Introduction
Breast cancer (BC) is the most frequently diagnosed
type of cancer and the leading cause of cancer-associated mortality
among women worldwide; ~1.7 million new diagnoses and 521,900 cases
of mortality are attributed to BC annually (1). However, despite extensive studies
focussing on BC, and the development of novel and less toxic
treatment regimens, the survival rates for patients with BC remain
low (2). Therefore, the
identification of more valuable and convenient biomarkers for early
diagnosis and survival prediction, as well as novel therapeutic
target genes for effective treatment, is urgently required.
Chaperonin-containing TCP1 subunit 6A (CCT6A) is the
ζ subunit of the chaperonin-containing TCP1 complex (CCT), which
comprises two identical rings stacked back to back, each containing
eight different homologous proteins (1–8 or α-θ); the protein
encoded by CCT6A functions as a molecular chaperone (3–5).
Previous studies have indicated that 5–10% of all newly synthesised
cytoplasmic proteins appear to pass this chaperone, suggesting it
has an essential role in cytoskeletal organisation and in the cell
cycle (6,7). As a subunit of the CCT family, CCT6A
is essential for the correct folding and
oligomerisation/polymerisation of native proteins, with actin and
tubulin being the major substrates (8). Furthermore, CCT6A has been identified
as a specific component of endogenous extracellular
signal-regulated kinase (ERK)1/2 signalling complexes (9) that possesses phosphorylation sites
for mitogen-activated protein kinase (MAPK)/ERK (10,11).
In addition, CCT6A and the MAPK signalling pathway serve a critical
role in matrix metalloproteinase-3-dependent regulation of neurite
outgrowth and neuronal migration in the developing brain (7).
Previous studies have revealed that CCT6A expression
is significantly increased in 10 human tumour cell lines and
drug-resistant human melanoma cell lines, suggesting that CCT6A may
have a critical role in tumorigenesis and drug resistance (12,13).
In addition, a recent experimental study on lung cancer indicated
that CCT6A overexpression significantly increases colony and tumour
sphere formation, as well as side population, and reduces
sensitivity to anoikis in A549 cells following transforming growth
factor (TGF)-β stimulation (14).
However, the functional significance of CCT6A in other types of
cancer, including BC, has yet to be investigated.
To determine the significance of CCT6A in BC, the
present study evaluated CCT6A expression in various tumours,
including BC, and analysed its association with survival,
clinicopathological parameters and related signalling pathways
using online datasets. The results revealed that various cancer
tissues exhibited significantly higher CCT6A expression compared
within noncancerous tissues, and increased CCT6A expression in BC
tissues was significantly associated with poor patient prognosis.
Furthermore, pathways based on associated genes indicated that
CCT6A was significantly associated with the cell cycle, and its
expression was significantly positively correlated with cyclin
(CCN)B2 and CCNA2 expression. Collectively, these findings
suggested that CCT6A may have an important role in BC
progression.
Materials and methods
Oncomine analysis
Gene expression array datasets from the online
cancer microarray database Oncomine (www.oncomine.org) (15) were used to evaluate CCT6A mRNA
expression in various tumour samples. CCT6A mRNA expression in BC
tissues was compared with that in normal control tissues using 10
datasets from 3 cohorts (16,17).
The fold change in CCT6A expression from different datasets was
presented using scatter plots. The datasets used in this study are
listed in Table I. Details
regarding standardised normalisation techniques and statistical
calculations are provided on the Oncomine website (18). The search parameters and filters
used to find the datasets were set as follows: P-value, 0.05; fold
change, 1.5; gene rank, 10%; analysis type, cancer vs. normal
analysis; data type, mRNA.
 | Table I.Oncomine analysis of
chaperonin-containing TCP1 subunit 6A expression in breast
cancer. |
Table I.
Oncomine analysis of
chaperonin-containing TCP1 subunit 6A expression in breast
cancer.
| Cohort no. | Cohort name | Data type | Sample (n) | Fold-change | P-value |
|---|
| 1 | Richardson Breast 2
Statistics (16) | mRNA | Ductal breast
carcinoma (40) vs. normal (7) | 2.209 |
3.97×10−10 |
| 2 | Curtis Breast
Statistics (17) | mRNA | Invasive breast
carcinoma (21) vs. normal (144) | 1.577 |
1.68×10−6 |
|
|
| mRNA | Tubular breast
carcinoma (67) vs. normal (144) | 1.672 |
1.27×10−21 |
|
|
| mRNA | Invasive lobular
breast carcinoma (148) vs. normal (144) | 1.571 |
1.42×10−31 |
|
|
| mRNA | Medullary breast
carcinoma (32) vs. normal (144) | 1.820 |
3.45×10−10 |
|
|
| mRNA | Invasive ductal and
invasive lobular breast carcinoma (90) vs. normal (144) | 1.611 |
1.59×10−22 |
|
|
| mRNA | Invasive ductal
breast carcinoma (1,556) vs. normal (144) | 1.709 |
4.41×10−61 |
|
|
| mRNA | Mucinous breast
carcinoma (46) vs. normal (144) | 1.544 |
4.24×10−11 |
| 3 | TCGA Breast
Statistics (TCGA) | mRNA | Invasive ductal
breast carcinoma (389) vs. normal (61) | 1.532 |
5.08×10−30 |
|
|
| mRNA | Intraductal
cribriform breast adenocarcinoma (3) vs. normal (61) | 1.599 |
1.46×10−4 |
Human Protein Atlas (HPA)
CCT6A expression was further evaluated using the
publicly available HPA (www.proteinatlas.org; version 18). The HPA is a
database containing images of tissue microarrays labelled with
antibodies against 11,250 human proteins (19). These tissue microarrays comprise
sections from 46 normal human tissues and 20 types of human cancer.
Analyses were performed using HPA images of breast sections
labelled with CCT6A antibody (cat. no. HPA042996; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany).
Kaplan-Meier plotter
The Kaplan-Meier plotter (www.kmplot.com) is an online database containing gene
expression data and survival information from 4,142 clinical
patients with BC (20,21). To further investigate the
prognostic value of CCT6A mRNA expression in BC, this study
analysed the association between CCT6A mRNA expression and
survival, including overall survival (OS), relapse-free survival
(RFS), distant metastasis-free survival (DMFS) and post progression
survival (PPS), in patients with BC. Samples from patients with BC
were divided into two groups according to median CCT6A expression
(high vs. low expression), after which the association between
CCT6A mRNA expression and survival was analysed using a
Kaplan-Meier survival plot together with hazard ratio, 95%
confidence interval and log-rank P-value.
Breast Cancer Gene-Expression Miner
v4.0 (bc-GenExMiner v4.0)
The bc-GenExMiner v4.0 (http://bcgenex.centregauducheau.fr/BC-GEM/GEM-Accueil.php?js=1)
contains 36 annotated genomic datasets and three statistical mining
functions (22,23). The bc-GenExMiner v4.0 was used to
compare target gene expression according to clinical criteria. The
present study analysed the association between CCT6A expression and
hormonal receptors, nodal status and other factors, in order to
assess the prognostic impact of this candidate gene in human BC and
to provide potential prognostic markers for BC.
R2 application
R2: Genomics Analysis and Visualisation Platform
(http://r2.amc.nl) (24) is a web-based genomics analysis and
visualisation application, which was used to investigate
CCT6A-related signalling pathways and genes. Pathway enrichment
analysis based on CCT6A-associated genes was performed using the R2
Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Finder and
the standard setting in The Cancer Genome Atlas (TCGA) collection
(BRCA was the TCGA ID and the search term in R2) (15). Furthermore, the correlation between
CCT6A expression and related gene expression was calculated using
the Pearson test based on the expression in BC cancer tissues and
noncancerous normal tissues. The related genes were identified
through the R2 Platform by searching ‘Find Correlated genes with a
single gene’ or ‘Correlate two genes’. The r-value, P-value and
T-value were calculated and provided by R2.
Statistical analysis
The expression of CCT6A in BC tissues and
noncancerous breast tissues was presented as the means ± 10% of
percentile of the median intensity. Comparison of CCT6A expression
between BC tissues and noncancerous breast tissues using the
Oncomine data was performed using Student's t-test. The association
between CCT6A mRNA expression and survival was analysed using
Kaplan-Meier survival plots, which were statistically compared
using the log-rank test on the Kaplan-Meier plotter website. Using
the R2 website, the correlation between CCT6A expression and the
expression of related genes was analysed using the Pearson test in
BC samples, and the r-, P- and T-values were calculated and
presented. The association between CCT6A and age, nodal status
(+/-), receptor statuses [estrogen receptor (ER)+ vs.
ER−, progesterone receptor (PR)+ vs.
PR−, human epidermal growth factor receptor 2
(HER2)+ vs. HER2−] and triple-negative status
was performed using a Welch's analysis of variance (ANOVA); if
global significant differences were found (for correlation analysis
of CCT6A with SBR and NPI grades), Dunnett-Tukey-Kramer's post-hoc
test was performed to compare the differences among groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
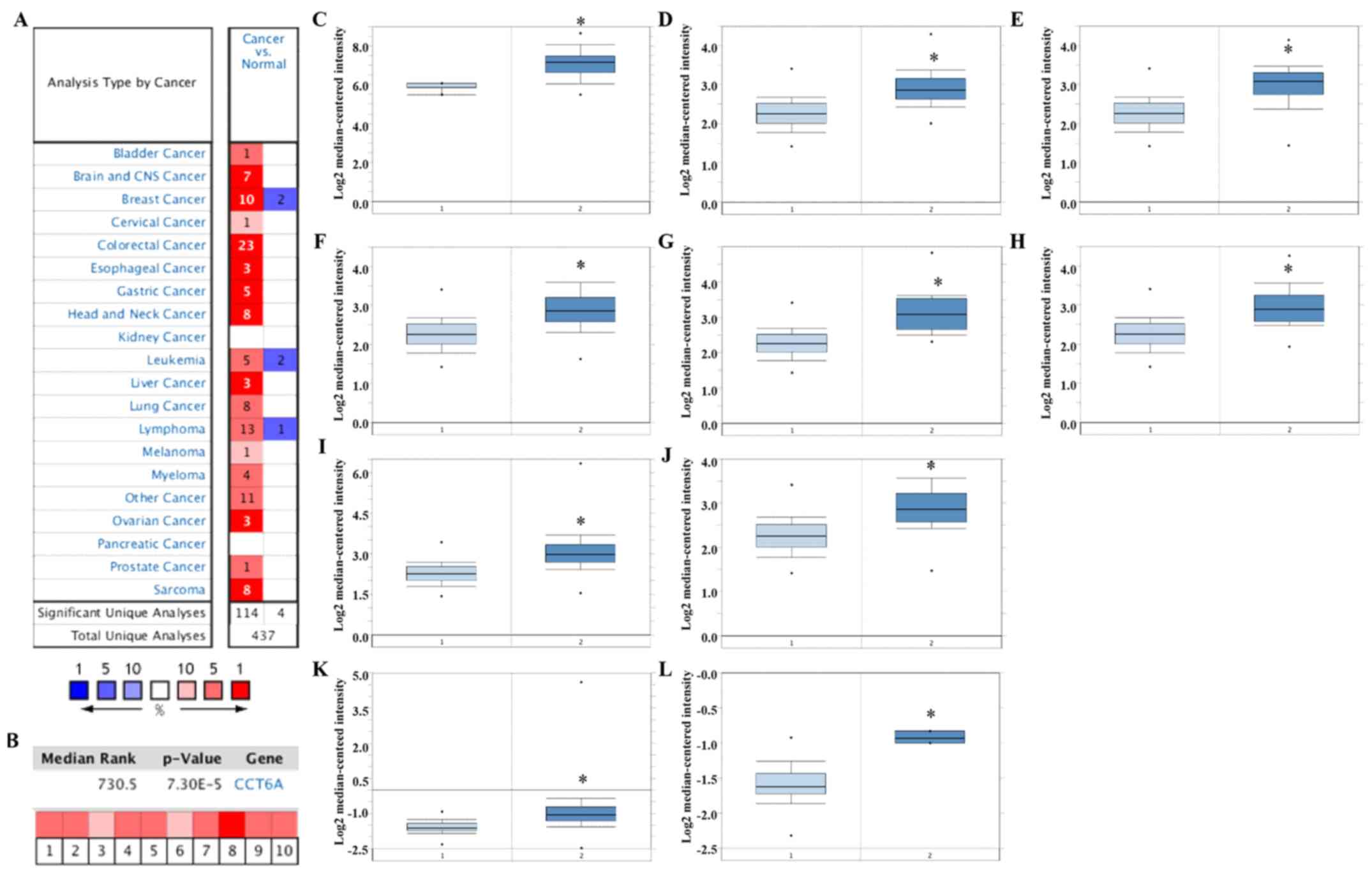
CCT6A upregulation in BC tissues
Analyses of mRNA expression in various tumour
samples from the Oncomine platform indicated that CCT6A expression
was significantly higher in tumour tissues compared with matched
normal tissues (Fig. 1A). In
particular, CCT6A was significantly upregulated in BC tissues
compared with in noncancerous breast tissues (Fig. 1B-L; P<0.05). Details of CCT6A
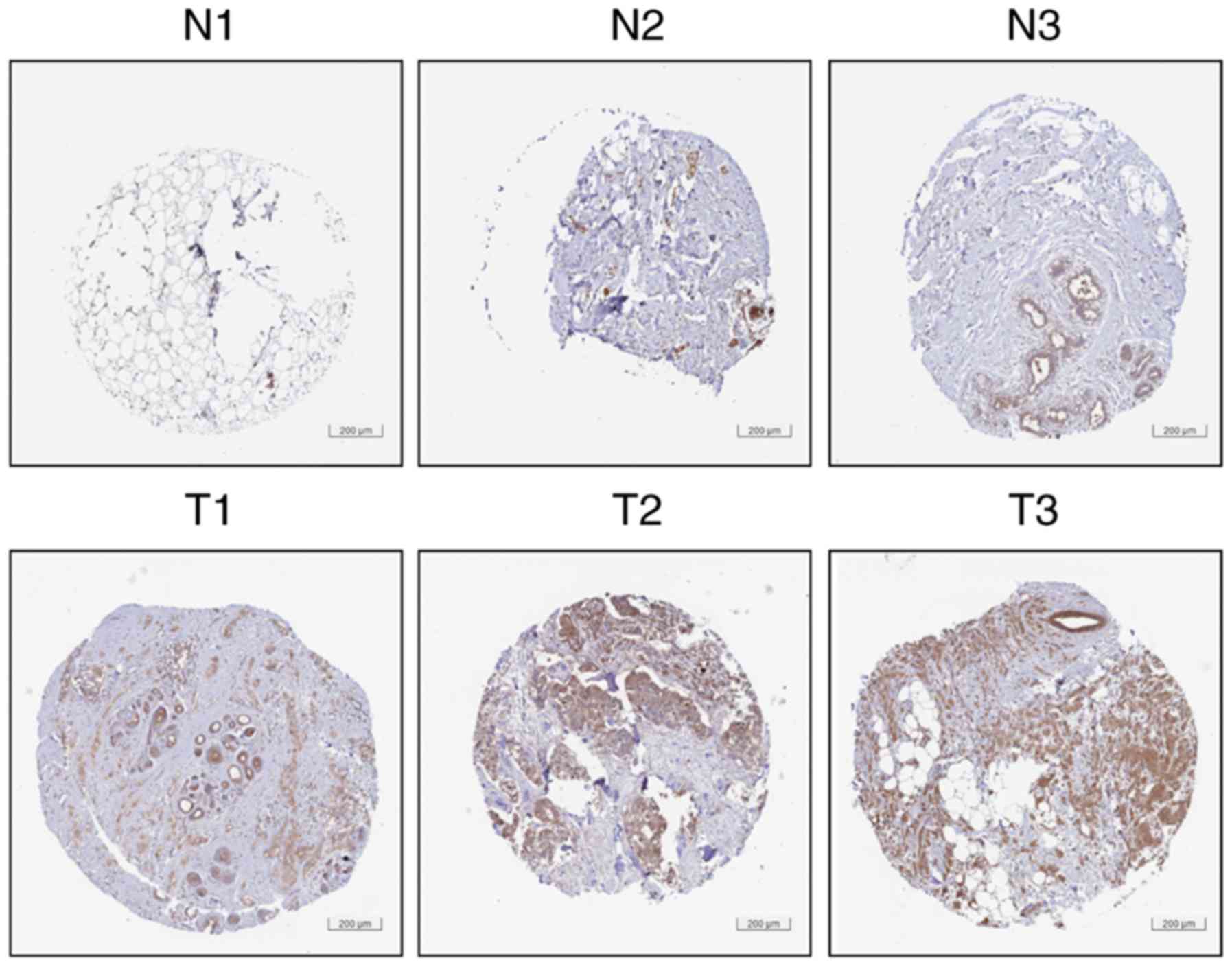
expression in these datasets are listed in Table I. Accordingly, as presented in
Fig. 2, HPA (version 18) data
analyses confirmed that BC tissues (images of T1-T3 available from
https://www.proteinatlas.org/ENSG00000146731-CCT6A/pathology/tissue/breast+cancer#img)
exhibited higher protein expression levels of CCT6A compared with
normal breast tissues (images of N1-N3 available from http://www.proteinatlas.org/ENSG00000146731-CCT6A/tissue/breast#img).
Collectively, the aforementioned analyses
demonstrated that the mRNA and protein expression levels of CCT6A
were higher in BC tissues than in normal breast tissues, thus
suggesting that CCT6A may act as an oncogene that promotes BC.
Association between CCT6A mRNA
expression and clinicopathological parameters in patients with
BC
Using Welch's ANOVA, CCT6A mRNA expression was
compared among groups of patients with various clinicopathological
parameters using bc-GenExMiner (Fig.
3 and Table II). In terms of
age, CCT6A mRNA expression was lower in the older group (>51
years; Fig. 3A; P=0.0007).
Furthermore, among patients with BC, those with positive nodal
status exhibited higher CCT6A mRNA expression than those with
negative nodal status (Fig. 3B;
P=0.0259). The ER and PR status of patients with BC was also
negatively associated with CCT6A mRNA expression (Fig. 3C and D; P<0.0001). However,
there was no difference in CCT6A expression between patients with
HER2− and HER2+ BC (Fig. 3E; P=0.0616). Triple-negative BC
(TNBC) is a type of BC characterised by ER−,
PR− and HER2−. CCT6A mRNA expression was
significantly upregulated in patients with TNBC (Fig. 3F; P<0.0001). The results also
indicated that patients with basal-like BC exhibited higher CCT6A
mRNA expression than those without basal-like BC (Fig. 3G; P<0.0001). With regards to
Scarff-Bloom-Richardson (SBR) grading (25), a more advanced SBR grade was
significantly associated with higher CCT6A mRNA expression
(Fig. 3H; P<0.0001) compared
with other SBR grades. Furthermore, a more advanced Nottingham
prognostic index (NPI) grade (26)
was associated with higher CCT6A mRNA expression (Fig. 3I; P<0.0001) compared with other
NPI grades.
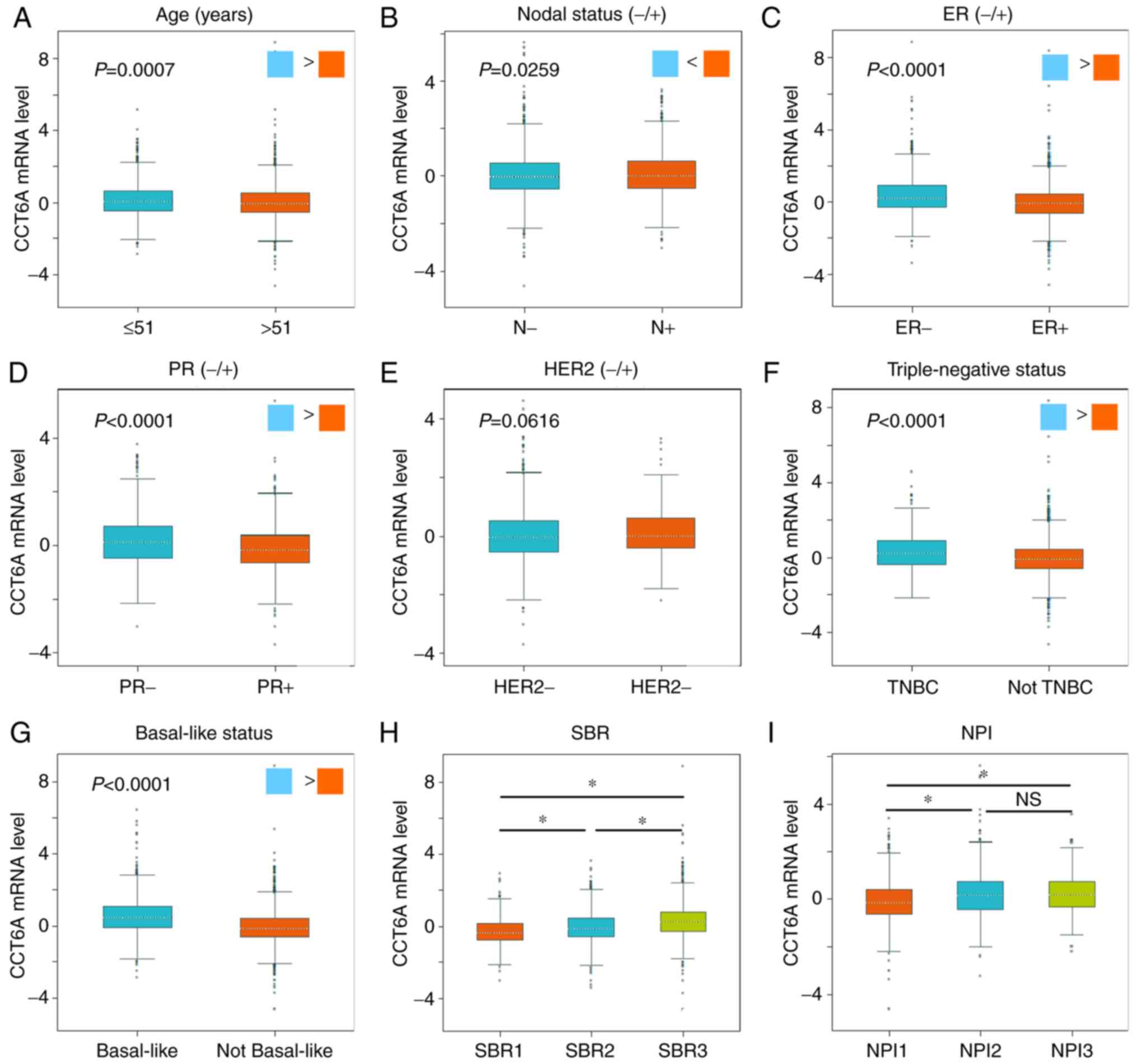 | Figure 3.Association between CCT6A mRNA
expression and clinicopathological parameters in patients with BC.
Association between CCT6A mRNA expression and (A) age, (B) nodal
status, (C) ER status, (D) PR status, (E) HER2 status, (F) TNBC,
(G) basal-like status, (H) SBR grade and (I) NPI grade. *P<0.05,
as analysed using Breast Cancer Gene-Expression Miner v4.0. BC,
breast cancer; CCT6A, chaperonin-containing TCP1 subunit 6A; ER,
estrogen receptor; HER2, human epidermal growth factor receptor 2;
IHC, immunohistochemistry; NPI, Nottingham prognostic index; NS,
not significant; SBR, Scarff-Bloom-Richardson; TNBC,
triple-negative breast cancer. |
 | Table II.Association between mRNA expression
of chaperonin-containing TCP1 subunit 6Aand clinicopathological
parameters of breast cancer. |
Table II.
Association between mRNA expression
of chaperonin-containing TCP1 subunit 6Aand clinicopathological
parameters of breast cancer.
| Variable | Case | mRNA
expression | P-value |
|---|
| Age (years) |
|
| 0.0007 |
|
≤51 | 1,392 | ↑ |
|
|
<51 | 2,210 | – |
|
| Nodal status |
|
| 0.0259 |
|
Negative | 2,493 | – |
|
|
Positive | 1,562 | ↑ |
|
| ER (IHC) |
|
| <0.0001 |
|
Negative | 1,446 | ↑ |
|
|
Positive | 3,766 | – |
|
| PR (IHC) |
|
| <0.0001 |
|
Negative | 804 | ↑ |
|
|
Positive | 1,249 | – |
|
| HER2 (IHC) |
|
| 0.0616 |
|
Negative | 1,409 | – |
|
|
Positive | 201 | – |
|
| Triple-negative
status |
|
| <0.0001 |
| Not
TNBC | 374 | ↑ |
|
|
TNBC | 3,857 | – |
|
| Basal-like
status |
|
| <0.0001 |
|
Basal-like | 1,060 | ↑ |
|
| Not
basal-like | 3,942 | – |
|
| SBR grade |
|
| <0.0001 |
|
SBR1 | 546 | – |
|
|
SBR2 | 1,431 | ↑ |
|
|
SBR3 | 1,317 | ↑ |
|
| NPI grade |
|
| <0.0001 |
|
NPI1 | 931 | – |
|
|
NPI2 | 727 | ↑ |
|
|
NPI3 | 125 | ↑ |
Higher CCT6A expression is associated
with poor survival in patients with BC
Using the Kaplan-Meier plotter, the association
between CCT6A expression and survival in patients with BC was
evaluated. Kaplan-Meier analysis revealed that high CCT6A
expression (Affymetrix microarrays, 201326-at and 201327_s_at;
Fig. 4A and B) in BC tissues was
significantly associated with shorter OS (P<0.05), RFS
(P<0.05), DMFS (P<0.05) and PPS (P<0.05) in patients with
BC, thus suggesting that high CCT6A expression may be predictive of
poor prognosis.
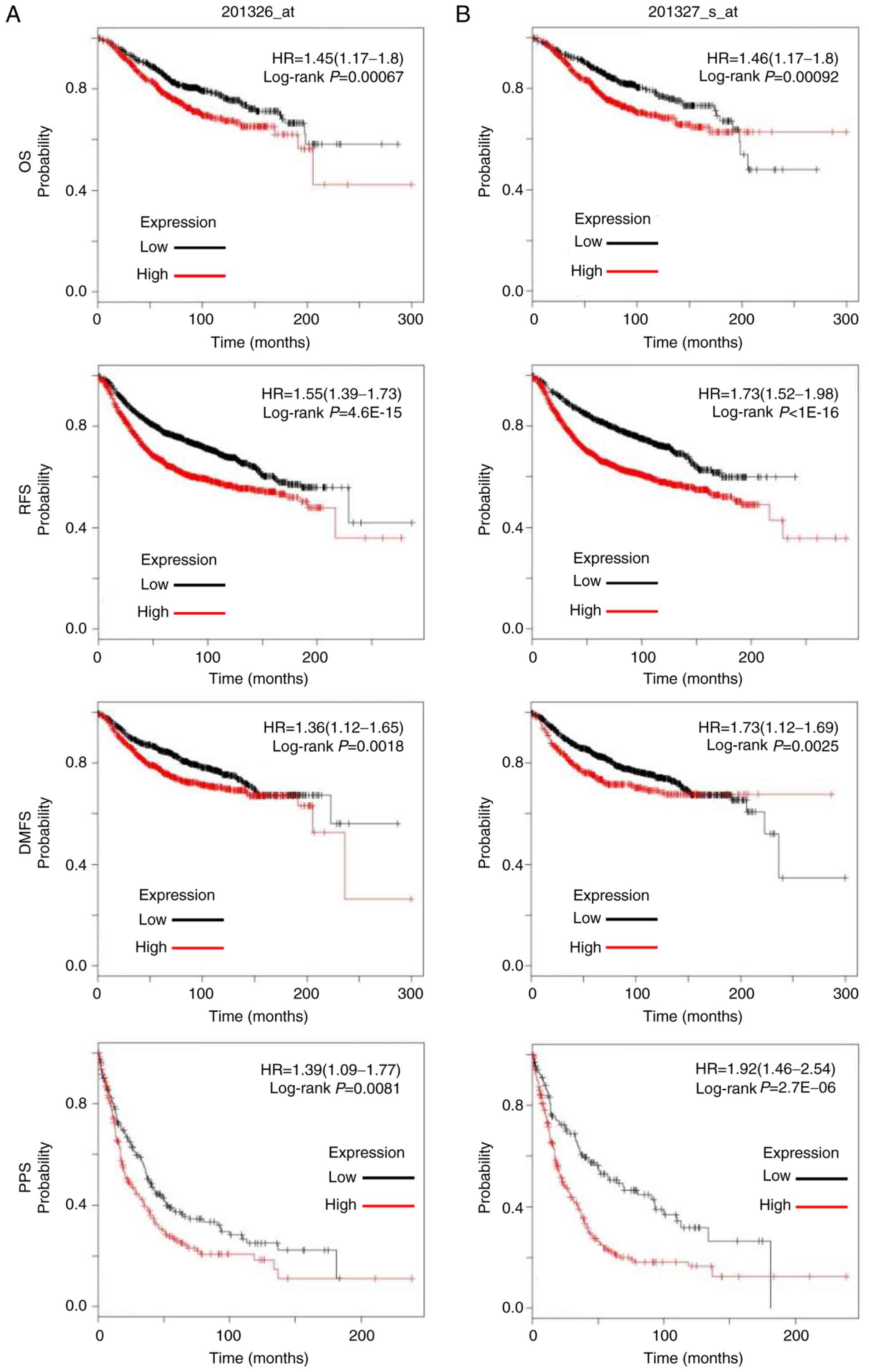 | Figure 4.Association between CCT6A expression
and survival in patients with BC. Association between CCT6A (A)
201326_at and (B) 201327_s_at expression and OS, RFS, DMFS and PPS
in patients with BC was analysed using log-rank tests based on
CCT6A expression in BC tissues from The Cancer Genome Atlas
dataset. Kaplan-Meier curves were plotted for CCT6A, and HRs and
95% confidence intervals are shown. BC, breast cancer; CCT6A,
chaperonin-containing TCP1 subunit 6A; DMFS, distant
metastasis-free survival; HR, hazard ratio; OS, overall survival;
RFS, disease-free survival; PPS, post progression survival. |
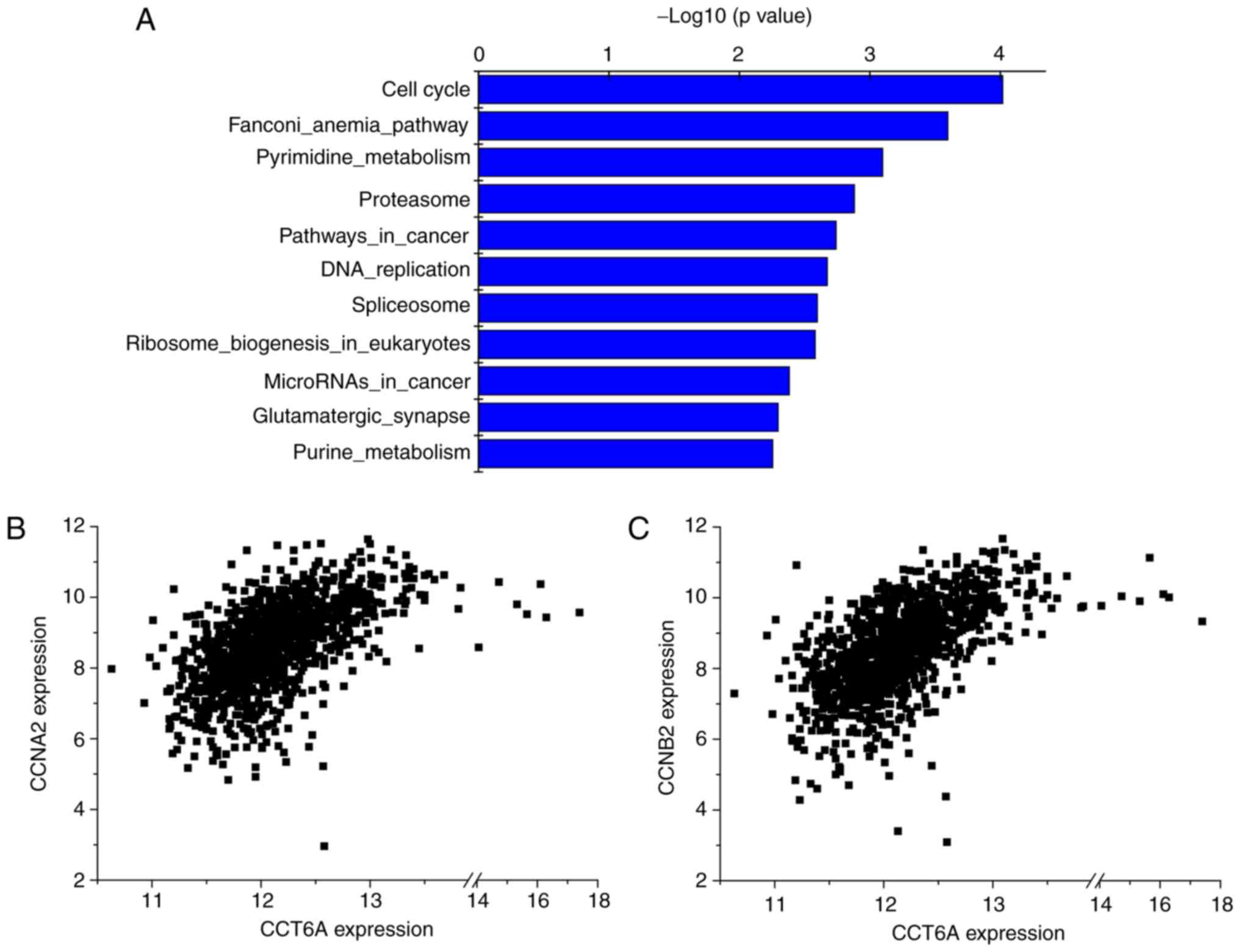
Possible involvement of CCT6A in the
cell cycle and its positive correlation with CCNB2 and CCNA2
To further investigate the signalling pathways in
which CCT6A might be involved, pathway enrichment analysis was
performed using the R2 KEGG Pathway Finder based on
CCT6A-associated genes in BC tissues from TCGA dataset (ID: BRCA).
Accordingly, the top 10 signalling pathways associated with CCT6A,
including the ‘cell cycle’, are presented in Fig. 5 and Table III. Among these correlated genes
involved in the cell cycle, CCT6A expression was significantly
positively correlated with CCNB2 (r-value=0.560;
P=1.0×10−91; T-value=22.389) and CCNA2 (r-value=0.532;
P=2.7×10−81; T-value=20.807) expression. These results
indicated that CCT6A may be involved in the cell cycle signalling
pathway.
 | Table III.Pathway enrichment analysis of
chaperonin-containing TCP1 subunit 6A-associated genes in TCGA. |
Table III.
Pathway enrichment analysis of
chaperonin-containing TCP1 subunit 6A-associated genes in TCGA.
| No. | Pathway | No. of input
genes | Total no. of
genes | Percentage (%) | P-value |
|---|
| 1 | Cell_cycle | 108 | 124 | 87.10 |
9.50×10−5 |
| 2 |
Fanconi_anemia_pathway | 49 | 52 | 94.20 |
2.50×10−4 |
| 3 |
Pyrimidine_metabolism | 88 | 102 | 86.30 |
7.90×10−4 |
| 4 | Proteasome | 41 | 44 | 93.20 |
1.30×10−3 |
| 5 |
Pathways_in_cancer | 311 | 397 | 78.30 |
1.80×10−3 |
| 6 |
DNA_replication | 34 | 36 | 94.40 |
2.10×10−3 |
| 7 | Spliceosome | 109 | 131 | 83.20 |
2.50×10−3 |
| 8 |
Ribosome_biogenesis_in_eukaryotes | 66 | 76 | 86.80 |
2.60×10−3 |
| 9 |
MicroRNAs_in_cancer | 122 | 149 | 81.90 |
4.10×10−3 |
| 10 |
Glutamatergic_synapse | 94 | 113 | 83.20 |
5.00×10−3 |
Discussion
BC is the most common malignant tumour among women
worldwide (1), and has been
reported to be associated with the overexpression of oncogenes
(27–29). Despite significant improvements in
the diagnosis, treatment and survival prediction of BC, the
identification of novel biomarkers for prognosis is urgently
required. Studies have revealed that CCT6A expression is increased
in 10 human tumour cell lines and drug-resistant human melanoma
cell lines (12,13). However, to the best of our
knowledge, its expression in clinical tumour samples has not been
evaluated previously. Using the online Oncomine dataset, the
present study demonstrated that the majority of tumour tissues,
including breast, lung, liver and colorectal cancer tissues,
exhibited significantly higher CCT6A expression compared within
adjacent noncancerous tissues. In the majority of the BC datasets
(10/12), BC tissues exhibited significantly higher CCT6A mRNA
expression compared with in noncancerous breast tissues; the same
observation was confirmed at the protein level by
immunohistochemistry (IHC) through HPA. These results indicated
that CCT6A may be among the overexpressed genes in tumour tissues
and may serve a critical role in tumorigenesis. However, the
protein expression levels of CCT6A in BC samples should be further
validated via western blot analysis and immunohistochemistry.
It has previously been reported that increased CCT6A
expression is significantly associated with poor survival in
patients who have high TGF-β levels in lung cancer tissues, but not
in those with low TGF-β levels, suggesting that CCT6A serves as a
valuable biomarker for lung cancer (14). Using the Kaplan-Meier plotter, it
was demonstrated that high CCT6A expression was significantly
associated with unfavourable OS, RFS, DMFS and PPS in all patients
with BC, indicating that CCT6A mRNA expression may serve as a
prognostic indicator in patients with BC. In addition, CCT6A
overexpression in cancer cells significantly shortens the survival
time in tumour-bearing mice, whereas CCT6A knockdown significantly
prolongs survival (14). Taken
together, these observations further support the role of CCT6A as
an essential biomarker for survival prediction in patients with
cancer, including BC.
BC is classified into distinct histological and
biological subtypes with different pathological, molecular and
clinical features (30).
Therefore, this study further investigated the association between
CCT6A expression, at both the mRNA and protein levels, and
clinicopathological parameters in patients with BC. The findings
indicated that high CCT6A expression was negatively associated with
ER and PR status, and positively associated with nodal, basal-like
and TNBC status. Furthermore, higher SBR and NPI grades were
significantly associated with higher CCT6A expression. HER2 is a
member of the epidermal growth factor receptor family (31), and is associated with poor outcome
in patients with BC. However, this study found no association
between CCT6A expression and HER2 status. Taken together, these
findings demonstrated that increased CCT6A expression may be
significantly associated with poor outcomes in patients with BC,
while possibly serving as a novel biomarker for the prognosis of
BC.
The evaluation of biological functions has revealed
that CCT6A overexpression significantly increases colony and tumour
sphere formation, increases side population and reduces sensitivity
to anoikis in A549 cells following TGF-β stimulation (14). However, to the best of our
knowledge, the biological functions and underlying mechanism of
CCT6A in BC remain to be evaluated. Accordingly, following the
analyses of signalling pathways associated with CCT6A based on
correlated genes, it was demonstrated that ‘cell cycle’ was among
the most enriched KEGG signalling pathways. In addition, CCT6A
expression was positively correlated with the expression of CCNB2
and CCNA2, two important cell cycle regulators, thus suggesting
that CCT6A may be involved in tumour growth mediated by the cell
cycle. However, further investigations on the biological function
and underlying mechanism of action of CCT6A in BC are required.
Future studies should aim to assess the role of CCT6A in growth,
metastasis and chemotherapeutic resistance in cancer, including BC,
which may improve understanding of the role of CCT6A in the
progression of cancer.
In conclusion, the present study revealed that BC
tissues exhibited significantly higher CCT6A expression compared
with in noncancerous breast tissues at the mRNA and protein levels.
In addition, increased CCT6A expression was significantly
associated with unfavourable survival in patients with BC.
Furthermore, CCT6A expression was negatively associated with ER and
PR status, positively correlated with nodal, basal-like and TNBC
status, and was increased in patients with advanced SBR and NPI
grades. CCT6A was also revealed to be involved in the cell cycle,
and its expression was positively correlated with CCNB2 and CCNA2
expression. Collectively, CCT6A may be a potential target in
BC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request. The images presented in Fig.
2 were provided by the Human Protein Atlas: BC tissues
(https://www.proteinatlas.org/ENSG00000146731-CCT6A/pathology/tissue/breast+cancer#img);
normal breast tissues (https://www.proteinatlas.org/ENSG00000146731-CCT6A/tissue/breast#img).
Authors' contributions
KH and YW conceived and designed the experiments. KH
and YZ performed the Oncomine analysis. YX and LH conducted the HPA
analysis. YZ and YX conducted the Breast Cancer Gene-Expression
Miner analysis. LH and YX conducted the survival analysis. YW
conducted the R2 KEGG Pathway Finder analysis. KH and YW wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu Y, Walia V and Elble RC: Loss of CLCA4
promotes epithelial-to-mesenchymal transition in breast cancer
cells. PLoS One. 8:e839432013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hynes G, Kubota H and Willison KR:
Antibody characterisation of two distinct conformations of the
chaperonin-containing TCP-1 from mouse testis. FEBS Lett.
358:129–132. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kubota H, Hynes G, Carne A, Ashworth A and
Willison K: Identification of six Tcp-1-related genes encoding
divergent subunits of the TCP-1-containing chaperonin. Curr Biol.
4:89–99. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rommelaere H, Van Troys M, Gao Y, Melki R,
Cowan NJ, Vandekerckhove J and Ampe C: Eukaryotic cytosolic
chaperonin contains t-complex polypeptide 1 and seven related
subunits. Proc Natl Acad Sci USA. 90:11975–11979. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yam AY, Xia Y, Lin HT, Burlingame A,
Gerstein M and Frydman J: Defining the TRiC/CCT interactome links
chaperonin function to stabilization of newly made proteins with
complex topologies. Nat Struct Mol Biol. 15:1255–1262. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Van Hove I, Verslegers M, Hu TT, Carden M,
Arckens L and Moons L: A proteomic approach to understand
MMP-3-driven developmental processes in the postnatal cerebellum:
Chaperonin CCT6A and MAP kinase as contributing factors. Dev
Neurobiol. 75:1033–1048. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brackley KI and Grantham J: Activities of
the chaperonin containing TCP-1 (CCT): Implications for cell cycle
progression and cytoskeletal organisation. Cell Stress Chaperones.
14:23–31. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
von Kriegsheim A, Baiocchi D, Birtwistle
M, Sumpton D, Bienvenut W, Morrice N, Yamada K, Lamond A, Kalna G,
Orton R, et al: Cell fate decisions are specified by the dynamic
ERK interactome. Nat Cell Biol. 11:1458–1464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiang B, Chatti K, Qiu H, Lakshmi B,
Krasnitz A, Hicks J, Yu M, Miller WT and Muthuswamy SK: Brk is
coamplified with ErbB2 to promote proliferation in breast cancer.
Proc Natl Acad Sci USA. 105:12463–12468. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roskoski R Jr: ERK1/2 MAP kinases:
Structure, function, and regulation. Pharmacol Res. 66:105–143.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tanic N, Brkic G, Dimitrijevic B,
Dedovic-Tanic N, Gefen N, Benharroch D and Gopas J: Identification
of differentially expressed mRNA transcripts in drug-resistant
versus parental human melanoma cell lines. Anticancer Res.
26:2137–2142. 2006.PubMed/NCBI
|
|
13
|
Myung JK, Afjehi-Sadat L,
Felizardo-Cabatic M, Slavc I and Lubec G: Expressional patterns of
chaperones in ten human tumor cell lines. Proteome Sci. 2:82004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ying Z, Tian H, Li Y, Lian R, Li W, Wu S,
Zhang HZ, Wu J, Liu L, Song J, et al: CCT6A suppresses SMAD2 and
promotes prometastatic TGF-β signaling. J Clin Invest.
127:1725–1740. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrateddata-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Richardson AL, Wang ZC, De Nicolo A, Lu X,
Brown M, Miron A, Liao X, Iglehart JD, Livingston DM and Ganesan S:
X chromosomal abnormalities in basal-like human breast cancer.
Cancer Cell. 9:121–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shin S, Kim Y, Chul Oh S, Yu N, Lee ST,
Rak Choi J and Lee KA: Validation and optimization of the Ion
Torrent S5 XL sequencer and Oncomine workflow for BRCA1 and BRCA2
genetic testing. Oncotarget. 8:34858–34866. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uhlen M, Oksvold P, Fagerberg L, Lundberg
E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S,
et al: Towards a knowledge-based Human Protein Atlas. Nat
Biotechnol. 28:1248–1250. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jézéquel P, Campone M, Gouraud W,
Guérin-Charbonnel C, Leux C, Ricolleau G and Campion L:
bc-GenExMiner: An easy-to-use online platform for gene prognostic
analyses in breast cancer. Breast Cancer Res Treat. 131:765–775.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jézéquel P, Frénel JS, Campion L,
Guérin-Charbonnel C, Gouraud W, Ricolleau G and Campone M:
bc-GenExMiner 3.0: New mining module computes breast cancer gene
expression correlation analyses. Database (Oxford).
2013:bas0602013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu Y, Shang R, Chen Y, Li J, Liang Z, Hu
J, Liu K and Chen C: Tumor suppressive ZBTB4 inhibits cell growth
by regulating cell cycle progression and apoptosis in Ewing
sarcoma. Biomed Pharmacother. 100:108–115. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Le Doussal V, Tubiana-Hulin M, Friedman S,
Hacene K, Spyratos F and Brunet M: Prognostic value of histologic
grade nuclear components of Scarff-Bloom-Richardson (SBR). An
improved score modification based on a multivariate analysis of
1262 invasive ductal breast carcinomas. Cancer. 64:1914–1921. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Galea MH, Blamey RW, Elston CW and Ellis
IO: The Nottingham prognostic index in primary breast cancer.
Breast Cancer Res Treat. 22:207–219. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li C, Du L, Ren Y, Liu X, Jiao Q, Cui D,
Wen M, Wang C, Wei G, Wang Y, et al: SKP2 promotes breast cancer
tumorigenesis and radiation tolerance through PDCD4 ubiquitination.
J Exp Clin Cancer Res. 38:762019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pucci S, Polidoro C, Greggi C, Amati F,
Morini E, Murdocca M, Biancolella M, Orlandi A, Sangiuolo F and
Novelli G: Pro-oncogenic action of LOX-1 and its splice variant
LOX-1Δ4 in breast cancer phenotypes. Cell Death Dis. 10:532019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
O'Toole SA, Selinger CI, Millar EK, Lum T
and Beith JM: Molecular assays in breast cancer pathology.
Pathology. 43:116–127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Michaut M, Chin SF, Majewski I, Severson
TM, Bismeijer T, de Koning L, Peeters JK, Schouten PC, Rueda OM,
Bosma AJ, et al: Integration of genomic, transcriptomic and
proteomic data identifies two biologically distinctsubtypes of
invasive lobular breast cancer. Sci Rep. 6:185172016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Slamon DJ, Clark GM, Wong SG, Levin WJ,
Ullrich A and McGuire WL: Human breast cancer: Correlation of
relapse and survival with amplification of the HER-2/neu oncogene.
Science. 235:177–182. 1987. View Article : Google Scholar : PubMed/NCBI
|