Introduction
Multiple myeloma (MM) is a plasma cell malignancy
for which there is currently no cure (1). Although the application of novel
drugs with different mechanisms has notably improved the survival
of patients with MM, the majority of patients eventually relapse,
highlighting the need for additional agents with novel mechanisms.
At present, DNA-damaging agents remain an important part of the
first-line treatment of MM. The PAD regimen of bortezomib combined
with doxorubicin and dexamethasone is associated with an excellent
anti-MM effect, and its first-line status remains unquestionable
(2). The combination of
DNA-damaging agents with novel drugs is also an important direction
for the treatment of MM.
Aberrations in epigenetic events are important
during the development of MM; thus, the interruption of these
processes may serve as an efficacious anti-MM strategy (3,4). It
has been well acknowledged that polycomb group (PcG) proteins take
an active part in silencing homeotic genes during embryonic
development (5). At the molecular
level, PcG proteins are classified into two groups, termed polycomb
repressive complexes (PRC1 and PRC2). Enhancer of zeste homolog 2
(EZH2) interacts with other PcG proteins, including suppressor of
zeste 12 and embryonic ectoderm development, all of which compose
PRC2 (5). It has been established
that overexpression of EZH2 is frequently observed in a variety of
human malignancies, including MM, and is associated with poor
prognosis (6). Moreover, EZH2
epigenetically represses the expression of tumor suppressor genes
through trimethylation of histone H3K27 to mediate cell
proliferation, invasion and metastasis (7). Thus, it appears that targeting EZH2
may be applicable to the treatment of MM.
A previous study reported that PcG proteins are not
only involved in epigenetic gene silencing, but also modulate the
outcomes of cancer cells in response to DNA damage (8). Specifically, PcG proteins have been
demonstrated to accumulate in areas of DNA double strands breaks
(DSBs). This is substantiated by the fact that the loss of PcG
genes decreases the ability of cells to repair DSBs and renders
them sensitive to ionizing radiation (9). However, the involvement of EZH2 in
the cellular response to DNA damage in MM cells has not been
extensively investigated.
The present study aimed to investigate the impact of
EZH2 inhibition on the anti-MM potency of the DNA-damaging agents
doxorubicin (DOX)/melphalan (MEL), and to preliminarily discuss the
possible synergistic mechanism from the perspectives of epigenetics
and the DNA damage response (DDR), with the intention of laying a
foundation for an improved understanding of the regulatory
mechanism of EZH2 in MM, and a rationale for developing novel
therapeutic strategies for MM.
Materials and methods
Cell culture and reagents
The human MM cell lines RPMI8226 and H929 (American
Type Culture Collection, Manassas, VA, USA) were cultured under
typical conditions (37°C with 5% CO2) in RPMI-1640
medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% fetal bovine serum (Zeta-Life Company, San
Francisco, CA, USA), penicillin (100 U/ml), and streptomycin (100
µg/ml). DOX and MEL (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
were kept in dimethyl sulfoxide (DMSO) at 50 and 10 mM stock
concentrations, respectively. GSK126 was kept in DMSO at a 50 mM
stock concentration. KU-55933 was kept in DMSO at a 50 mM stock
concentration.
Cell proliferation assay
Cells were seeded into 96-well plates at
1.5×104 cells/well for 48 h. MTT (Sigma-Aldrich; Merck
KGaA) was used to determine cell proliferation. Cells were
incubated in 10% MTT reagent at 37°C for 4 h and dissolved in 150
µl DMSO (Sigma-Aldrich; Merck KGaA) for 10 min. Subsequently, the
absorbance at 490 nm was measured using a microplate reader. All
the experiments were performed three times.
Cell cycle and apoptosis assay
For the cell cycle assay, cells were gathered at 48
h, washed in ice-cold PBS and fixed in ice-cold 70% ethanol
overnight. Following removal of the ethanol, the cells were stained
in the dark with 50 µg/ml propidium iodide (PI; Sigma-Aldrich;
Merck KGaA) and 100 µg/ml RNase for 30 min at room temperature. The
stained cells were examined for DNA content by Epics XL flow
cytometry (Beckman Coulter, Inc., Brea, CA, USA). The result was
analyzed by ModFit LT software (version 3.1, Verity Software House
Inc., Topsham, ME, USA).
To identify apoptosis, flow cytometry analysis was
performed with an Annexin V-Phycoerythrin (PE) Apoptosis Detection
kit (BD Biosciences, San Jose, CA, USA). Cells were stained with 5
µl Annexin V-PE, 5 µl 7-aminoactinomycin D (AAD), and 500 µl 1X
binding buffer for 15 min, and subsequently examined by Epics XL
flow cytometry. The result was analyzed by CXP software (version
2.1, Beckman Coulter, Inc., Brea, CA, USA).
Western blotting
Cells were harvested and lysed with
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Haimen, China) for 10 min on ice. Supernatant from
every tube was gathered and moved to a new tube. The concentration
of proteins in cell lysates was measured using a BCA assay.
Identical amounts (20 µg) of protein lysates were separated on
8–15% by SDS-PAGE gels (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) and transferred to a polyvinylidene fluoride membrane (EMD
Millipore, Billerica, MA, USA). Membranes were blocked in 5% skim
milk for 1 h at room temperature. Membranes were incubated with
primary antibodies against cleaved caspase-3 (1:500; cat. no. 9664;
Cell Signaling Technology, Boston, MA, USA), serine-protein kinase
ATM (ATM; 1:5,000; cat. no. ab32420; Abcam, Cambridge, MA, USA),
phosphorylated ATM (pATM; 1:5,000; cat. no. ab81292), EZH2
(1:1,000; cat. no. ab191080), cleaved poly (ADP-ribose) polymerase
(cleaved PARP; 1:1,000; ab74290), cellular tumor antigen p53 (p53;
1:1,000; cat. no. ab32389), γ-histone H2AX (γ-H2AX; 1:1,000; cat.
no. ab2893), GAPDH (1:3,000; cat. no. CW0101; CWBIO, Beijing,
China), H3K27me3 (1:10,000; cat. no. 07-449; Millipore, Temecula,
CA, USA) and total histone H3 (1:5,000; cat. no. ab1791) at 4°C
overnight. Membranes were washed 3 times with TBS-T and were
incubated with peroxidase-conjugated goat anti-rabbit IgG (1:4,000;
cat. no. ZB-2301, OriGene Technologies, Inc., Beijing, China) for 1
h at room temperature. Immunoblotting was detected using an
Enhanced Chemiluminescence Western Blot kit (EMD Millipore). All
western blotting data were confirmed in a minimum of three
independent biological experiments.
Statistical analysis
GraphPad Prism 5 (GraphPad Software, Inc., La Jolla,
CA, USA) was utilized for all the data analyses. Data are presented
as the mean ± standard deviation. Differences between multiple
groups were confirmed using one-way analysis of variance followed
by Tukey's post hoc test. The significance of differences between
two groups was determined by Student's t-test. Data were obtained
from at least three independent experiments. P<0.05 was
considered to indicate a statistically significant difference. The
impacts of synergy and antagonism between GSK126 and DOX or MEL
were examined using Calcusyn software (version 2.0; Biosoft, Great
Shelford, UK). The combination index (CI) was calculated via the
Chou-Talalay method (10). Drug
synergism and antagonism were established by CI, which
quantitatively established additivity (CI=1), synergy (CI<1) and
antagonism (CI>1). The resulting values were utilized in the
construction of a plot of CI values over a range of affected
fractions (Fa-CI plot) (10,11).
Results
Pharmacological inhibition of EZH2
sensitizes MM cells to DNA-damaging agents
First, the present study assessed whether
pharmacological inhibition of EZH2 with GSK126 was able to enhance
DNA-damaging agent-induced synergistic lethality in MM cells in
vitro. RPMI8226 and H929 cells were treated with DMSO, GSK126,
DNA-damaging agents or combinations of GSK126 (2.5, 5, and 10 µM)
with DNA-damaging agents (0.05, 0.1 or 0.2 µM DOX; or 5, 10 or 20
µM of MEL) for 48 h. Cell proliferation was assessed by MTT assay.
The results indicated that the combination treatment yielded a
stronger anti-proliferative effect compared with either drug alone.
CI plot analysis using GSK126/DOX or GSK126/MEL at fixed
concentration ratios of 1:50 and 1:2 indicated that co-treatment
with GSK126 and DNA-damaging agents inhibited cell proliferation
synergistically at the majority of concentrations, as indicated by
the CI (Fig. 1). This experiment
established the phenomenon of the synergistic effect of combination
GSK126 with DNA-damaging agents against MM cells. To examine the
mechanism of action of GSK126 against MM cells in vivo, the
concentration with the most pronounced synergistic effect was
selected. Subsequent experiments in vitro were performed
with GSK126 at 5 µM, which is below its half-maximal inhibitory
concentration for each of the MM cell lines.
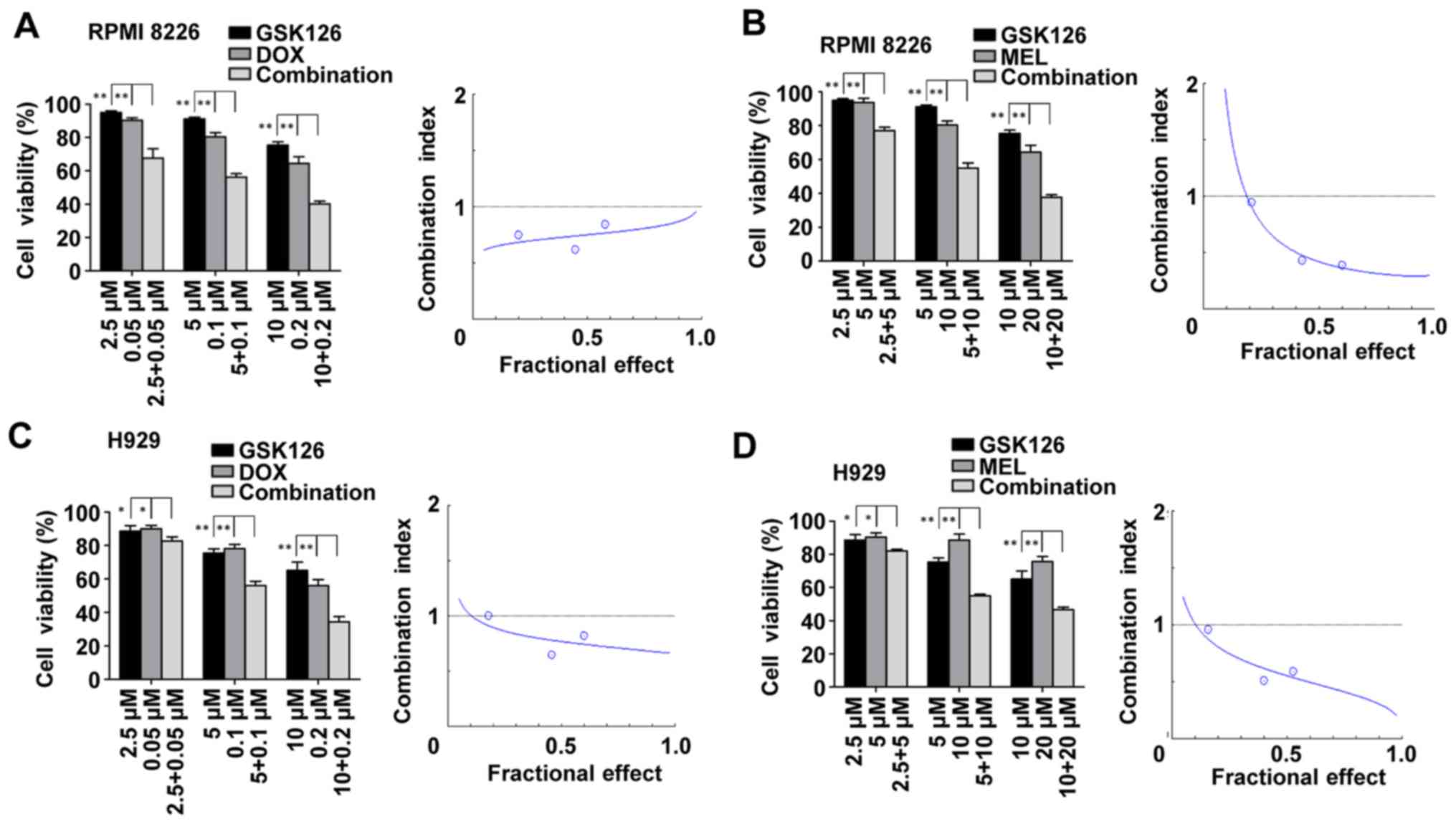 | Figure 1.Enhancer of zeste homolog inhibitor
GSK126 sensitizes MM cells to DOX- and MEL-induced cell death. (A)
RPMI8226 cells were treated with DOX at the indicated
concentrations in the presence or absence of GSK126 (2.5, 5 and 10
µM) for 48 h and subjected to MTT-based assay. Dose-effect curves
were produced using the commercially available software, Calcusyn
2.0. (B) RPMI8226 cells were treated with MEL at the indicated
concentrations in the presence or absence of GSK126 (2.5, 5 and 10
µM) for 48 h and subjected to MTT-based assay. Dose-effect curves
were produced using the commercially available software, Calcusyn
2.0. (C) H929 cells were treated with DOX at the indicated
concentrations in the presence or absence of GSK126 (2.5, 5 and 10
µM) for 48 h and subjected to MTT-based assay. Dose-effect curves
were produced using the commercially available software, Calcusyn.
(D) H929 cells were treated with MEL at the indicated
concentrations in the presence or absence of GSK126 (2.5, 5 and 10
µM) for 48 h and subjected to MTT-based assay. Dose-effect curves
were produced using the commercially available software, Calcusyn
2.0. Data are representative of a minimum of three independent
experiments. *P<0.05, **P<0.01. DOX, doxorubicin; MEL,
melphalan. |
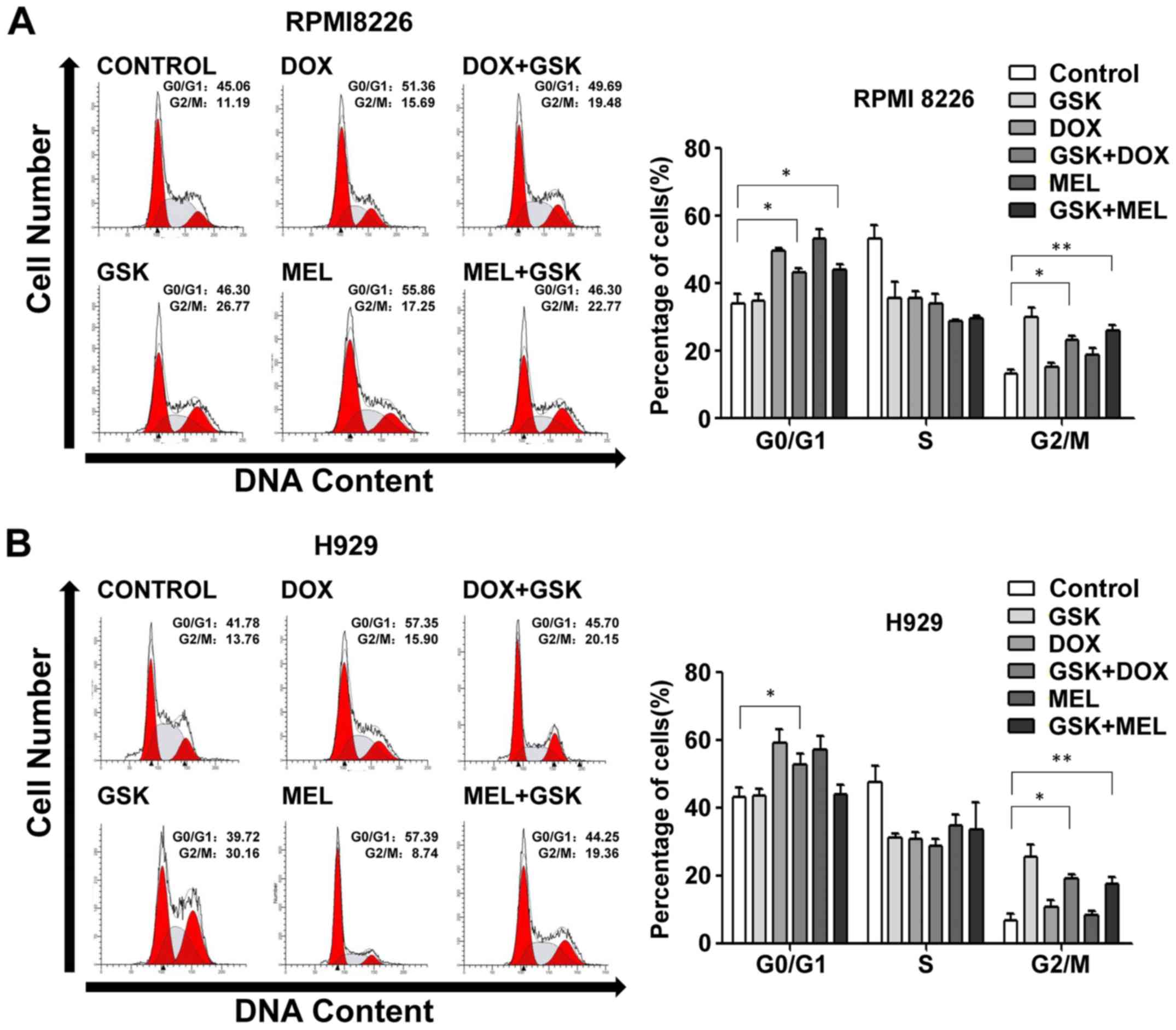
Pharmacological inhibition of EZH2
influences DNA-damaging agent-induced cell cycle
redistribution
Cell cycle regulation serves an important role in
cell proliferation; thus, the present study investigated whether
functional inhibition of EZH2 affected cell cycle progression. Flow
cytometry analysis following PI staining was used to assess the
cell cycle distribution induced by GSK126, DNA-damaging agents or
combination treatment. It was evident that EZH2 inhibition resulted
in arrest at the G2/M phase and DNA-damaging agents induced G0/G1
phase exit in RPMI8226 and H929 cells (Fig. 2). Notably, combination treatment
simultaneously induced G0/G1 and G2/M phase arrest in RPMI8226 and
H929 cells (Fig. 2), suggesting
that GSK126 and DNA-damaging agents might serve an independent role
in cell cycle progression without crosstalk effects.
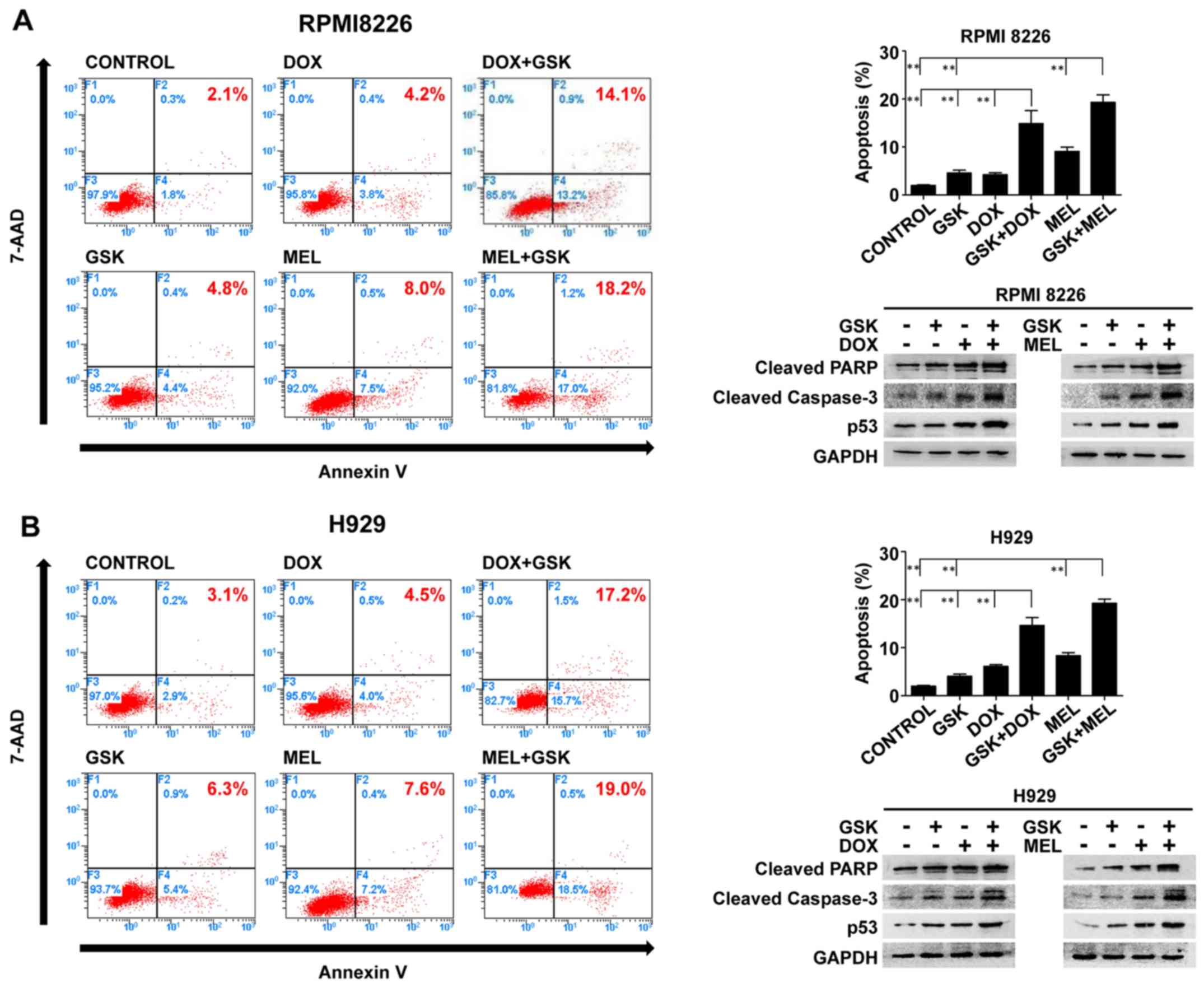
Pharmacological inhibition of EZH2
augments DNA-damaging agent-induced apoptosis in MM cells
To further illustrate the direction of cell fate
that was triggered by these combination regimens, Annexin
V-PE/7-AAD apoptosis detection was conducted to quantitatively
determine the apoptosis of MM cells. The apoptosis-promoting
effects were observed. GSK126 and DNA-damaging agents triggered an
increase in apoptosis relative to the control group in RPMI8226 or
H929 cells. Moreover, GSK126 markedly enhanced DNA-damaging
agent-induced apoptosis compared with that of either single agent
group (Fig. 3).
To further verify the apoptosis induced by single
agents or combination treatment, the present study evaluated the
expression of cleaved PARP, p53 and cleaved caspase-3 proteins by
western blotting performed on whole-cell lysates from control and
treated MM cells, presented in Fig.
3. In accordance with the findings of the flow cytometry, the
combination treatment increased the expression levels of cleaved
PARP, p53 and cleaved caspase-3 compared with GSK126 and
DNA-damaging agents alone in RPMI8226 and H929 cells. These results
demonstrated that apoptosis was the principal fate of MM cells in
the model of the synergistic inhibition of combined application of
GSK126 and DNA-damaging agents, suggesting that EZH2 is an
important factor affecting the cellular response to chromosomal
lethality.
Pharmacological inhibition of EZH2
weakens the accompanying increase in H3K27me3 induced by
DNA-damaging agents and further heightens DNA damage in this
apoptotic model of MM cells
A number of previous studies have reported on the
inextricable association between histone methylation and the DDR
(3,9,10).
Treatment of MM cells with increasing concentrations of GSK126 for
48 h revealed decreases in H3K27me3 expression (Fig. 4A). EZH2 and H3K27me3 were
upregulated following treatment with different concentrations of
DNA-damaging agents over the course of 48 h in MM cells (Fig. 4B), suggesting that the concomitant
hypermethylation of histones following DNA damage may be involved
in the DDR. To further understand the scope of the function of
EZH2/H3K27me3 in the DDR, the present study focused on the
alteration in the expression of EZH2/H3K27me3 and DDR components
under combination treatment. The results revealed that in this
synergistic inhibition model of MM cells, EZH2-inhibited cells
exhibited a decrease in DNA-damaging agent-induced histone
hypermethylation, and hyperactivation of DNA-damaging agent-induced
ATM and H2AX phosphorylation (Fig.
4C). These results indicated that GSK126 likely amplifies
DNA-damaging agent-triggered DNA damage signaling by generating
histone hypomethylation, thereby serving a synergistic anti-MM role
with DNA-damaging agents.
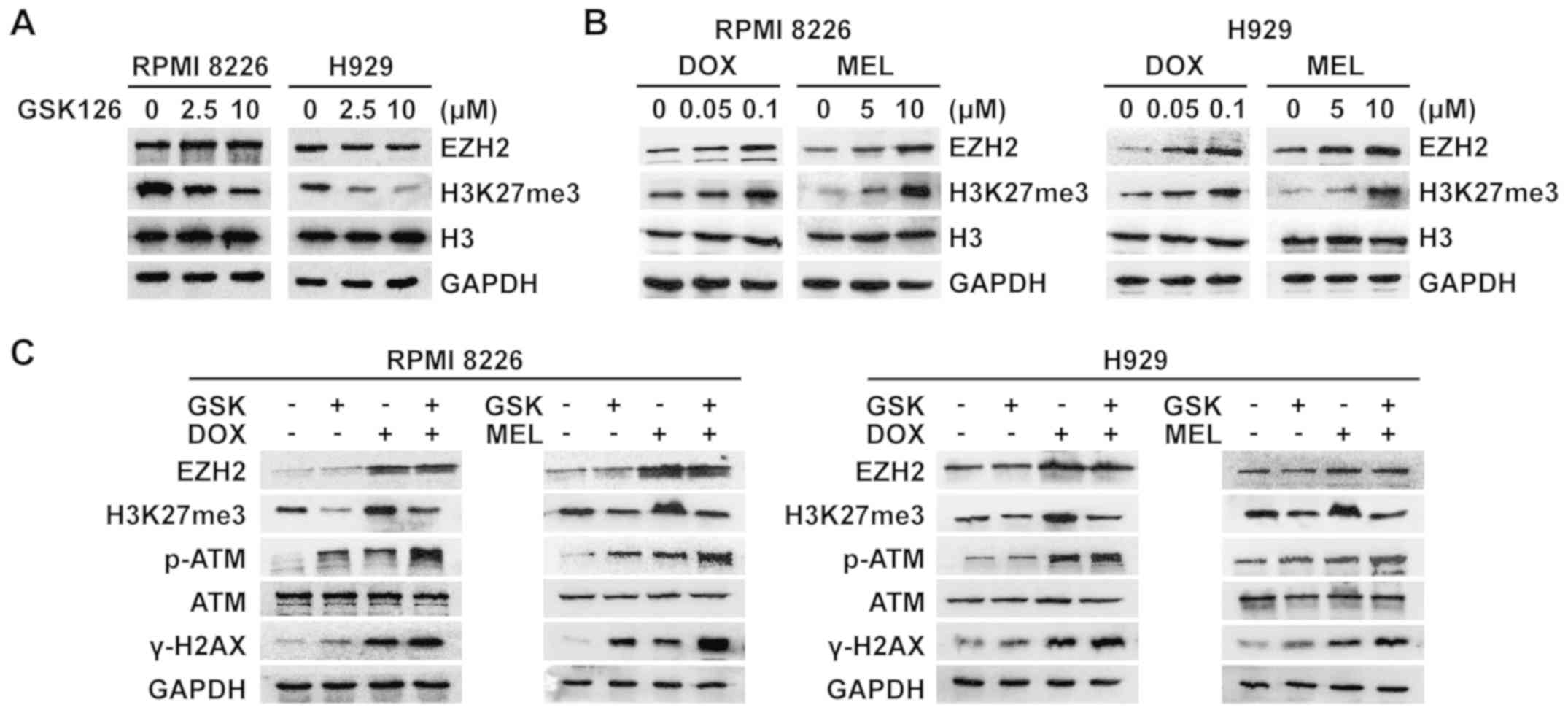 | Figure 4.GSK126 weakens the accompanying
increase in H3K27me3 induced by DNA-damaging agents, and further
heightens DNA damage in this apoptotic model of MM cells. (A)
RPMI8226 and H929 cells were treated with different concentrations
of GSK126 for 48 h. Whole-cell lysates were utilized to evaluate
EZH2 and H3K27me3. Total histone H3 and GAPDH were utilized as the
loading control. (B) RPMI8226 and H929 cells were treated with
different concentrations of DNA-damaging agents for 48 h.
Whole-cell lysates were utilized to evaluate EZH2 and H3K27me3.
Total histone H3 and GAPDH were utilized as the loading control.
(C) RPMI8226 and H929 cells were treated with GSK126 (5 µM), DOX
(0.1 µM), MEL (10 µM) or a combination treatment for 48 h.
Whole-cell lysates were utilized to evaluate EZH2, H3K27me3, p-ATM,
ATM and γ-H2AX. GAPDH served as the loading control. DOX,
doxorubicin; MEL, melphalan; EZH2, enhancer of zeste homolog 2; p,
phosphorylated; ATM, serine-protein kinase ATM; H2AX, histone H2AX;
H3K27me3, trimethylated histone H3K27. |
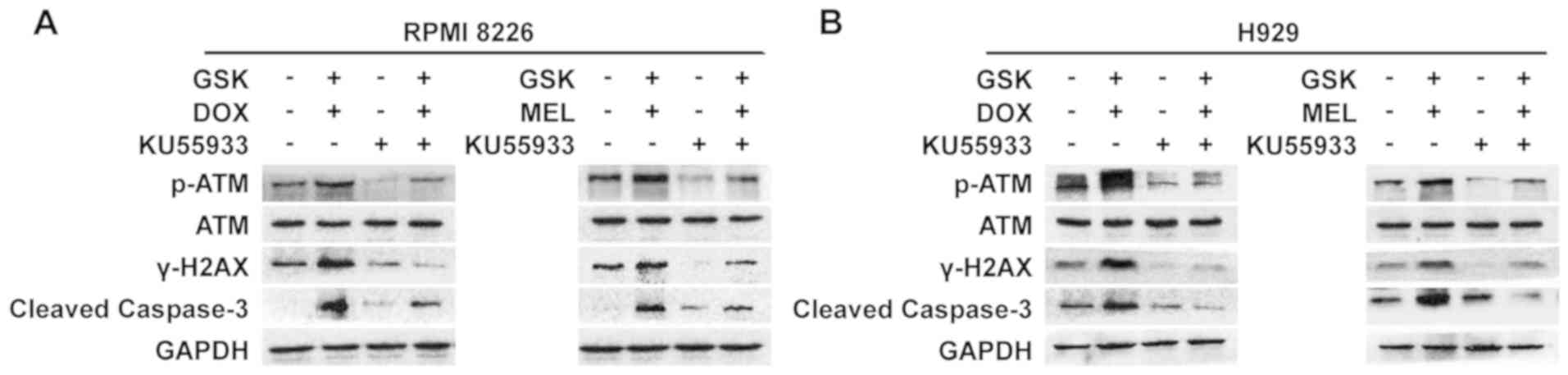
Inhibition of ATM activity interrupts
ATM-dependent DNA damage signaling and caspase-3 cleavage
To thoroughly understand the mechanism by which
GSK126 enhanced the anti-MM effect of DNA-damaging agents by
affecting DNA damage signaling, the ATM inhibitor KU-55933 was
added to suppress ATM activity in the synergistic effect model,
followed by detection of the expression of proteins associated with
DNA damage and cleaved caspase-3. The results demonstrated that the
loss of ATM kinase activity impaired the upregulation of ATM
phosphorylation and the expression of downstream γ-H2AX in this
synergistic effect model, which resulted in a decrease in cleaved
caspase-3, indicating the remission of apoptosis in MM cells
(Fig. 5). These results further
indicated that the synergistic anti-MM effect mediated by
pharmacological inhibition of EZH2 was largely due to regulation of
the ATM-dependent DDR.
Discussion
The incurability and likely recurrence of MM have
driven the emergence of novel drugs in recent years; additionally,
the appearance of more therapeutics also reflects the difficulty of
finding a cure for MM (11,12).
The development of novel drugs for MM is ongoing. Studies have
demonstrated that high expression of EZH2 in MM cells indicates a
poor outcome, and targeting EZH2 may induce apoptosis in MM cells
(13,14). In this way, EZH2 is a promising
drug target for MM therapy. The present study focused on EXH2 to
examine the synergistic anti-MM effect of pharmacological
inhibition of EZH2 with DNA-damaging agents, and the associated
mechanisms were preliminarily discussed.
Growing evidence has also demonstrated that EZH2 is
overexpressed in numerous cancer types, and its overexpression is
associated with cancer resistance and poor prognosis. Due to the
role of epigenetic regulators in gene silencing and chromatin
structure modulation, studies have identified a function for PcG
proteins in the cellular response to DNA damage that indicates that
PcG genes directly and indirectly control different aspects of the
DDR (15–17). The PcG gene EZH2 may impact DSB
repair indirectly via suppression of the homologous recombination
enzyme DNA repair protein RAD51 homolog 1 and of its paralogs, and
through the modulation of breast cancer type 1 susceptibility
protein in human uterine fibroids (18). Meanwhile, it remains unknown how
EZH2 mediates DDR signaling and what its role in transcriptional
silencing at DNA breaks in MM cells may be, thus epigenetic
regulation may effectively enhance the anti-MM effect of
DNA-damaging agents. The present results confirmed this conjecture
that GSK126, an inhibitor of EZH2, combined with DNA-damaging
agents synergistically induced apoptosis in MM cells.
It was observed that DOX and MEL induced
upregulation of EZH2 and its downstream molecule H3K27me3.
Combining an EZH2 inhibitor with DOX or MEL triggered the
hyperphosphorylation of ATM and H2AX and further directed DNA
damage by amplifying the apoptotic response. The results
demonstrated that ATM inactivity induced by KU-55933 interrupted
ATM-dependent DNA damage signaling and caspase-3 cleavage, further
indicating that the synergistic anti-MM effect mediated by the
pharmacological inhibition of EZH2 was at least partially due to
the ATM-dependent DDR. These results were consistent with a
previous report that EZH2 inhibition by DZNep or GSK126, combined
with other DNA-damaging agents (including etoposide or cisplatin),
modifies the cellular response to apoptosis (19).
It is known that the DNA damage reaction consists of
the identification of the DNA lesion, transduction of the damage
signal, and the establishment of conditions that induce DNA repair
(20). The DDR-associated protein
ATM detects DSBs and stimulates the DNA-damage checkpoint, causing
cell cycle arrest and an increase in the concentration of proteins
associated with the repair of DNA damage to maintain the stability
of the genome (21). It was
hypothesized that EZH2 may be involved in DNA damage repair and
influence the accumulation of other key DDR proteins to the DNA
damage foci. GSK126 alters the chromatin status and genomic
stability, which subsequently affects the initiation and
amplification of DDR signaling in addition to the accompanying DNA
repair (22). The inhibition of
EZH2 may cause a decrease in DNA repair activity, leading to more
marked cell apoptosis. Another possibility for ATM signaling
activation via EZH2 inhibition is the induction of oxidative
stress. Based on previous studies, mitochondrial damage is
substantially increased by a combination of etoposide and GSK126.
It is also associated with EZH2-H3K27me3-induced gene silencing of
Bcl-2 homologous antagonist/killer and apoptosis regulator BAX
(23,24). The inhibition of EZH2 leads to
microRNA (miR)-29b upregulation together with the suppression of
miR-29b downstream pro-survival targets, including SP1, MCL-1 and
cyclin-dependent kinase 6. Inhibition of miR-29b markedly reduces
the sensitivity of MM cells to small molecule EZH2-inhibitors
indicating that tumor suppressor miRs may also be involved in
epigenetic alterations (25).
However, the precise mechanism underlying GSK126-induced ATM
activation in MM cells is beyond the scope of the present study,
and future studies are required.
In conclusion, the present study preliminarily
demonstrated that pharmacological inhibition of EZH2 by GSK126
enhanced the anti-MM effect of DNA-damaging agents, partially by
decreasing H3K27me3 and via involvement in the regulation of the
ATM-dependent DDR. With the toxic effect of chemotherapy, any
sensitizer that is able to effect even a mild DNA damage reaction,
triggering an apoptotic process, may be able to improving the
efficacy of DNA-damaging chemotherapeutic agents and limit their
toxic effects (26). The present
study ultimately presented a novel and effective method of treating
patients with MM.
Acknowledgements
Not applicable.
Funding
This study was supported by the Social Development
Science and Technology Fund of Shaanxi Province (grant no.
2016SF-071) and the Natural Science Foundation of Shaanxi Province
(grant no. 2017JM8025).
Availability of data and materials
All data generated or analyzed in this study are
included in this manuscript.
Authors' contributions
GG and XC conceived and designed the experiment. LX
and HT designed and performed the experiments, analyzed the data
and drafted the manuscript. KW, YZ and JF performed the experiments
and analyzed the data. HD, YJ and CC acquired and interpreted the
data. GG gave final approval of the article. All authors have
reviewed the final version of the manuscript and approved it for
publication.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pourabdollah M and Chang H: Coexistence of
multiple myeloma and Paget disease of bone. Blood. 130:11732017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mai EK, Bertsch U, Durig J, Kunz C, Haenel
M, Blau IW, Munder M, Jauch A, Schurich B, Hielscher T, et al:
Phase III trial of bortezomib, cyclophosphamide and dexamethasone
(VCD) versus bortezomib, doxorubicin and dexamethasone (PAd) in
newly diagnosed myeloma. Leukemia. 29:1721–1729. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Issa ME, Takhsha FS, Chirumamilla CS,
Perez-Novo C, Vanden Berghe W and Cuendet M: Epigenetic strategies
to reverse drug resistance in heterogeneous multiple myeloma. Clin
Epigenetics. 9:172017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pawlyn C, Kaiser MF, Davies FE and Morgan
GJ: Current and potential epigenetic targets in multiple myeloma.
Epigenomics. 6:215–228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sanulli S, Justin N, Teissandier A,
Ancelin K, Portoso M, Caron M, Michaud A, Lombard B, da Rocha ST,
Offer J, et al: Jarid2 methylation via the PRC2 complex regulates
H3K27me3 deposition during cell differentiation. Mol Cell.
57:769–783. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pawlyn C, Bright MD, Buros AF, Stein CK,
Walters Z, Aronson LI, Mirabella F, Jones JR, Kaiser MF, Walker BA,
et al: Overexpression of EZH2 in multiple myeloma is associated
with poor prognosis and dysregulation of cell cycle control. Blood
Cancer J. 7:e5492017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cao R, Wang L, Wang H, Xia L,
Erdjument-Bromage H, Tempst P, Jones RS and Zhang Y: Role of
histone H3 lysine 27 methylation in Polycomb-group silencing.
Science. 298:1039–1043. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ito T, Teo YV, Evans SA, Neretti N and
Sedivy JM: Regulation of cellular senescence by polycomb chromatin
modifiers through distinct DNA damage- and histone
methylation-dependent pathways. Cell Rep. 22:3480–3492. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zeidler M, Varambally S, Cao Q, Chinnaiyan
AM, Ferguson DO, Merajver SD and Kleer CG: The polycomb group
protein EZH2 impairs DNA repair in breast epithelial cells.
Neoplasia. 7:1011–1019. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ha K, Lee GE, Palii SS, Brown KD, Takeda
Y, Liu K, Bhalla KN and Robertson KD: Rapid and transient
recruitment of DNMT1 to DNA double-strand breaks is mediated by its
interaction with multiple components of the DNA damage response
machinery. Hum Mol Genet. 20:126–140. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kuroda J: Therapeutic approach for
relapsed/refractory multiple myeloma: The logic and practice.
Rinsho Ketsueki. 58:2058–2066. 2017.(In Japanese). PubMed/NCBI
|
|
12
|
Varga C, Maglio M, Ghobrial IM and
Richardson PG: Current use of monoclonal antibodies in the
treatment of multiple myeloma. Br J Haematol. 181:447–459. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuser-Abali G, Gong L, Yan J, Liu Q, Zeng
W, Williamson A, Lim CB, Molloy ME, Little JB, Huang L and Yuan ZM:
An EZH2-mediated epigenetic mechanism behind p53-dependent tissue
sensitivity to DNA damage. Proc Natl Acad Sci USA. 115:3452–3457.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Cheng P, Pavlyukov MS, Yu H, Zhang
Z, Kim SH, Minata M, Mohyeldin A, Xie W, Chen D, et al: Targeting
NEK2 attenuates glioblastoma growth and radioresistance by
destabilizing histone methyltransferase EZH2. J Clin Invest.
127:3075–3089. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu Z, Lee ST, Qiao Y, Li Z, Lee PL, Lee
YJ, Jiang X, Tan J, Aau M, Lim CZ and Yu Q: Polycomb protein EZH2
regulates cancer cell fate decision in response to DNA damage. Cell
Death Differ. 18:1771–1779. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vissers JH, van Lohuizen M and Citterio E:
The emerging role of Polycomb repressors in the response to DNA
damage. J Cell Sci. 125:3939–3948. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bouwman P and Jonkers J: The effects of
deregulated DNA damage signalling on cancer chemotherapy response
and resistance. Nat Rev Cancer. 12:587–598. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Q, Nair S, Laknaur A, Ismail N,
Diamond MP and Al-Hendy A: The polycomb group protein EZH2 impairs
DNA damage repair gene expression in human uterine fibroids. Biol
Reprod. 94:692016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu H, Li W, Yu X, Gao F, Duan Z, Ma X,
Tan S, Yuan Y, Liu L, Wang J, et al: EZH2-mediated Puma gene
repression regulates non-small cell lung cancer cell proliferation
and cisplatin-induced apoptosis. Oncotarget. 7:56338–56354.
2016.PubMed/NCBI
|
|
20
|
Zhou BB and Elledge SJ: The DNA damage
response: Putting checkpoints in perspective. Nature. 408:433–439.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan S, Sorrell M and Berman Z: Functional
interplay between ATM/ATR-mediated DNA damage response and DNA
repair pathways in oxidative stress. Cell Mol Life Sci.
71:3951–3967. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang Y, Wang X, Niu X, Wang X, Jiang R,
Xu T, Liu Y, Liang L, Ou X, Xing X, et al: EZH2 suppresses the
nucleotide excision repair in nasopharyngeal carcinoma by silencing
XPA gene. Mol Carcinog. 56:447–463. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gursoy-Yuzugullu O, Carman C, Serafim RB,
Myronakis M, Valente V and Price BD: Epigenetic therapy with
inhibitors of histone methylation suppresses DNA damage signaling
and increases glioma cell radiosensitivity. Oncotarget.
8:24518–24532. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kirk JS, Schaarschuch K, Dalimov Z,
Lasorsa E, Ku S, Ramakrishnan S, Hu Q, Azabdaftari G, Wang J, Pili
R and Ellis L: Top2a identifies and provides epigenetic rationale
for novel combination therapeutic strategies for aggressive
prostate cancer. Oncotarget. 6:3136–3146. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stamato MA, Juli G, Romeo E, Ronchetti D,
Arbitrio M, Caracciolo D, Neri A, Tagliaferri P, Tassone P and
Amodio N: Inhibition of EZH2 triggers the tumor suppressive miR-29b
network in multiple myeloma. Oncotarget. 8:106527–106537. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu Q: Restoring p53-mediated apoptosis in
cancer cells: New opportunities for cancer therapy. Drug Resist
Updat. 9:19–25. 2006. View Article : Google Scholar : PubMed/NCBI
|