Introduction
Congenital heart disease (CHD) is a structural and
functional defect induced by the abnormal development of
cardiovascular blood vessels. CHD is the most common type of birth
defect and the leading cause of fetal mortality (1). CHD frequently leads to miscarriage,
stillbirth and severe cardiopathy following birth, which seriously
endangers neonatal health (2,3).
Ventricular septal defect (VSD) is the most common type of CHD,
comprising 40% of all cardiac abnormalities (4). At present, it is hypothesized that
CHD is primarily induced by a combination of genetic and
environmental factors, leading to cardiac malformation at the
embryonic stage; only 2–5% of CHD cases result from environmental
factors alone (5–7). Previous studies have revealed certain
genes associated with cardiac developmental abnormalities,
including Notch homolog 1, heart- and neural crest
derivatives-expressed protein 2 (HAND2) and GATA (8); however, the precise mechanisms
underlying the roles of these genes in the regulation of embryonic
heart development are yet to be determined. Therefore, further
study of the mechanisms underlying embryonic heart formation and
development is required to develop novel strategies for the
prevention of CHD.
Long noncoding RNAs (lncRNAs) comprise a group of
transcripts that are >200 nucleotides in length. LncRNAs do not
encode proteins as they lack a specific open reading frame
(9). Functionally, lncRNAs
regulate gene expression by influencing chromatin remodeling,
transcriptional regulation, and post-transcriptional modification
(10,11). Previous studies have demonstrated
that lncRNAs exhibit important roles in the development of the
cardiovascular system and the pathogenesis of associated disorders,
including myocardial infarction, heart failure, dilated
cardiomyopathy and VSD (12,13).
For example, lncRNA cardiac hypertrophy-related factor induces
cardiac hypertrophy via competitive binding to microRNA (miR)-489
(14), whereas lncRNA braveheart
(Bvht) promotes stem cell differentiation during cardiac
development (15).
LncRNA uc.457 is located at 22q11.21 (chr22:
19395909-19396119) and is 211 bp in length. Our previous study
reported that uc.457 is differentially expressed in children with
VSD (16); however, its specific
role in cardiac development remains unclear. P19 cells are
pluripotent stem cells that can be cultured in vitro. The
self-replication ability and differentiation potential of P19 cells
enable them to differentiate into cardiomyocyte-like cells when
induced by a low concentration of dimethyl sulfoxide (DMSO)
(17). P19 cells have been widely
utilized in the study of cardiac development (18).
In the present study, differentiated P19 cells were
employed to investigate the effects of differential expression of
uc.457 on the proliferation and differentiation of cardiomyocytes,
providing novel insight into the mechanisms of cardiac
development.
Materials and methods
Cell culture and transfection
Mouse embryonic carcinoma P19 cells were obtained
from the American Type Culture Collection (Manassas, VA, USA).
Cells were cultured in α-minimal essential medium (α-MEM; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin and 100 mg/ml streptomycin. Cells were
maintained in a 5% CO2 incubator at 37°C. Cells were
seeded in 6-well plates at a density of 5×104 cells/ml.
The medium was replaced by complete medium without antibiotics 6 h
before transfection. Plasmids (pGPU6/GFP/Neo-uc.457 and
pGPU6/GFP/Neo-siRNA-uc.457) were constructed by Shanghai GenePharma
Co., Ltd., (Shanghai, China). The sequences of the 3 siRNAs were as
follows: uc457-siRNA: 5′-GGGCCTTATCTTTCTAATTAC-3′; siNC:
5′-GTTCTCCGAACGTGTCACGT-3′; siGAPDH: 5′-GTATGACAACAGCCTCAAG-3′.
Cells were seeded in 6-well culture plates and grown to 70–80%
confluence before transfection. Transfection was conducted
according to the Lipofectamine® 2000 DNA transfection
reagent protocol (Life Technologies; Thermo Fisher Scientific,
Inc.). Then, 24 h after transfection, the confluence was
approximately 90% and the cells were removed to grow in flasks
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The cells were
also collected to confirm uc.457 silencing efficiency and
overexpression by reverse transcription-quantitative polymerase
chain reaction (RT-qPCR).
Cardiomyocyte differentiation
P19 cells were seeded in a 10-cm culture dish at a
density of 1×106 cells/ml. Cardiomyocyte differentiation
was induced using α-MEM containing 1% DMSO at 37°C. On the 4th day
post-induction, 10 embryonic bodies were seeded in 6-well plates.
Autonomously beating cardiomyocyte-like cell masses were first
observed on the 8th day, and were largely present by the 10th
day.
Cell Counting Kit-8 (CCK-8) assay
P19 cells were seeded in 96-well plates at a density
of 3×104 cells/ml. CCK-8 reagent (10 µl; Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) was added to each
well for 4 consecutive days. Following incubation for a further 2 h
at 37°C, the optical density of each well was measured at 450 nm
using a microplate reader (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
RNA extraction and RT-qPCR
Total RNA was isolated from cultured P19 cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.) and RT
was performed using a PrimeScript RT Reagent kit (Takara Bio, Inc.,
Otsu, Japan) according to the manufacturer's protocols. RNA
concentration was determined using a spectrophotometer (Hitachi,
Ltd., Tokyo, Japan) and an ABI Prism 7500 cycler. qPCR was
performed using the following thermocycling conditions: 95°C for 30
sec, 95°C for 5 sec and 60°C for 30 sec, for a total of 40 cycles.
Gene expression was measured in triplicate. The data was analyzed
using the 2−ΔΔCq method (19). The relative gene expression levels
were quantified based on the Ct and normalized to a reference gene,
GAPDH. The primers used were as follows: Uc.457, forward
5′-CCTTTGCAGGCTTTGCGTG-3′, reverse, 5′-CCGCACGGGGCCTTATCTT-3′;
histone cell cycle regulation defective homolog A (HIRA), forward
5′-CTGGACACTGGGTACTCACTC-3′, reverse,
5′-AACTGGCTAACTGACAACAGAAG-3′; natriuretic peptide A (NPPA),
forward 5′-GCTTCCAGGCCATATTGGAG-3′, reverse,
5′-GGGGGCATGACCTCATCTT-3′; cardiac muscle troponin T (cTnT),
forward 5′-CAGAGGAGGCCAACGTAGAAG-3′, reverse,
5′-CTCCATCGGGGATCTTGGGT-3′; myocyte-specific enhancer factor 2C
(Mef2c), forward 5′-ATCCCGATGCAGACGATTCAG-3′, reverse,
5′-AACAGCACACAATCTTTGCCT-3′; GAPDH, forward
5′-CTGCGACTTCAACAGCAACT-3′, reverse,
5′-GAGTTGGGATAGGGCCTCTC-3′.
Western blotting
P19 cells were lysed using cell lysis buffer
(Nanjing KeyGen Biotech Co., Ltd., Nanjing, China), agitated on ice
for 30 min and centrifuged (14,000 × g) at 4°C for 15 min. The
total protein concentration was calculated using a bicinchoninic
acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.).
Extracted proteins (50 µg of total protein per lane) were separated
via 10% SDS-PAGE and subsequently transferred onto polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA). The
membranes were blocked with 5% Bovine Serum Albumin (BSA;
Sigma-Aldrich; Merck KGaA) in TBST (50 mM tris-buffered saline, pH
7.5, 150 mM NaCl, 0.05% Tween-20). The membrane was incubated at
4°C overnight in 5% BSA in TBST containing primary antibodies to
one of the following: HIRA (1:200; ab20655; Abcam, Cambridge, UK),
NPPA (1:200; ab180649; Abcam), CTnT (1:250; ab209813; Abcam), Mef2c
(1:200; ab211493; Abcam) and β-actin (1:1,000; ab124964; Abcam).
The membrane was washed 5 times with TBST for 5 min each wash.
Following washing, the membrane was incubated with horseradish
peroxidase (HRP) conjugated goat anti-rabbit secondary antibody
(ab6721; Abcam) at 1:5,000 dilution for 1 h at room temperature,
then washed with TBST. Western blot analysis was performed
according to standard procedures.
Bioinformatics analysis
The University of California, Santa Cruz (UCSC)
Genome Browser (http://genome.ucsc.edu/) was employed to identify
transcription factor (TF) binding sites, TF partners, histone
modification and DNase I hypersensitive sites (indicative of open
chromatin) in the uc.457 region.
Statistical analysis
Each experiment was performed at least three times.
SPSS version 16.0 software (SPSS, Inc., Chicago, IL, USA) was used
for data analysis. Data were presented as the mean ± standard
deviation. Continuous variables were analyzed by Student's t-tests.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Biological predictions of uc.457
The UCSC database was employed to predict the
potential biological functions of uc.457. It was revealed that
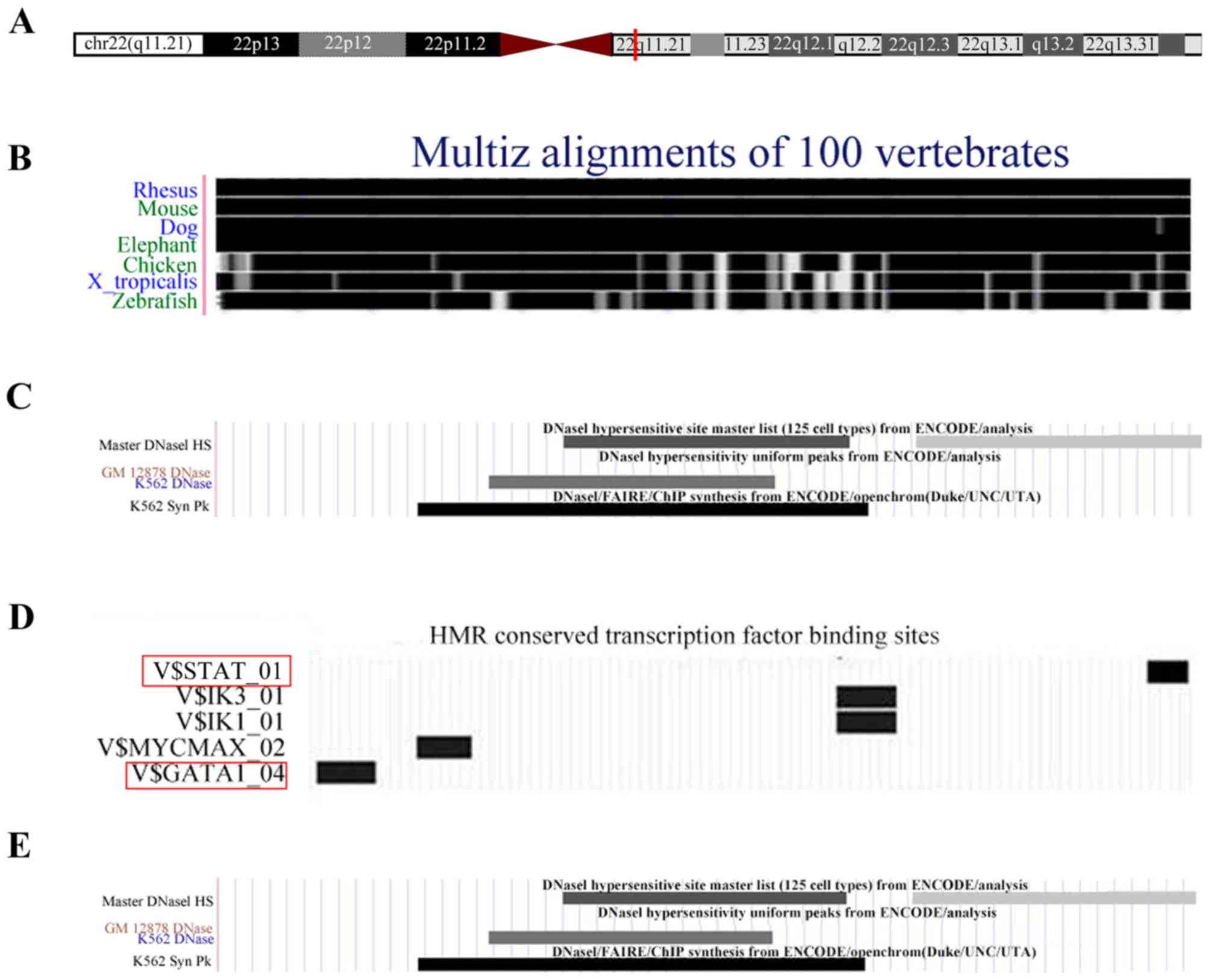
uc.457 is located at 22q11.21 (chr22: 19395909-19396119) and is 211
bp in length (Fig. 1A). In
addition, uc.457 is highly conserved among vertebrates, with
identical sequences in humans, rhesus monkeys and mice (Fig. 1B). By analyzing the DNase I
footprint, TFs and histone modifications, it was determined that
uc.457 contains a binding site for cis-acting elements
(Fig. 1C). Uc.457 was also
predicted to be regulated by signal transducer and activator of
transcription (STAT) and GATA-binding factor 1 (GATA1; Fig. 1D). There were numerous histone
modifications identified within the chromosomal region of uc.457,
including H3K4me1, H3K9ac, H3K27ac, H3K27me3 and H3K36me3 (Fig. 1E).
Uc.457 overexpression suppresses the
proliferation of P19 cells
To determine the effects of uc.457 on cardiac
development, uc.457 overexpression plasmids and small interfering
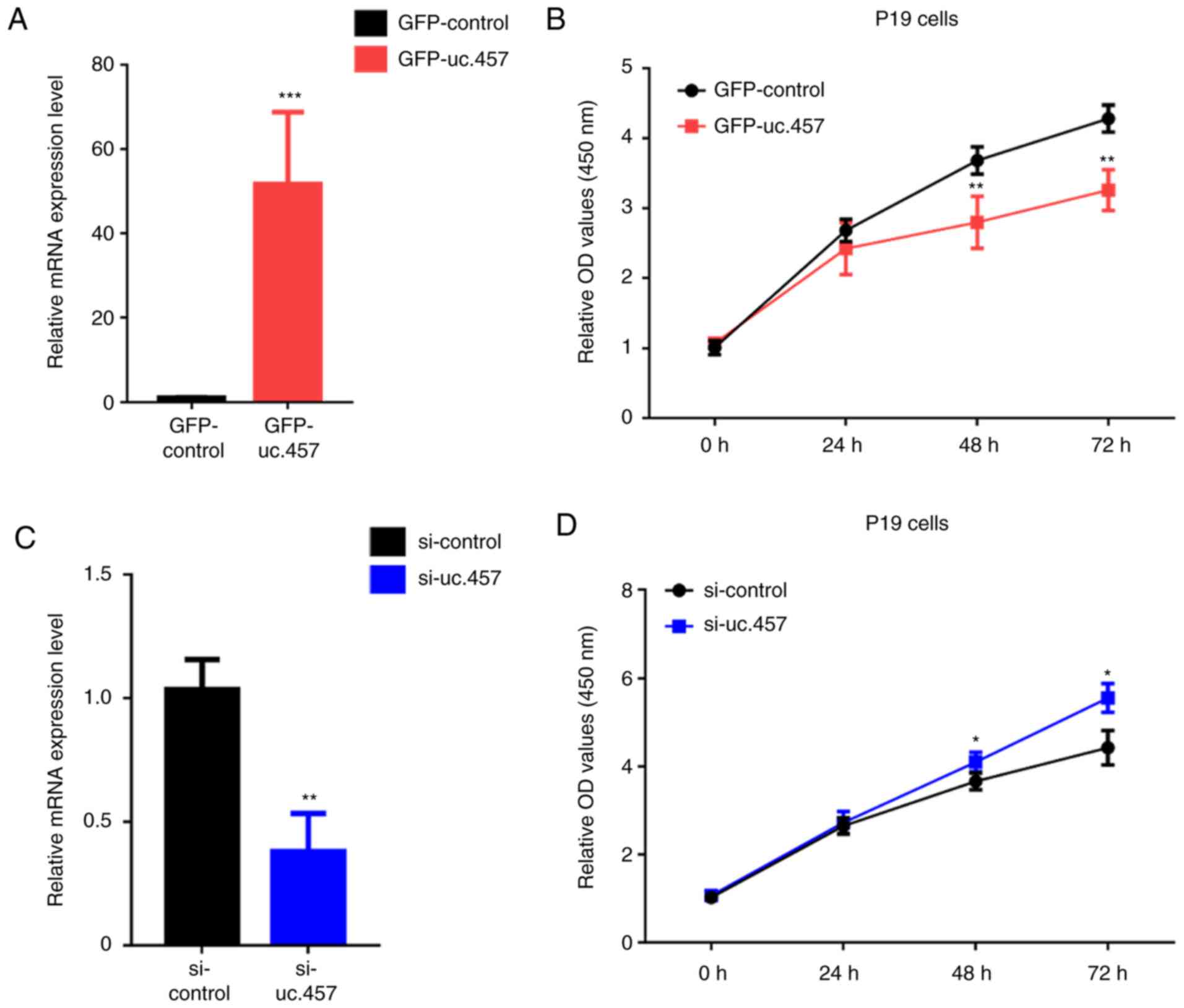
RNA (siRNA) against uc.457 were generated. Overexpression of uc.457
significantly inhibited the proliferation of P19 cells in a
time-dependent manner, with significant decreases in absorbance
observed at 48 and 72 h compared with transfection with green
fluorescent protein (GFP)-control (pGPU6/GFP/Neo; Fig. 2A and B). Conversely, siRNA-mediated
knockdown of uc.457 significantly increased the proliferation of
cells compared with the control (Fig.
2C and D).
Uc.457 overexpression inhibits the
differentiation of P19 cells
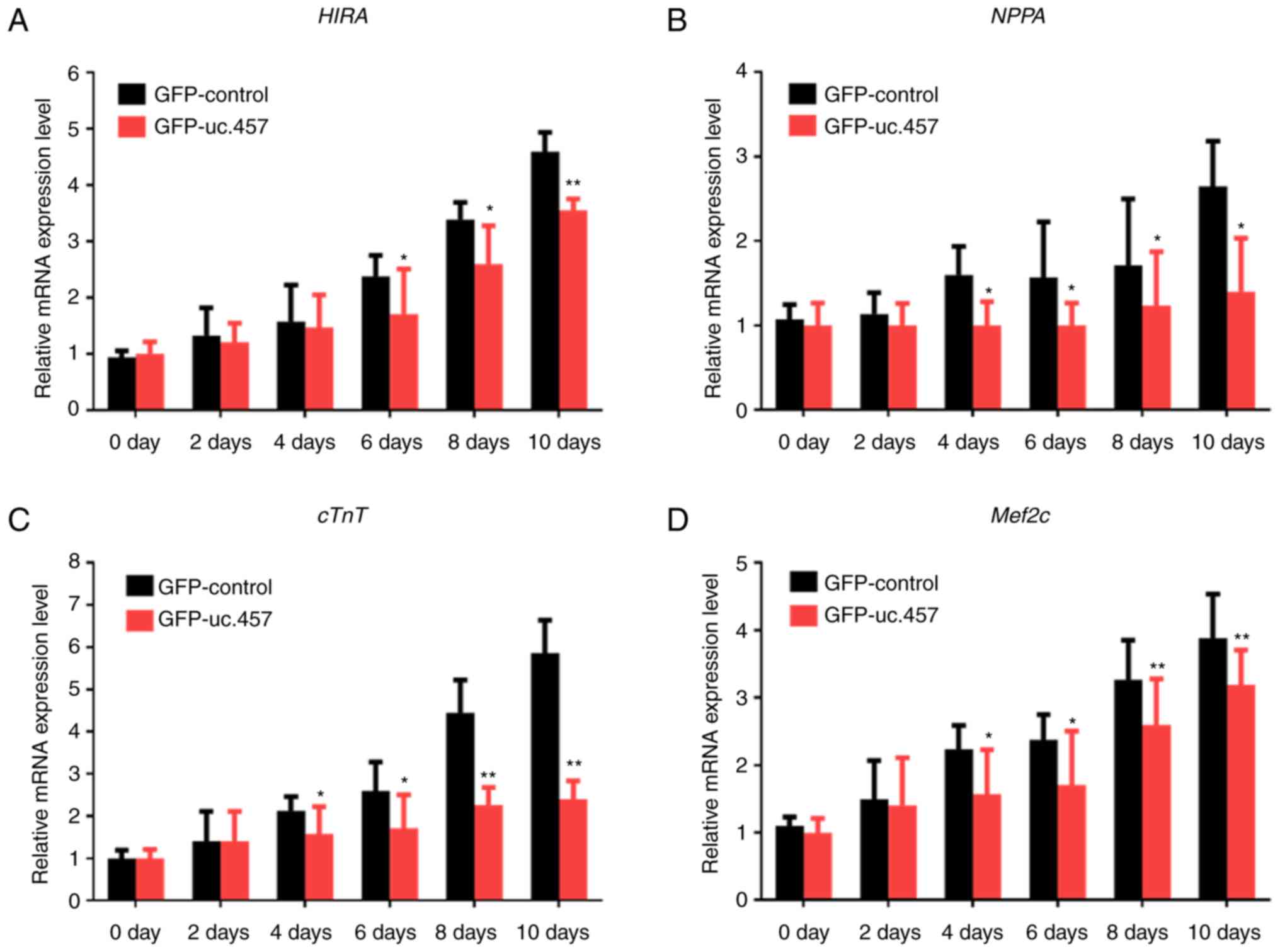
The mRNA expression levels of the cardiomyocyte
maturation-associated genes HIRA, NPPA, cTnT and Mef2c were
markedly increased over time in differentiating P19 cells, reaching
a peak on the 10th day (Fig. 3);
however, overexpression of uc.457 notably suppressed. Significant
reductions in expression following uc.457 were observed from day 4
for NPPA and cTnT, and day 6 for HIRA and Mef2c compared with the
control.
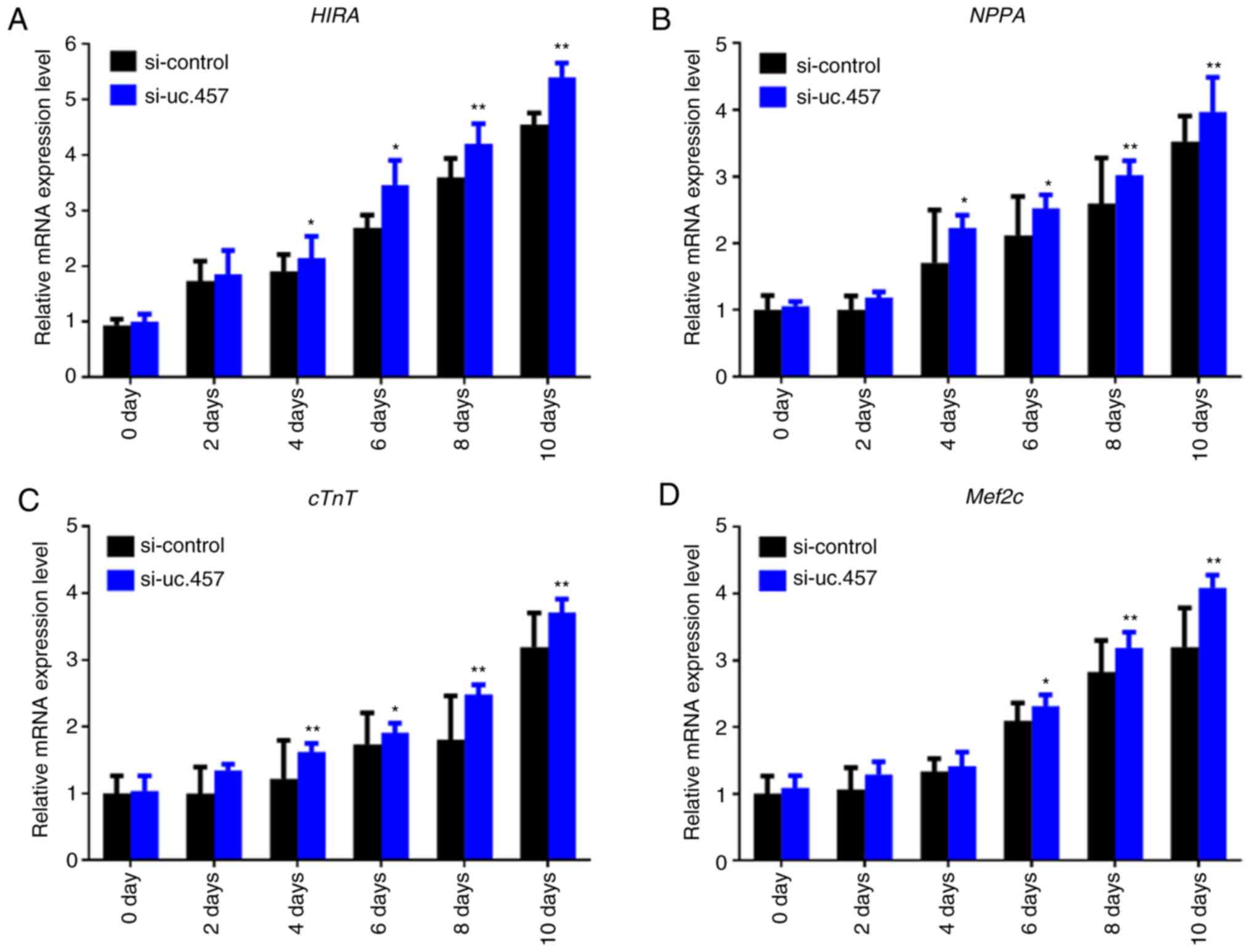
Uc.457 knockdown promotes the
differentiation of P19 cells
To further investigate the effects of uc.457 on the
differentiation of P19 cells, the mRNA expression levels of HIRA,
NPPA, cTnT and Mef2c were determined following knockdown of uc.457.
It was revealed that transfection with uc.457 siRNA (si-uc.457)
significantly upregulated the expression of HIRA, NPPA and cTnT
from 4 days, and Mef2c from 6 days in P19 cells compared with in
control siRNA (si-control)-transfected cells (Fig. 4).
Uc.457 regulates the expression of
genes associated with cardiomyocyte differentiation
Western blot analysis demonstrated that the protein
expression of HIRA, NPPA, cTnT, and Mef2c increased in a
time-dependent manner under conditions of cell differentiation
(Fig. 5). Overexpression of uc.457
in P19 cells resulted in a marked downregulation of these proteins,
whereas si-uc.457 transfection was associated with upregulated
expression.
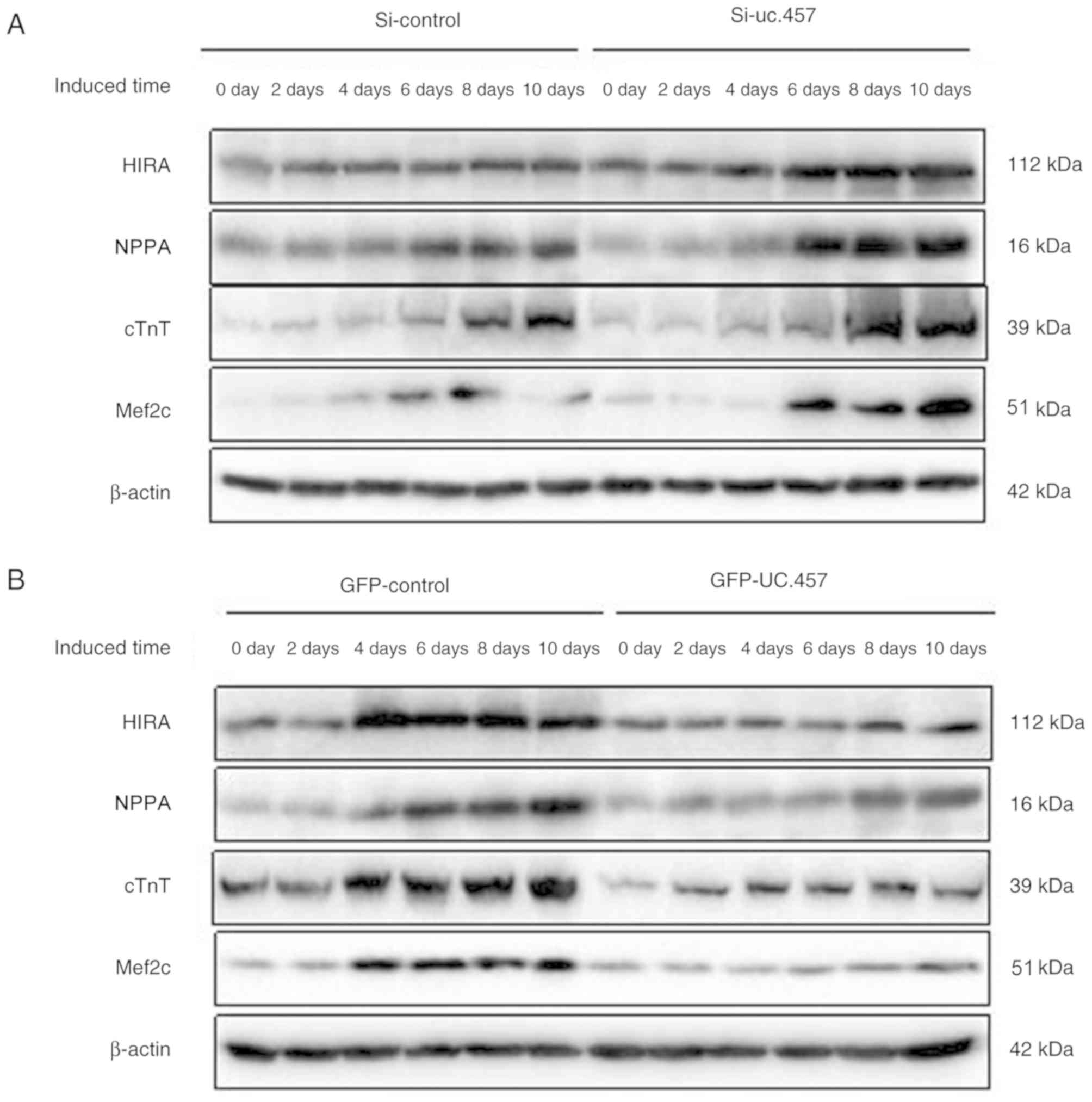 | Figure 5.Uc.457 regulates the expression of
genes associated with cardiomyocyte differentiation. (A) Expression
levels of HIRA, NPPA, cTnT, and Mef2C following si-uc.457 or
si-control transfection. (B) Expression levels of HIRA, NPPA, cTnT
and Mef2c following GFP-uc.457 or GFP-control plasmid transfection.
cTnT, cardiac muscle troponin T; GFP, green fluorescent protein;
HIRA, histone cell cycle regulation defective homolog A; Mef2c,
myocyte-specific enhancer factor 2C; NPPA, natriuretic peptide A;
si, small interfering RNA. |
Discussion
CHD is the most common type of congenital
malformation in infants and young children, and is the leading
cause of mortality in members of this population with
non-infectious diseases (20).
Cardiac development involves the migration, differentiation,
proliferation and apoptosis of various types of cells, including
cardiomyocytes, endocardial cells and cardiac neural crest cells
(7). The development of CHD
involves a precise and complex network of various TFs, cell
adhesion molecules and signaling molecules (21,22).
A previous study demonstrated that genetic defects are the leading
cause of cardiac dysplasia (23);
GATA4, homeobox protein NKX2.5 and HAND2 have been identified to be
associated with abnormal heart development (24,25).
Post-transcriptional regulation also serves an important role in
heart development; noncoding RNAs are notably involved in
post-transcriptional regulation (26).
It has been demonstrated the important roles of
lncRNAs in embryonic and cardiovascular development (27). In the process of stem cell
differentiation, LncRNA Bvht stimulates the differentiation of
cardiac cells by activating a gene network that promotes the
formation of pluripotent cardiac progenitor cells from cardiac
mesoderm (15). Furthermore, Bvht
interacts with polycomb repressive complex 2, which in turn
regulates the differentiation of cell lineages and cardiac
formation (28). Additionally,
lncRNA TERMINATOR regulates the differentiation state of
pluripotent stem cells and regulates the function of the
cardiovascular development-associated lncRNA ALIEN, whereas lncRNA
PUNISHER attenuates the function of cardiac endothelial cells
(29). Grote et al
(30) found that the
tissue-specific lncRNA Fendrr is an essential regulator of heart
and body wall development in the mouse.
Our previous study identified numerous
differentially expressed lncRNAs in heart tissues from patients
with VSD (16). Subsequent
bioinformatics analyses predicted that these lncRNAs were closely
associated with cardiac development, apoptosis and proliferation;
however, their biological functions were not verified. In the
present study, the biological functions of uc.457 were investigated
using an in vitro model. Via bioinformatics analysis, it was
revealed that uc.457 is highly conserved among vertebrates and
contains a binding site for the TFs, STAT (associated with the
growth, survival and apoptosis of cardiomyocytes) and GATA1
(associated with cardiac development). In addition, the chromosomal
region of uc.457 is enriched with H3K4me1, H3K9ac and H3K27ac
histone modifications. Various studies have reported that histone
lysine methylation is closely associated with heart development and
CHD (31,32). In the present study, P19 cells
overexpressing uc.457 exhibited reduced proliferation compared with
the control, whereas uc.457 silencing increased proliferation,
suggesting that uc.457 may affect cardiac development by regulating
cell proliferation.
Additionally, during the differentiation of P19
cells into cardiomyocytes, it was revealed that overexpression of
uc.457 downregulated the expression of HIRA, NPPA, cTnT and Mef2c.
Song et al (33) reported
that lncRNAs and cardiomyogenesis-associated RNAs act
synergistically during cardiac differentiation; for example,
co-expression of Mef2c and lncRNA uc.167 partially reversed the
suppressive effects of lncRNA uc.167 on the proliferation,
apoptosis, and differentiation of P19 cells. Long intergenic
non-protein coding RNA, muscle differentiation 1 sequesters miR-135
to regulate the expression of Mef2c, serving a role in the
activation of muscle-specific gene expression (34). As an H3.3 chaperone, HIRA is a
negative regulator of histone gene expression (35). It is highly conserved among various
species and is differentially expressed during embryonic
development; HIRA is important for embryonic survival and
physiological development (35).
It has been reported that mutations in HIRA prevent the axial
rotation of embryos and normal heart looping, leading to CHD
(36). HIRA is also located on the
long arm of human chromosome 22, specifically at 22q11.2 (37). Therefore, it was considered that
uc.457 may affect the differentiation of cardiac precursors into
mature cardiomyocytes via an unknown mechanism.
P19 cells are characterized by a single genetic
background with a simple method to induce differentiation compared
with human embryonic stem cells, mesenchymal stem cells and induced
pluripotent stem cells (15).
Additional cell lines such as H9C2 cells and in vivo
experiments are required to further determine the roles of uc.457
in cardiac development.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81870167
and 81570209) and the Nanjing Medical Science and Technique
Development Foundation (grant no. Ykk15148).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
QZ performed the majority of the experiments and
wrote the manuscript. ZC and ZY analyzed the data. CZ and LQ
designed the study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sadowski SL: Congenital cardiac disease in
the newborn infant: Past, present, and future. Crit Care Nurs Clin
North Am. 2137–48. (vi)2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Srivastava D: Making or breaking the
heart: From lineage determination to morphogenesis. Cell.
126:1037–1048. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van der Linde D, Konings EE, Slager MA,
Witsenburg M, Helbing WA, Takkenberg JJ and Roos-Hesselink JW:
Birth prevalence of congenital heart disease worldwide: A
systematic review and meta-analysis. J Am Coll Cardiol.
58:2241–2247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Trojnarska O, Grajek S, Katarzyński S and
Kramer L: Predictors of mortality in adult patients with congenital
heart disease. Cardiol J. 16:341–347. 2009.PubMed/NCBI
|
|
5
|
Blue GM, Kirk EP, Sholler GF, Harvey RP
and Winlaw DS: Congenital heart disease: Current knowledge about
causes and inheritance. Med J Aust. 197:155–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bruneau BG: The developmental genetics of
congenital heart disease. Nature. 451:943–948. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Olson EN: Gene regulatory networks in the
evolution and development of the heart. Science. 313:1922–1927.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
de la Pompa JL and Epstein JA:
Coordinating tissue interactions: Notch signaling in cardiac
development and disease. Dev Cell. 22:244–254. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Frank S, Aguirre A, Hescheler J and Kurian
L: A lncRNA perspective into (Re)Building the heart. Front Cell Dev
Biol. 4–128. 2016.PubMed/NCBI
|
|
11
|
Shen S, Jiang H, Bei Y, Xiao J and Li X:
Long non-coding RNAs in cardiac remodeling. Cell Physiol Biochem.
41:1830–1837. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ounzain S, Crippa S and Pedrazzini T:
Small and long non-coding RNAs in cardiac homeostasis and
regeneration. Biochim Biophys Acta. 1833:923–933. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Di Mauro V, Barandalla-Sobrados M and
Catalucci D: The noncoding-RNA landscape in cardiovascular health
and disease. Noncoding RNA Res. 3:12–19. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang K, Liu F, Zhou LY, Long B, Yuan SM,
Wang Y, Liu CY, Sun T, Zhang XJ and Li PF: The long noncoding RNA
CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res.
114:1377–1388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Klattenhoff CA, Scheuermann JC, Surface
LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey
L, Haas S, et al: Braveheart, a long noncoding RNA required for
cardiovascular lineage commitment. Cell. 152:570–583. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song G, Shen Y, Zhu J, Liu H, Liu M, Shen
YQ, Zhu S, Kong X, Yu Z and Qian L: Integrated analysis of
dysregulated lncRNA expression in fetal cardiac tissues with
ventricular septal defect. PLoS One. 8:e774922013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
van der Heyden MA, van Kempen MJ, Tsuji Y,
Rook MB, Jongsma HJ and Opthof T: P19 embryonal carcinoma cells: A
suitable model system for cardiac electrophysiological
differentiation at the molecular and functional level. Cardiovasc
Res. 58:410–422. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wen J, Xia Q, Lu C, Yin L, Hu J, Gong Y,
Yin B, Monzen K, Yuan J, Qiang B, et al: Proteomic analysis of
cardiomyocytes differentiation in mouse embryonic carcinoma P19CL6
cells. J Cell Biochem. 102:149–160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hoelscher SC, Doppler SA, Dreßen M, Lahm
H, Lange R and Krane M: MicroRNAs: Pleiotropic players in
congenital heart disease and regeneration. J Thorac Dis. 9 (Suppl
1):S64–S81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nemer M: Genetic insights into normal and
abnormal heart development. Cardiovasc Pathol. 17:48–54. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bajolle F, Zaffran S and Bonnet D:
Genetics and embryological mechanisms of congenital heart diseases.
Arch Cardiovasc Dis. 102:59–63. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Andersen TA, Troelsen Kde L and Larsen LA:
Of mice and men: Molecular genetics of congenital heart disease.
Cell Mol Life Sci. 71:1327–1352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garg V, Kathiriya IS, Barnes R,
Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS,
Hirayama-Yamada K, Joo K, et al: GATA4 mutations cause human
congenital heart defects and reveal an interaction with TBX5.
Nature. 424:443–447. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pabst S, Wollnik B, Rohmann E, Hintz Y,
Glänzer K, Vetter H, Nickenig G and Grohé C: A novel stop mutation
truncating critical regions of the cardiac transcription factor
NKX2-5 in a large family with autosomal-dominant inherited
congenital heart disease. Clin Res Cardiol. 97:39–42. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sallam T, Sandhu J and Tontonoz P: Long
noncoding RNA discovery in cardiovascular disease: Decoding form to
function. Circ Res. 122:155–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zangrando J, Zhang L, Vausort M, Maskali
F, Marie PY, Wagner DR and Devaux Y: Identification of candidate
long non-coding RNAs in response to myocardial infarction. BMC
Genomics. 15:4602014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kurian L, Aguirre A, Sancho-Martinez I,
Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Krause MN, Nelles
DA, et al: Identification of novel long noncoding RNAs underlying
vertebrate cardiovascular development. Circulation. 131:1278–1290.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Grote P, Wittler L, Hendrix D, Koch F,
Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M and
Herrmann BG: The tissue-specific lncRNA Fendrr is an essential
regulator of heart and body wall development in the mouse. Dev
Cell. 24:206–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ang SY, Uebersohn A, Spencer CI, Huang Y,
Lee JE, Ge K and Bruneau BG: KMT2D regulates specific programs in
heart development via histone H3 lysine 4 di-methylation.
Development. 143:810–821. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cloos PA, Christensen J, Agger K and Helin
K: Erasing the methyl mark: Histone demethylases at the center of
cellular differentiation and disease. Genes Dev. 22:1115–1140.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song G, Shen Y, Ruan Z, Li X, Chen Y, Yuan
W, Ding X, Zhu L and Qian L: LncRNA-uc.167 influences cell
proliferation, apoptosis and differentiation of P19 cells by
regulating Mef2c. Gene. 590:97–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Szenker E, Lacoste N and Almouzni G: A
developmental requirement for HIRA-dependent H3.3 deposition
revealed at gastrulation in Xenopus. Cell Rep. 1:730–740. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dilg D, Saleh RN, Phelps SE, Rose Y,
Dupays L, Murphy C, Mohun T, Anderson RH, Scambler PJ and Chapgier
AL: HIRA is required for heart development and directly regulates
Tnni2 and Tnnt3. PLoS One. 11:e01610962016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ju ZR, Wang HJ, Ma XJ, Ma D and Huang GY:
HIRA gene is lower expressed in the myocardium of patients with
tetralogy of fallot. Chin Med J (Engl). 129:2403–2408. 2016.
View Article : Google Scholar : PubMed/NCBI
|