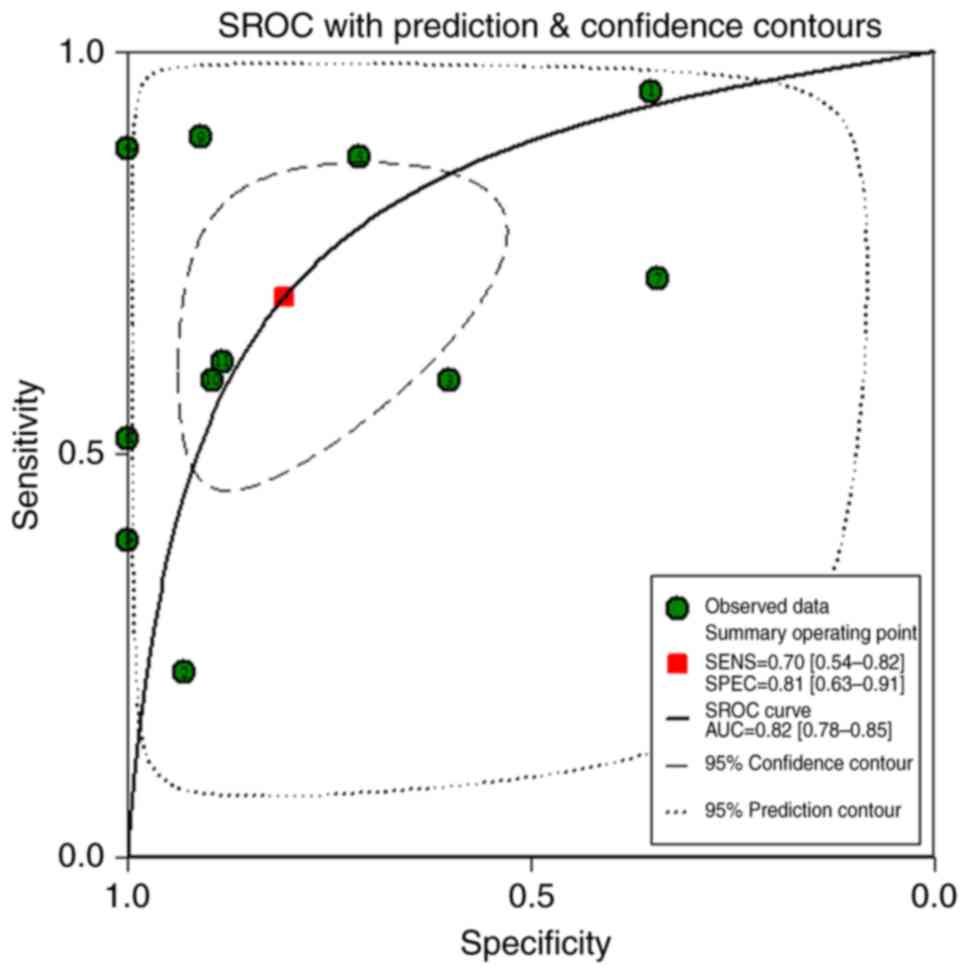
The AUC of sROC reached 0.82 with a sensitivity of 0.70 (0.54–0.82)
and a specificity of 0.81 (0.63–0.91), which implied that
miR-203a-3p could be used to distinguish between BC tissue and
adjacent breast tissue (Figs. 11
and 12).
GO enrichment, KEGG pathway analyses
and PPI network
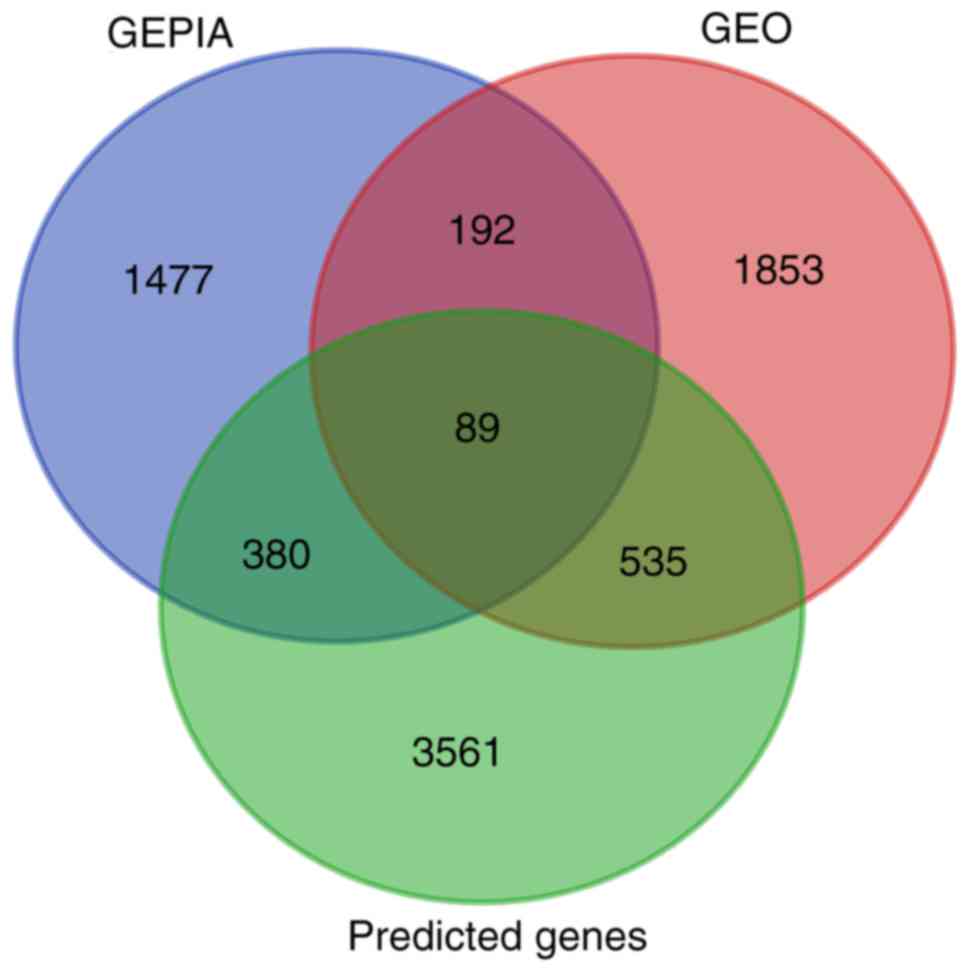
Online prediction tools were used to acquire a total
of 4,565 predicted target genes, which had to appear at least four
times in searches to qualify. Meanwhile, 2,669 DEGs from GEO and
2,138 DEGs from TCGA were acquired. Of these DEGs, 89 genes
intersected with the predicted target genes (Fig. 13). The result of GO enrichment
analysis indicated that the overlapped genes were associated with
‘plasma membrane integrity’, ‘cell surface receptor linked signal
transduction’ and ‘3′,5′-cyclic nucleotide phosphodiesterase
activity’ (Fig. 14; Table II). In addition, a pathway termed
‘purine metabolism’ was identified to be closely associated with
miR-203a-3p expression in BC via its target genes, including
phosphodiesterase 1C (PDE1C), adenylate cyclase 5 (ADCY5),
phosphodiesterase 1A (PDE1A), phosphodiesterase 5A (PDE5A) and
phosphodiesterase 8B (PDE8B; Table
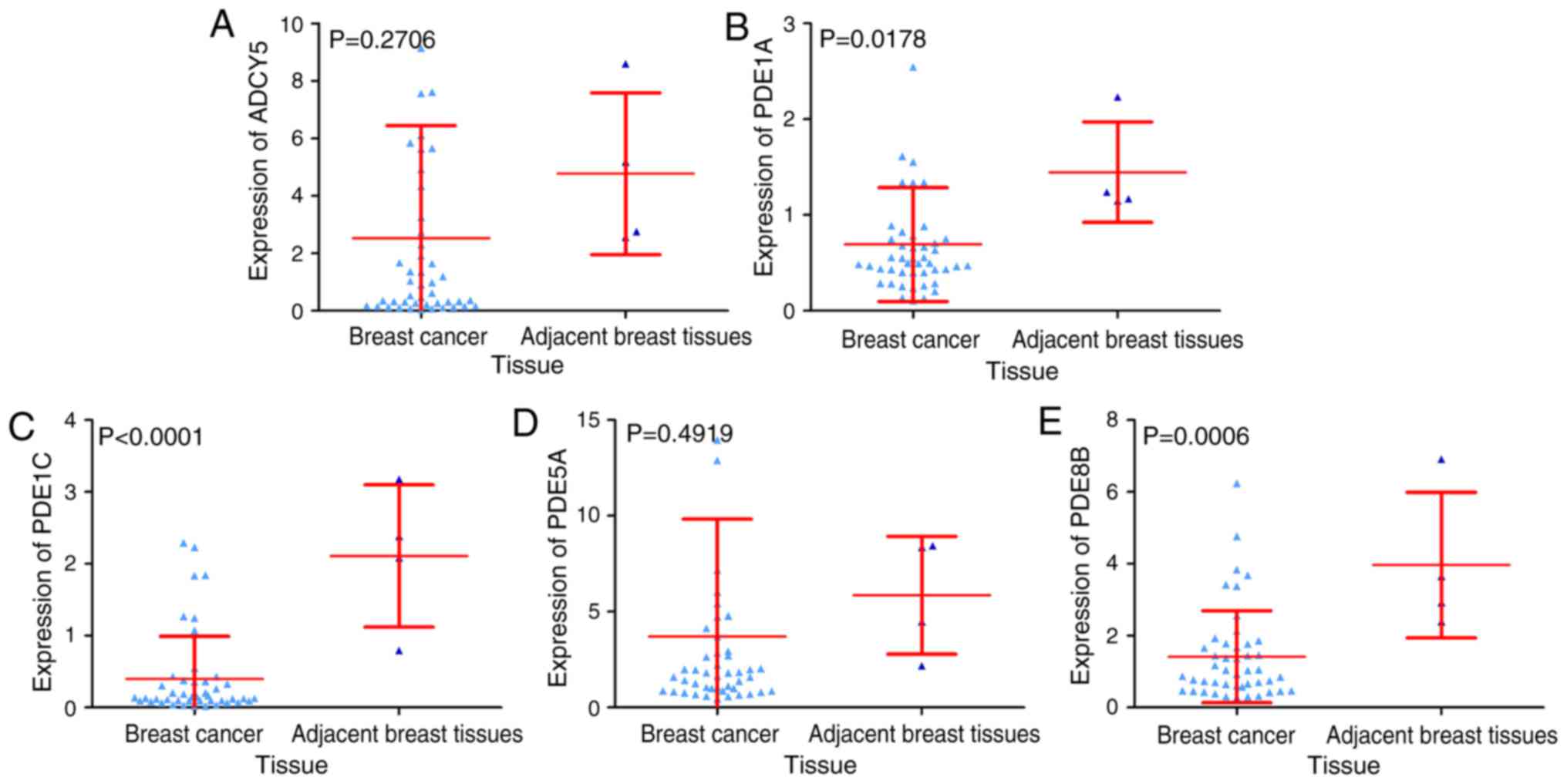
III). Notably, the expression of three genes (PDE1A, PDE1C and
PDE8B) was significantly reduced in BC tissue compared with
adjacent breast tissue. The other genes demonstrated a reduced
trend in BC tissue compared to adjacent breast tissue, but no
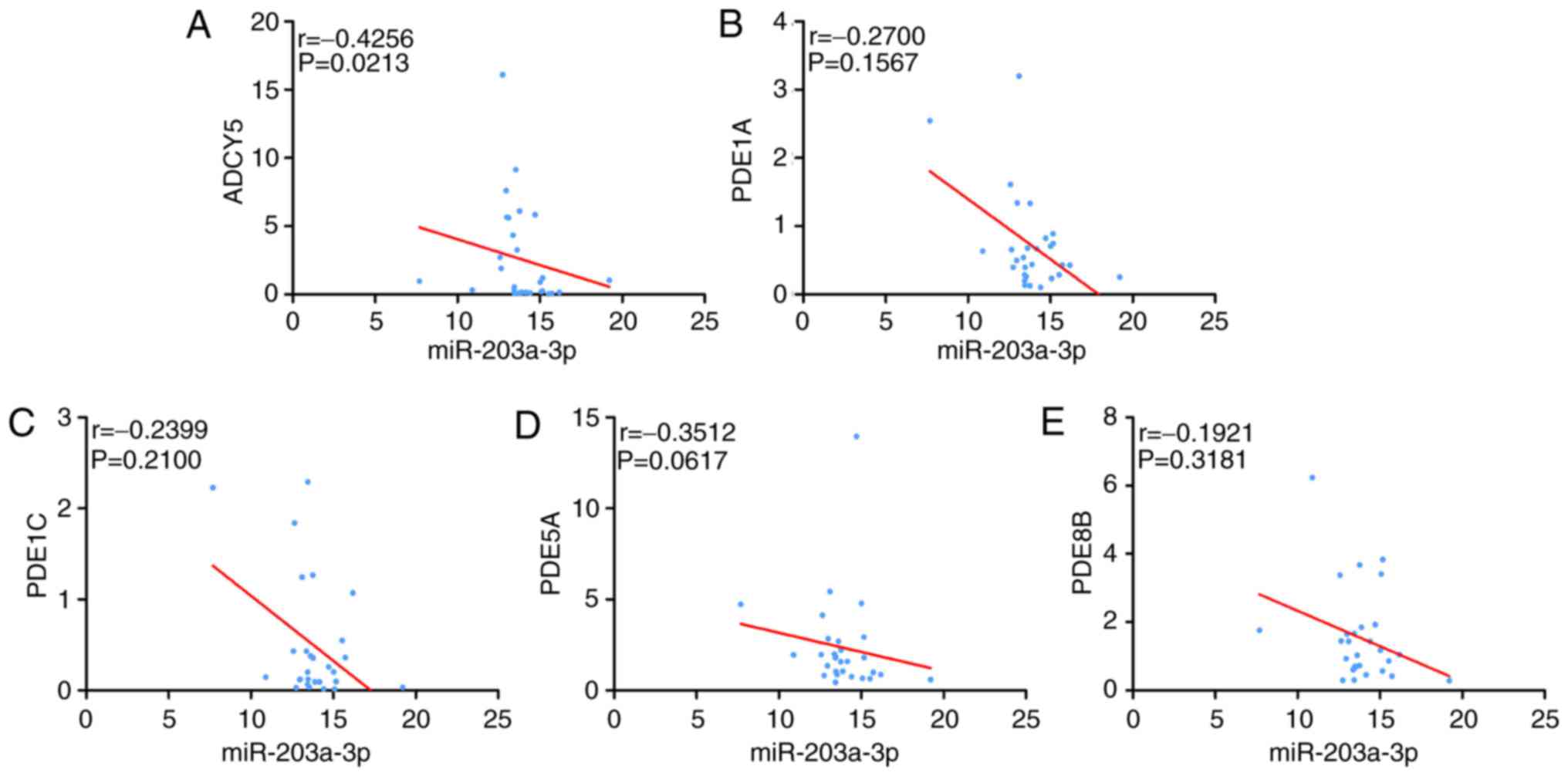
statistical significance was observed (Fig. 15). Spearman's correlation analysis
identified that ADCY5 was negatively correlated with miR-203a-3p. A
minor negative correlation was identified between the other four
genes and miR-203a-3p, but no statistical significance was observed
(Fig. 16). The ROC demonstrated
that all these genes could be used to distinguish between BC tissue
and adjacent breast tissue (Fig.
17). Through the PPI network, four hub genes were identified:
Epidermal growth factor receptor, ADCY5, metalloproteinase
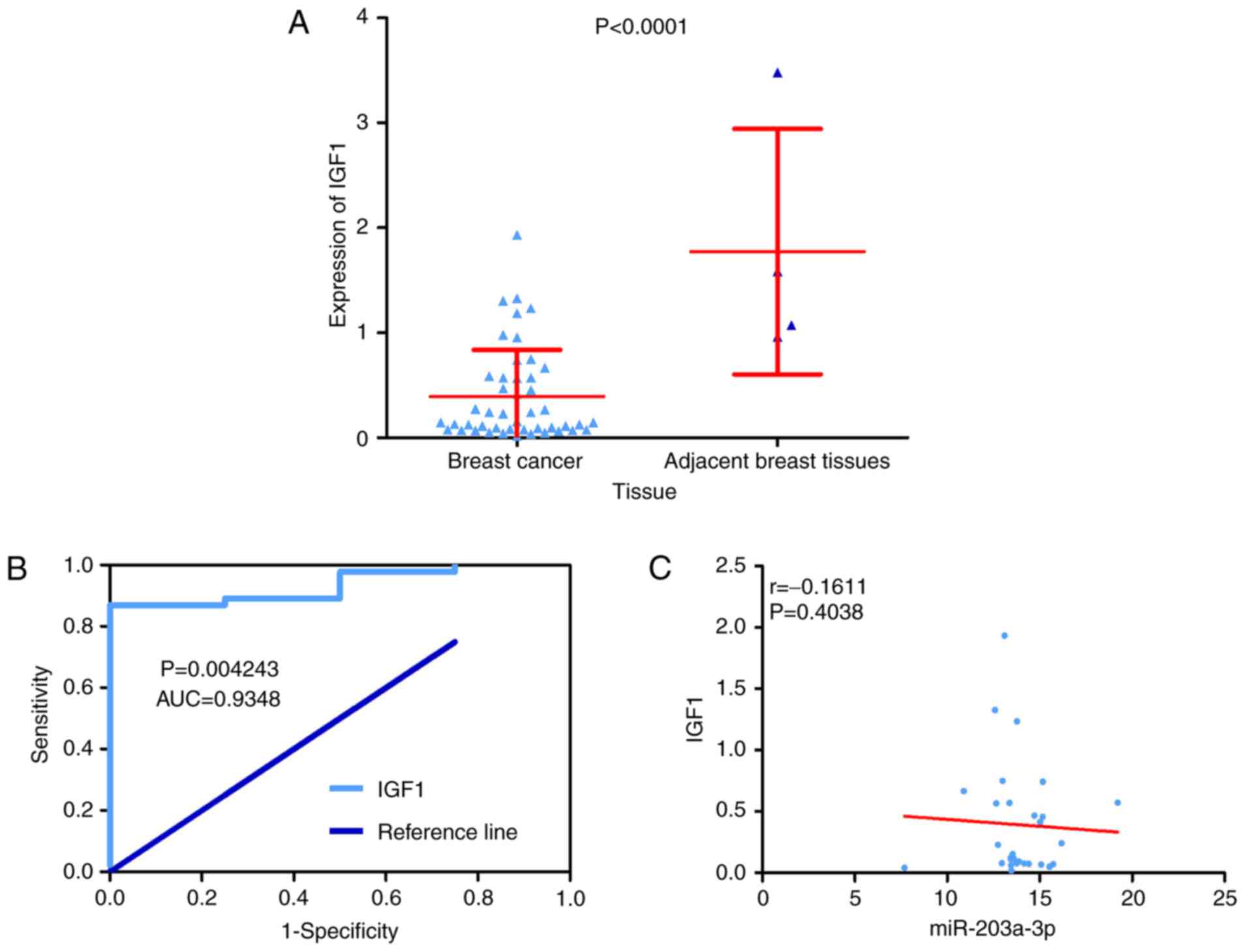
inhibitor 3 and insulin-like growth factor 1 (IGF1; Fig. 18). Depending on data from TCGA, it
was identified that only IGF1 was predominantly decreased in BC
tissue compared with adjacent breast tissue (Figs. 15A, 19 and 20A). As the expression of the hub genes
should be decreased in BC tissue compared with adjacent breast
tissue, IGF1 was identified as the hub gene of miR-203a-3p in BC.
Furthermore, IGF1 exhibited a distinction between BC tissue and
adjacent breast tissue with an AUC of ROC that reached 0.9348
(Fig. 20B). Additionally, a
slight negative correlation was identified between IGF1 and
miR-203a-3p according to the Spearman's correlation analysis;
however, the correlation was not statistically significant
(r=−0.1611; P=0.4038; Fig.
20C).
 | Table II.GO enrichment of the 89 overlapped
genes. |
Table II.
GO enrichment of the 89 overlapped
genes.
| GO ID | Term | Count | Ontology | P-value |
|---|
| GO:0007166 | Cell surface
receptor linked signal transduction | 22 | BP |
3.877×10−4 |
| GO:0007167 | Enzyme linked
receptor protein signaling pathway | 8 | BP |
2.061×10−3 |
| GO:0019932 |
Second-messenger-mediated signaling | 6 | BP |
7.787×10−3 |
| GO:0030335 | Positive regulation
of cell migration | 4 | BP |
1.140×10−2 |
| GO:0030334 | Regulation of cell
migration | 5 | BP |
1.194×10−2 |
| GO:0009725 | Response to hormone
stimulus | 7 | BP |
1.247×10−2 |
| GO:0007242 | Intracellular
signaling cascade | 14 | BP |
1.281×10−2 |
| GO:0050806 | Positive regulation
of synaptic transmission | 3 | BP |
1.367×10−2 |
| GO:0040017 | Positive regulation
of locomotion | 4 | BP |
1.477×10−2 |
| GO:0051272 | Positive regulation
of cell motion | 4 | BP |
1.477×10−2 |
| GO:0005887 | Integral to plasma
membrane | 16 | CC |
6.200×10−4 |
| GO:0031226 | Intrinsic to plasma
membrane | 16 | CC |
7.800×10−4 |
| GO:0044459 | Plasma membrane
part | 22 | CC |
1.800×10−3 |
| GO:0016021 | Integral to
membrane | 37 | CC |
1.200×10−2 |
| GO:0044433 | Cytoplasmic vesicle
part | 5 | CC |
1.400×10−2 |
| GO:0005886 | Plasma
membrane | 28 | CC |
2.100×10−2 |
| GO:0031224 | Intrinsic to
membrane | 37 | CC |
2.100×10−2 |
| GO:0031091 | Platelet α
granule | 3 | CC |
3.200×10−2 |
| GO:0030659 | Cytoplasmic vesicle
membrane | 4 | CC |
3.200×10−2 |
| GO:0012506 | Vesicle
membrane | 4 | CC |
4.000×10−2 |
| GO:0004114 |
3′,5′-cyclic-nucleotide phosphodiesterase
activity | 4 | MF |
3.300×10−4 |
| GO:0004112 | Cyclic-nucleotide
phosphodiesterase activity | 4 | MF |
3.740×10−4 |
| GO:0008081 | Phosphoric diester
hydrolase activity | 5 | MF |
1.330×10−3 |
| GO:0004117 |
Calmodulin-dependent cyclic-nucleotide
phosphodiesterase activity | 2 | MF |
1.700×10−2 |
| GO:0003690 | Double-stranded DNA
binding | 4 | MF |
1.788×10−2 |
| GO:0003779 | Actin binding | 6 | MF |
3.819×10−2 |
| GO:0008144 | Drug binding | 3 | MF |
4.182×10−2 |
| GO:0043566 | Structure-specific
DNA binding | 4 | MF |
4.981×10−2 |
| GO:0004714 | Transmembrane
receptor protein tyrosine kinase activity | 3 | MF |
5.588×10−2 |
| GO:0060090 | Molecular adaptor
activity | 3 | MF |
5.588×10−2 |
 | Table III.KEGG pathway of the 89 overlapped
genes. |
Table III.
KEGG pathway of the 89 overlapped
genes.
| ID | Term | Count | P-value |
|---|
| hsa00230: | Purine
metabolism | 5 |
9.189×10−3 |
| hsa04020: | Calcium signaling
pathway | 5 |
1.483×10−2 |
| hsa05214: | Glioma | 3 |
4.645×10−2 |
| hsa05218: | Melanoma | 3 |
5.755×10−2 |
| hsa05212: | Pancreatic
cancer | 3 |
5.899×10−2 |
| hsa04914: |
Progesterone-mediated oocyte
maturation | 3 |
8.051×10−2 |
| hsa05215: | Prostate
cancer | 3 |
8.540×10−2 |
| hsa04666: | Fc γR-mediated
phagocytosis | 3 |
9.545×10−2 |
| hsa00230: | Purine
metabolism | 5 |
9.189×10−3 |
Discussion
Previous studies have identified that miR-203a-3p is
significantly associated with various cancers; a trend of
miR-203a-3p elevation has been detected in hepatocellular (33) and colorectal (34) carcinoma. By contrast, downregulated
miR-203a-3p expression was detected in gastric cancer (35), prostate cancer (36), non-small-cell lung carcinoma
(37) and esophageal cancer
(38). However, only one study has
identified the expression and potential functions of miR-203a-3p in
BC; Gomes et al (21)
reported that the expression of miR-203a-3p was markedly
upregulated in 109 BC samples compared with matched normal breast
samples and also identified that upregulated expression of
miR-203a-3p was established in five clinic pathological
characteristics groups: Tumor size ≤18.5 mm, HER2-negative,
PR-positive, ER-positive and high Ki-67 index groups.
Since the sample size of the study by Gomes et
al (21) was not large or
varied enough, the current study combined data from three projects
with a larger sample size to ensure the accuracy of the results. It
was identified that the expression of precursor miR-203a was
significantly elevated in 1,077 BC tissue samples compared with 104
adjacent breast tissue samples in TCGA project data. In the UCSC
Xena project, the expression of miR-203a-3p was significantly
increased in 756 BC tissue cases compared with 76 adjacent breast
tissue cases. In addition, an elevated trend was detected in BC
tissues compared with adjacent breast tissue in three GEO
microarrays. The outcome of the comprehensive meta-analysis
indicated that the expression of miR-203a-3p trended toward
overexpression in 2,444 BC tissue cases compared with 559 adjacent
breast tissue cases. Additionally, ROC and sROC suggested that
miR-203a-3p could be used to distinguish between BC tissue and
adjacent breast tissue. It was detected that upregulated
miR-203a-3p was associated with age (<60-year-old patients),
PR-negative BC tissue and ER-negative BC tissue. Regarding the
prognosis value of miR-203a-3p in BC, no prognostic value was
observed. Taken together, it was hypothesized that miR-203a-3p may
enhance the development and oncogenesis of BC.
GO enrichment and KEGG pathway analyses were
conducted to identify the potential molecular mechanism of the role
of miR-203a-3p in BC. The predicted miR-203a-3p target genes were
significantly enriched in three biological processes: ‘Plasma
membrane’, ‘cell surface receptor linked signal transduction’ and
‘3′,5′-cyclic nucleotide phosphodiesterase activity’. Therefore, it
was hypothesized that miR-203a-3p may influence BC via the above
processes. In addition, a pathway termed ‘purine metabolism’ was
closely associated with miR-203a-3p target genes. The expression
and ROCs of the pathway-related genes were assessed; the expression
of three genes (PDE1C, PDE1A and PDE8B) was significantly decreased
in BC tissue compared with adjacent breast tissue and the
expression of other genes (PDE5A and ADCY5) was marginally reduced
in BC tissue compared with adjacent breast tissue, but the change
was not statistically significant. ROCs from these five genes
indicated that each was able to distinguish BC from adjacent normal
tissue. In addition, it was detected that ADCY5 expression was
negatively correlated with miR-203a-3p expression. Taken together,
the findings indicate that miR-203a-3p may be involved in purine
metabolism in BC by targeting ADCY5, PDE1C, PDE1A, PDE5A and
PDE8B.
Finally, the hub gene IGF1 was selected for further
investigation. IGF1 is regarded as a vital gene in regulating cell
differentiation, apoptosis and proliferation in BC. IGF1
polymorphisms may enhance the risk for BC (39). De Santi et al (40) demonstrated that IGF1 is comprised a
pro-form and mature form. The IGF1 pro-form enhances cell
proliferation in BC via IGF1 receptor. The current study evaluated
the expression and diagnostic ability of IGF1 in BC tissue and
adjacent breast tissue. It was identified that the expression of
IGF1 was reduced in BC tissue compared with adjacent breast tissue
and IGF1 could be used to distinguish BC tissue; however, the
negative correlation between IGF1 and miR-203a-3p expression was
not statistically significant. The findings of the present study
suggested that miR-203a-3p may be involved in certain pivotal
processes in BC by targeting IGF1.
Although certain findings were acquired from the
comprehensive meta-analysis and bioinformatics analyses, there are
limitations of the current study. The heterogeneity test indicated
that there was significant heterogeneity in the included studies;
although an attempt was made to solve this. Unfortunately, the
level of I2 was still >50% following the omission of
the source of heterogeneity. It was hypothesized that the following
factors may have resulted in the significant heterogeneity: i) The
GEO microarrays were acquired from different countries with four
microarrays obtained from Spain (GSE32922, GSE44124, GSE48088 and
GSE58606), two microarrays obtained from USA (GSE22981, GSE44281)
and GSE31309, GSE37407 and GSE40525 were acquired from Germany,
Sweden and Israel, respectively; ii) the approaches for determining
the expression of miR-203a-3p were different across the different
studies. Various platforms were conducted to analyze GEO
microarrays. Furthermore, in vitro or in vivo
experiments to support the hypothesis of the present study were not
performed, which is a major limitation. Thus, in vitro and
in vivo studies should be performed in the near future.
In general, the present study established that the
expression of miR-203a-3p was markedly elevated in BC tissue
compared with adjacent breast tissue. Thus, it is hypothesized that
miR-203a-3p may enhance the development and oncogenesis of BC. In
addition, the target gene IGF1 was identified as a hub gene of
miR-203a-3p in BC while the expression of IGF1 was significantly
reduced in BC tissue compared with adjacent breast tissue.
Acknowledgements
Not applicable.
Funding
The present study was funded by Guangxi Zhuang
Autonomous Region University Student Innovative Plan (grant no.
201710598065).
Availability of data and materials
The data and materials of the present study are
available from the corresponding authors on reasonable request.
Authors' contributions
CF, JZha, RH and JM collected and analyzed the data.
KC and JZho conceived the study and wrote the manuscript. All
authors read the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jalali C, Ghaderi B, Amini S, Abdi M and
Roshani D: Association of XRCC1 Trp194 allele with risk of breast
cancer, and Ki67 protein status in breast tumor tissues. Saudi Med
J. 37:624–630. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liang Z and Xi Y: MicroRNAs mediate
therapeutic and preventive effects of natural agents in breast
cancer. Chin J Nat Med. 14:881–887. 2016.PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shou D, Wen L, Song Z, Yin J, Sun Q and
Gong W: Suppressive role of myeloid-derived suppressor cells
(MDSCs) in the microenvironment of breast cancer and targeted
immunotherapies. Oncotarget. 7:64505–64511. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xing L, He Q, Wang YY, Li HY and Ren GS:
Advances in the surgical treatment of breast cancer. Chin Clin
Oncol. 5:342016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hennequin C, Barillot I, Azria D,
Belkacémi Y, Bollet M, Chauvet B, Cowen D, Cutuli B, Fourquet A,
Hannoun-Lévi JM, et al: Radiotherapy of breast cancer. Cancer
Radiother. 20 Suppl:S139–S146. 2016.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ejlertsen B: Adjuvant chemotherapy in
early breast cancer. Dan Med J. 63:B52222016.PubMed/NCBI
|
|
8
|
Zhang X, Ren D, Guo L, Wang L, Wu S, Lin
C, Ye L, Zhu J, Li J, Song L, et al: Thymosin beta 10 is a key
regulator of tumorigenesis and metastasis and a novel serum marker
in breast cancer. Breast Cancer Res. 19:152017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Z and Kang Y: Emerging therapeutic
targets in metastatic progression: A focus on breast cancer.
Pharmacol Ther. 161:79–96. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song JL, Chen C, Yuan JP and Sun SR:
Progress in the clinical detection of heterogeneity in breast
cancer. Cancer Med. 5:3475–3488. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen H and Li Z: miRNAs in NMDA
receptor-dependent synaptic plasticity and psychiatric disorders.
Clin Sci (Lond). 130:1137–1146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Su W, Aloi MS and Garden GA: MicroRNAs
mediating CNS inflammation: Small regulators with powerful
potential. Brain Behav Immun. 52:1–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou W, Zou B, Liu L, Cui K, Gao J, Yuan S
and Cong N: MicroRNA-98 acts as a tumor suppressor in
hepatocellular carcinoma via targeting SALL4. Oncotarget.
7:74059–74073. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang L, Liang H, Wang Y, Gao S, Yin K, Liu
Z, Zheng X, Lv Y, Wang L, Zhang CY, et al: MiRNA-203 suppresses
tumor cell proliferation, migration and invasion by targeting Slug
in gastric cancer. Protein Cell. 7:383–387. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao Y, Feng B, Han S, Lu L, Chen Y, Chu X,
Wang R and Chen L: MicroRNA-129 in human cancers: From
tumorigenesis to clinical treatment. Cell Physiol Biochem.
39:2186–2202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ren W, Li C, Duan W, Du S, Yang F, Zhou J
and Xing J: MicroRNA-613 represses prostate cancer cell
proliferation and invasion through targeting Frizzled7. Biochem
Biophys Res Commun. 469:633–638. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hao W, Luo W, Bai M, Li J, Bai X, Guo J,
Wu J and Wang M: MicroRNA-206 inhibited the progression of
glioblastoma through BCL-2. J Mol Neurosci. 60:531–538. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liang AL, Zhang TT, Zhou N, Wu CY, Lin MH
and Liu YJ: MiRNA-10b sponge: An anti-breast cancer study in
vitro. Oncol Rep. 35:1950–1958. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Du S, Li H, Sun X, Li D, Yang Y, Tao Z, Li
Q and Liu K: MicroRNA-124 inhibits cell proliferation and migration
by regulating SNAI2 in breast cancer. Oncol Rep. 36:3259–3266.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang L, Sun H, Wang X, Hou N, Zhao L, Tong
D, He K, Yang Y, Song T, Yang J and Huang C: EGR1 mediates miR-203a
suppress the hepatocellular carcinoma cells progression by
targeting HOXD3 through EGFR signaling pathway. Oncotarget.
7:45302–45316. 2016.PubMed/NCBI
|
|
21
|
Gomes BC, Martins M, Lopes P, Morujão I,
Oliveira M, Araújo A, Rueff J and Rodrigues AS: Prognostic value of
microRNA-203a expression in breast cancer. Oncol Rep. 36:1748–1756.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu H and Zhang J: Decreased expression of
TFAP2B in endometrial cancer predicts poor prognosis: A study based
on TCGA data. Gynecol Oncol. 149:592–597. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nasif D, Campoy E, Laurito S, Branham R,
Urrutia G, Roqué M and Branham MT: Epigenetic regulation of ID4 in
breast cancer: Tumor suppressor or oncogene? Clin Epigenetics.
10:1112018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Lan Q and Lin J: Identification
of key gene modules for human osteosarcoma by co-expression
analysis. World J Surg Oncol. 16:892018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang YW, Zhang W and Ma R: Bioinformatic
identification of chemoresistance-associated microRNAs in breast
cancer based on microarray data. Oncol Rep. 39:1003–1010.
2018.PubMed/NCBI
|
|
26
|
Hui HX, Hu ZW, Jiang C, Wu J, Gao Y and
Wang XW: ZNF418 overexpression protects against gastric carcinoma
and prompts a good prognosis. Onco Targets Ther. 11:2763–2770.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu N, Yan J, Han T, Zou J and Shen W:
Integrated assessment of differentially expressed plasma microRNAs
in subtypes of nonsyndromic orofacial clefts. Medicine (Baltimore).
97:e112242018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan L, Zhan C, Wu J and Wang S: Expression
profile analysis of head and neck squamous cell carcinomas using
data from the cancer genome atlas. Mol Med Rep. 13:4259–4265. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou L, Lin Z, Ni Y, Wu Y, Chen D, Song L,
Huang X, Hu H and Yang D: Microarray expression profiling and gene
ontology analysis of long non-coding RNAs in spontaneously
hypertensive rats and their potential roles in the pathogenesis of
hypertension. Mol Med Rep. 13:295–300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li GM, Zhang CL, Rui RP, Sun B and Guo W:
Bioinformatics analysis of common differential genes of coronary
artery disease and ischemic cardiomyopathy. Eur Rev Med Pharmacol
Sci. 22:3553–3569. 2018.PubMed/NCBI
|
|
31
|
Zhou W, Ma CX, Xing YZ and Yan ZY:
Identification of candidate target genes of pituitary adenomas
based on the DNA microarray. Mol Med Rep. 13:2182–2186. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu F, Gao F, Liu Y, Wang Z, Zhuang X, Qu
Z, Ma H, Liu Y, Fu C, Zhang Q and Duan X: Bioinformatics analysis
of molecular mechanisms involved in intervertebral disc
degeneration induced by TNF-α and IL-1β. Mol Med Rep. 13:2925–2931.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huo W, Du M, Pan X, Zhu X, Gao Y and Li Z:
miR-203a-3p.1 targets IL-24 to modulate hepatocellular carcinoma
cell growth and metastasis. FEBS Open Bio. 7:1085–1091. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kara M, Yumrutas O, Ozcan O, Celik OI,
Bozgeyik E, Bozgeyik I and Tasdemir S: Differential expressions of
cancer-associated genes and their regulatory miRNAs in colorectal
carcinoma. Gene. 567:81–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu W, Dong Z, Liang J, Guo X, Guo Y, Shen
S, Kuang G and Guo W: Downregulation of potential tumor suppressor
mir-203a by promoter methylation contributes to the invasiveness of
gastric cardia adenocarcinoma. Cancer Invest. 34:506–516. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Riemann A, Reime S and Thews O:
Hypoxia-related tumor acidosis affects MicroRNA expression pattern
in prostate and breast tumor cells. Adv Exp Med Biol. 977:119–124.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin QH, Zhang KD, Duan HX, Liu MX, Wei WL
and Cao Y: ERGIC3, which is regulated by miR-203a, is a potential
biomarker for non-small cell lung cancer. Cancer Sci.
106:1463–1473. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Y, Dong Z, Liang J, Guo Y, Guo X, Shen
S, Kuang G and Guo W: Methylation-mediated repression of potential
tumor suppressor miR-203a and miR-203b contributes to esophageal
squamous cell carcinoma development. Tumour Biol. 37:5621–5632.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Costa-Silva DR, Barros-Oliveira MD, Borges
RS, Tavares CB, Borges US, Alves-Ribeiro FA, Silva VC and Silva BB:
Insulin-like growth factor 1 gene polymorphism and breast cancer
risk. An Acad Bras Cienc. 88:2349–2356. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
De Santi M, Annibalini G, Barbieri E,
Villarini A, Vallorani L, Contarelli S, Berrino F, Stocchi V and
Brandi G: Human IGF1 pro-forms induce breast cancer cell
proliferation via the IGF1 receptor. Cell Oncol (Dordr).
39:149–159. 2016. View Article : Google Scholar : PubMed/NCBI
|