Introduction
Prostate cancer is the second leading cause of
cancer-associated mortality in males worldwide (1). Nowadays, conventional drugs such as
cis-platinum and mitomycin are usually selected for use as
auxiliaries in chemotherapy or radiotherapy for prostate carcinoma
treatment (2,3). Several studies have directly or
indirectly implied that patients with terminal cancer could be
treated in the future using drugs targeting critical endogenous
factors such as Her2/Neu, epidermal growth factor or tumor necrosis
factor-α (2–5). Improved target selection of
anti-cancer drugs is very important for clinical research. BLM RecQ
like helicase (BLM) is a member of the RecQ helicase family that
has a pivotal role in genetic recombination, transcription, DNA
replication and DNA repair. BLM gene defects can cause Bloom
syndrome, accompanied by cancer predisposition (6–9). A
recent study indicated that knockdown of BLM impairs the normal
proliferation and metabolism of cancer cells (5). Drugs targeting BLM have been used to
treat cancer (5,10–14).
However, the majority of anti-cancer drugs are ineffective due to
poor target specificity, and certain regulation mechanisms, such as
the post-transcriptional control pathway of BLM gene expression,
remain unclear, which limits the application of drugs targeting BLM
for carcinoma therapy in the future. Therefore, it is urgent to
identify endogenous factors that control BLM gene expression for
subsequent cancer therapy research.
MicroRNAs (miRNAs) are small, noncoding RNA
molecules that inhibit gene expression by post-transcriptional
interaction with the 3′ untranslated region (3′-UTR) of target
mRNAs. miRNAs are ~20 nucleotides in length and are highly
conserved short single-stranded RNA molecules (15–17).
Increasing evidence has revealed the critical roles of miRNAs in
proliferation, colony formation, migration, invasion and DNA
metabolism (18–20). miRNAs also have potential to be
applied in cancer therapies as a novel drugs, and certain miRNAs
that regulate BLM expression may be important for prostate
carcinoma treatment. However, which miRNAs alter BLM helicase
expression remains unclear. Recent studies have suggested miRNAs
are involved in proliferation, differentiation, invasion,
migration, cell cycle and apoptosis by acting on BLM mRNA (21,22).
Identifying miRNAs that target BLM is very useful and will expand
the understanding of the relationship among miRNAs, BLM and cancer,
as well as provide a prospective clinical treatment strategies for
patients with prostate cancer by targeting BLM. However, one gene
may be regulated by multiple miRNAs, so that the synergy or
antagonism of miRNAs in post-transcriptional suppression process
has huge potential benefits for novel miRNA and drug combination
treatments. Therefore, understanding the miRNA interactions is
important for further studies.
In the present study, prostate cancer cell line PC3
was selected for use in investigating the post-transcriptional
regulation BLM gene expression by miRNA, and to understand whether
potential anti-cancer drugs targeting BLM could be used in the
future. Five miRNA were selected as candidates using online
software and it was determined whether these have a negative effect
on with BLM gene expression levels. miRNA mimics were overexpressed
in PC3 cells and the BLM mRNA expression levels were subsequently
detected. The miRNA seed sequence recognizes the target gene
3′-UTR, and a dual luciferase vector was constructed to detect
miRNA effects on the target gene. Finally, western blot analysis
was performed to monitor BLM protein expression levels following
candidate miRNA overexpression. To determine the functional effects
of multiple miRNAs in BLM gene expression, a Box-Behnken Design
(BBD) experiment was designed to demonstrate the mutual effects
between miRNAs. Proliferation, colony formation, migration and
invasion assays provided further evidence for the hypothesis that
miRNAs cooperatively regulate BLM gene expression and impact the
normal proliferation and metabolism of cancer cells.
Materials and methods
miRNA screening
The tools, including miRSystem (mirsystem.cgm.ntu.edu.tw; version 21), TargetScan
(www.targetscan.org; version 7.2),
miRanda (www.microrna.org; version 2010), PicTar
(www.pictar.org; version 2007), DIANA-MicroT-CDS
(diana.imis.athena-innovation.gr; version 5.0), mirTarBase
(mirtarbase.mbc.nctu.edu.tw; version
7.0) and DAVID (david.ncifcrf.gov; version 6.8), were used to identify
miRNAs that potentially bind to the BLM 3′-UTR (Fig. 1). The TargetScan V7.2, mirTarBase
and DAVID V6.8 had been jointly used to predict candidate roles in
cells and further screen the potential that miRNAs regulate BLM
expression. Finally, the five miRNAs were selected as candidates by
logical estimate.
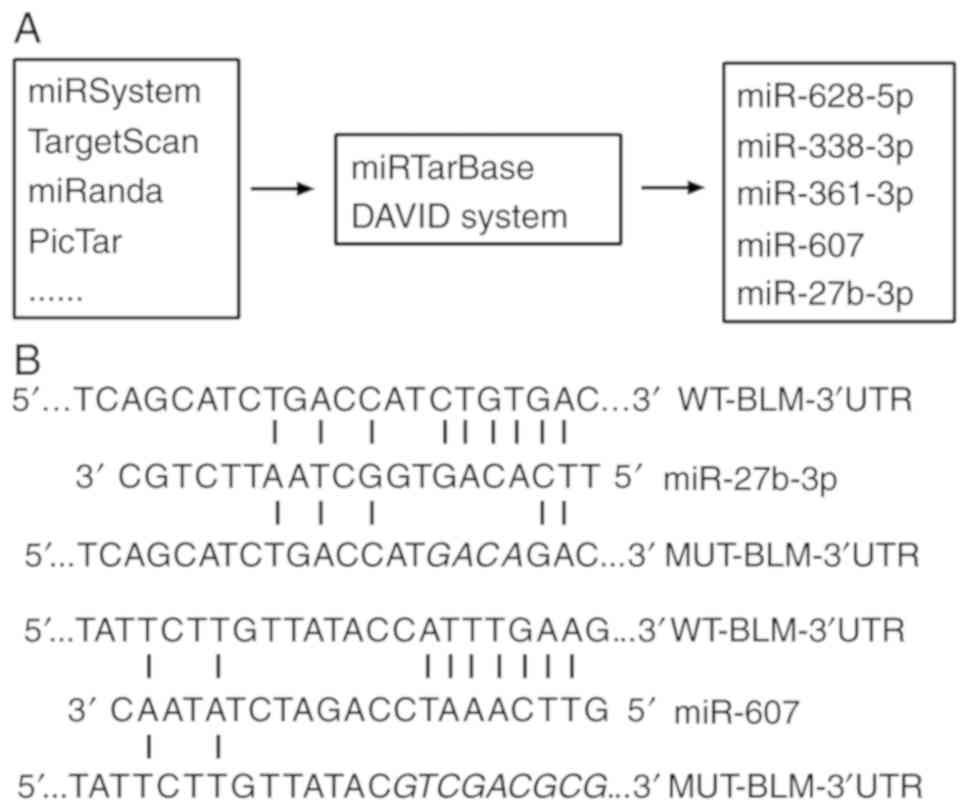 | Figure 1.miR screening method and designed
mutation or deletion sites. (A) Several prediction and function
analysis software programs were used to screen candidate miRs,
including miR-27b-3p, miR-607, miR-361-3p, miR-628-5p and
miR-338-3p as candidates. (B) miR-27b-3p and miR-607 interaction
sites with BLM 3′-UTR and mutational sites where four interaction
bases of miR-27b-3p are changed and edited sequence where the
interaction region of miR-607 is deleted. Predicted miR binding
sites of BLM 3′-UTR are displayed. miR, microRNA; WT, wild type;
BLM, BLM, BLM RecQ like helicase; UTR, untranslated region; MUT,
mutated. |
Cell culture and transfection
The human RWPE-2 normal prostate epithelial cell
line and PC3 prostate cancer cell line were obtained from the Key
Laboratory of Animal Genetics, Breeding and Reproduction (Guizhou
University, Guiyang, China). RWPE-2 and PC3 were maintained using
Dulbecco's modified Eagle's medium/F-12 medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) in addition with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 1% penicillin/streptomycin. Vector plasmids were
constructed by a series of digestion and connection experiments.
SalI, Xhol and T4 ligase (Thermo Fisher Scientific,
Inc.) had were used within the process of vector construction. The
constructed plasmids (Promega Corporation, Madison, WI, USA), and
miRNA mimics (negative control; hsa-miR-27b-3p mimics; hsa-miR-607
mimics; hsa-miR-628-5p mimic; miR-338-3p mimic; hsa-miR-361-3p
mimics; GenePharma Co., Ltd., Shanghai, China) were transfected
into PC3 cells using Lipofectamine® 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.) when cells were ~60% confluent,
with 2,500 ng plasmid and 5 µmol/l miRNA used per well of a
6-well-plate, according to the manufacturer's protocol. The
transfection efficiency of miRNA mimics was monitored by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
after 24 h.
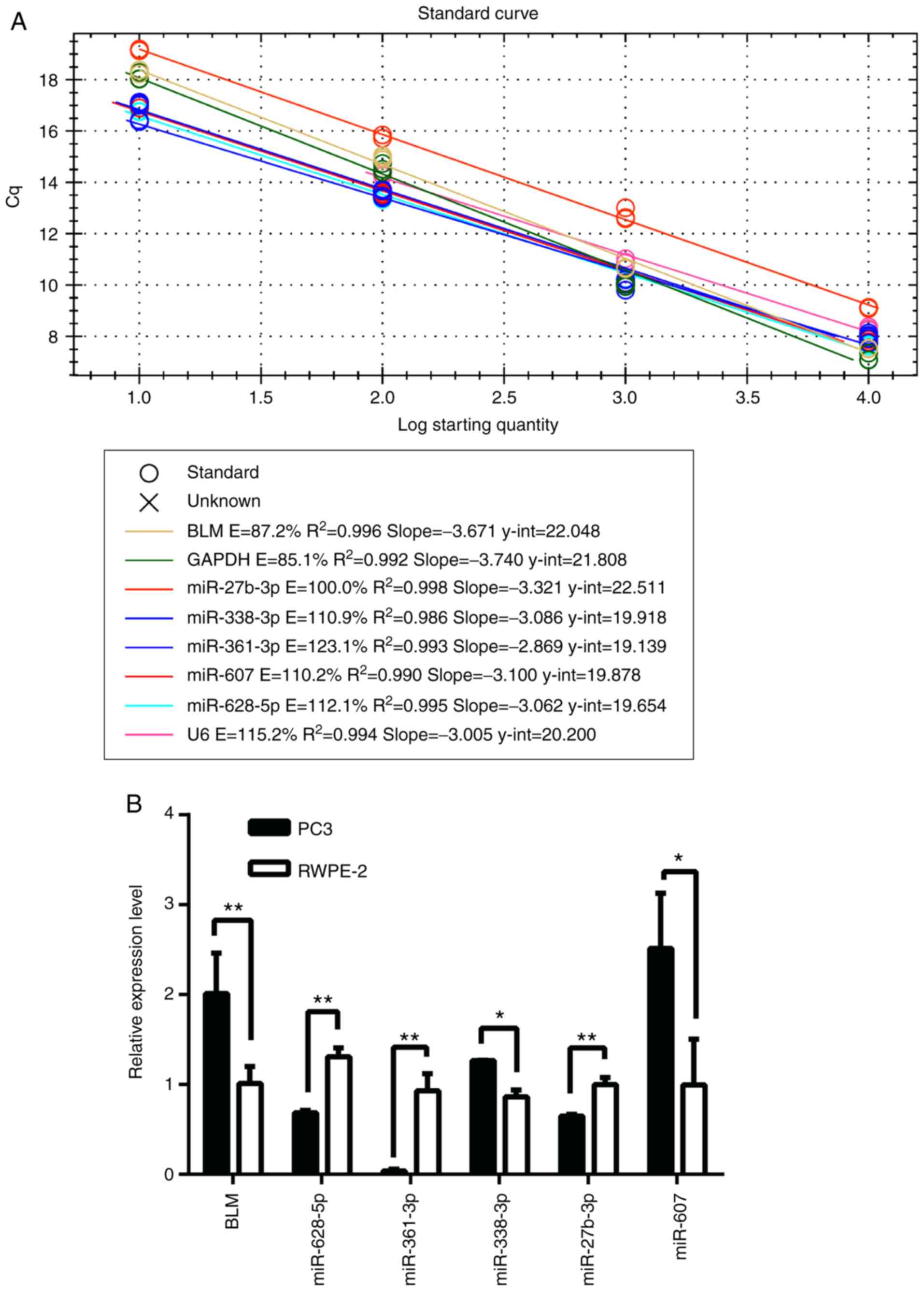
RNA isolation and RT-qPCR
Total RNA was extracted by using TRIzol reagent
(Thermo Fisher Scientific, Inc.) and RNA quality had been monitored
by ultraviolet spectrophotometry (Thermo Fisher Scientific, Inc.).
Designed stem-loop primers (23)
and common primers, and 2 µg RNA were used for reverse
transcription of miRNAs and BLM, respectively, using RevertAid
First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Inc.).
qPCR analyses were performed using CFX-96 Real-Time PCR Systems and
SYBR Green Mix (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Reverse and qPCR primers designed by software Primer5.0 and
Oligo7.0 are shown in Tables I and
II. miRNA transfectional
efficiency is shown in Table III
Relative expression levels were normalized to U6 or GAPDH, and
calculated using the Pfaffl's method (calculated relative
expression by using each PCR efficiency and of genes and Cq
values).
 | Table I.miR reverse primers. |
Table I.
miR reverse primers.
| Gene name | Sequence (5′ to
3′) |
|---|
| Stem loop
sequence |
GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACC |
| RT-miR-27b-3p |
GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCGCAGAACTT |
| RT-miR-338-3p |
GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCCAACAAAATC |
| RT-miR-361-3p |
GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCAAATCAGAATC |
| RT-miR-607 |
GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCGTTATAGATCT |
| RT-miR-628-5p |
GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCCCTCTAGTAA |
| R-U6 |
AACGCTTCACGAATTTGCGT |
 | Table II.Quantitative polymerase chain
reaction primers. |
Table II.
Quantitative polymerase chain
reaction primers.
| Gene name | Amplified length
(bp) | Sequence (5′ to
3′) |
|---|
| F-miR-27b-3p | 76 |
GCGGCATTCACAGTGGCT |
| F-miR-338-3p | 78 |
CGGCATCTCCAGCATCACT |
| F-miR-361-3p | 77 |
CGGCATCCCCCAGGTGT |
| F-miR-607 | 78 |
CAGGCATCGTTCAAATCC |
| F-miR-628-5p | 78 |
GGCGGCAATGCTGACATAT |
|
miR-universal-R |
|
CAAAGCAGGGTCCGAGGTATC |
| F-QBLM | 245 |
AAGCGACATCAGGAGCCAAT |
| R-QBLM |
|
GAAGAACTATCACCCCCCAGC |
| F-GAPDH | 307 |
CGGAGTCAACGGATTTGGTCGTAT |
| R-GAPDH |
|
AGCCTTCTCCATGGTGGTGAAGAC |
| F-U6 | 93 |
CTCGCTTCGGCAGCACA |
| R-U6 |
|
AACGCTTCACGAATTTGCGT |
 | Table III.Transfectional efficiency detection
of miR mimic. |
Table III.
Transfectional efficiency detection
of miR mimic.
| miR | Fold increase vs.
NC | P-value |
|---|
| miR-628-5p | 360.21 | P<0.001 |
| miR-361-3p | 4,802.11 | P<0.001 |
| miR-338-3p | 36.147 | P<0.001 |
| miR-607 | 29,0972.79 | P<0.001 |
| miR-27b-3p | 14.58 | P<0.001 |
Ratio=(1+Etarget)cqTcontrol-cqTsample(1+Ereference)cqRsample-cqRcontrol
Ratio, relative expression ratio of target gene;
Etarget, efficiency of target gene; Ereference, efficiency of
reference gene; cqTsample, quantitative PCR cycle values of target
gene in an unknown sample amplification satisfying the threshold;
cqTcontrol, quantitative PCR cycle values of target gene in a
control sample amplification satisfying the threshold; cqRsample,
quantitative PCR cycle values of reference gene in an unknown
sample amplification satisfying the threshold; cqRcontrol,
quantitative PCR cycle values of reference gene in a control sample
amplification satisfying the threshold.
Plasmids
The pmir-GLO dual luciferase reporter vector
(Promega Corporation), which expresses Firefly and Renilla
luciferase, was used for a reporter gene assay analyzing the
potential targeting BLM region of miRNA. The BLM gene 3′-UTR,
mutational and deleted fragments were amplified using designed
primers (Table IV) for the PCR
reaction and subcloned into the pmir-GLO dual luciferase reporter
vector to obtained reporter vectors PGL-UTR-WT, PGL-UTR-MUT and
PGL-UTR-DEL. All reconstruction vectors had been verified via
enzyme digestion analysis and Sanger sequencing. Table IV displays relative amplification
primers of fragments.
 | Table IV.Primers for fragment
amplification. |
Table IV.
Primers for fragment
amplification.
| Primer name | Amplified length
(bp) | Sequence (5′ to
3′) |
|---|
| F-BLM-3′-UTR | 264 |
CCGCTCGAGAAGCGACATCAGGAGCCAAT |
| R-BLM-3′-UTR |
|
ACGCGTCGACGAAGAACTATCACCCCCCAGC |
| 27b-mutation-F | 113 |
CTGACCATGACAGACTATAAAGCTGTTAT |
| 27b-mutation-R | 173 |
CTTTATAGTCTGTCATGGTCAGATGCT |
| 607-det-R | 201 |
ACGCGTCGACGTATAACAAGAATA |
Dual luciferase reporter assay
At 24 h after co-transfection with the miRNA mimic
and constructed reporter vectors, PC3 cells were analyzed for
luciferase activity using the Dual-Glo® Luciferase Assay
kit (Promega Corporation) following manufacturer's protocol.
PGL-UTR-WT and each miRNA were co-transfected into PC3 cells.
PGL-UTR-MUT and miRNA (miR)-27b-3p, or PGL-UTR-DEL and miR-605 were
co-transfected to further verify miRNA targeting sites of BLM. All
data detected using a microplate reader are presented as relative
firefly luciferase activity normalized to Renilla luciferase
activity (Luc/Rluc).
Western blot analysis
Total proteins were extracted from cells using lysis
buffer (Solarbio Science & Technology Co., Ltd., Beijing,
China) containing protease inhibitors at 24 h post-transfection
with the miRNA mimic. Protein concentration was determined using a
BCA Protein Assay kit (Solarbio Science & Technology Co., Ltd.)
and 10 mg of each protein sample was used for western blotting.
Total protein had been stacked and separated by polyacrylamide gel
electrophoresis by 6% stacking gel and 10% separating gel. Protein
was transferred to PVDF (0.22 µm). The membrane was blocked using
5% defatted milk in TBS Tween (TBST) at 37°C for 1.5 h. Specific
monoclonal BLM (cat. no. ab5409; 1:1,000 dilution) and β-actin
(cat. no. ab6276; 1:5,000 dilution) antibodies (Abcam, Cambridge,
UK) in 5% defatted milk TBST were incubated with membranes at 4°C
for 12 h. Goat anti-mouse horseradish peroxidase-conjugated IgG
(cat. no. ab97040) was used as the secondary antibody and incubated
with the membrane at 37°C for 1.5 h. When residual secondary
antibodies were removed via TBST washing, the blot was detected
using ImageLab software (version 2.0; Bio-Rad Laboratories, Inc.)
after enzyme substrate reactions with BeyoECL Star Kit (Beyotime
Institute of Biotechnology, Haimen, China).
BBD
In the present study, Box-Behnken factorial design
including 17 runs, 3 factors and 3 levels had been applied to
explore the interaction of miR-27b-3p and miR-607 in regulating BLM
gene. The relative Luc/Rluc activities were detected at 24 h after
miRNA and PGL-UTR-WT vector co-transfection into PC3 and were used
to calculate the interaction effects between miRNAs using
Design-Expert 8.0 software (Stat-Ease, Inc., Minneapolis, MN, USA).
Designed doses of miRNAs and designed methods were displayed in
tables and each factor was coded by three levels: Low (−1; 0
µmol/l), medium (0; 5 µmol/l) and high (+1; 10 µmol/l; Tables V and VI).
 | Table V.Coded and real values of variables in
the Box-Behnken design. |
Table V.
Coded and real values of variables in
the Box-Behnken design.
|
|
| Level (µmol/l) |
|---|
|
|
|
|
|---|
| Symbol | Variable | −1 | 0 | +1 |
|---|
| A | miR-607 | 0 | 5 | 10 |
| B | miR-27b-3p | 0 | 5 | 10 |
| C | miR-338-3p | 0 | 5 | 10 |
 | Table VI.Box-Behnken design with code
values. |
Table VI.
Box-Behnken design with code
values.
| Experiment
number | A, miR-607 | B, miR-27b-3p | C, miR-338-3p |
|---|
| 1 | −1 | −1 | 0 |
| 2 | 1 | −1 | 0 |
| 3 | −1 | 1 | 0 |
| 4 | 1 | 1 | 0 |
| 5 | −1 | 0 | −1 |
| 6 | 1 | 0 | −1 |
| 7 | −1 | 0 | 1 |
| 8 | 1 | 0 | 1 |
| 9 | 0 | −1 | −1 |
| 10 | 0 | 1 | −1 |
| 11 | 0 | −1 | 1 |
| 12 | 0 | 1 | 1 |
| 13 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 |
Cell proliferation and colony
formation assays
Cell proliferation was measured using the MTT
method. Cells were seeded (10,000 cells/well), transfected with 2.5
µmol/l miRNA mimic in 12-well plates and incubated for various
times. MTT incubation for 4 h, DMSO added to dissolve formazan,
medium was removed and sample absorbency was detected at 490 nm.
The growth curves over 3 days were calculated using the mean values
of four wells. In the colony formation assay, PC3 cells transfected
with miRNA were seeded at 5,000 cells/well in 12-well-plates and
cell colonies in each well were counted following Giemsa staining
after 5 days. Methanol (100%) was used to fix cells for 10 min at
room temperature before Giemsa staining (1 mg/ml) for 10 min at
room temperature. The numbers of colonies were counted using
Gel-Pro32 software (version 4.0; Media Cybernetics, Inc.,
Rockville, MD, USA).
Cell invasion and migration
assays
Transwell chambers (Corning Inc., Corning, NY, USA)
where the two chambers were separated by a Matrigel-coated
polycarbonate membrane (Solarbio Science & Technology Co.,
Ltd.; size: 8 µm) were used to analyze cell invasion at 12 h after
transfection. The upper chamber was filled with serum-free medium
and the lower chamber was filled with complete medium with 10% FBS.
Cells (5,000 per well) were added into the upper chamber and
invaded cells were counted after 12 h. Cells were stained with 0.1%
crystal violet for 15 min at room temperature after 4%
paraformaldehyde fixation for 10 min in the lower chamber. Cell
migration was monitored via wound healing assay and Transwell
chamber assays. Briefly, when transfected cells were 80–90%
confluent, a wound was created using a pipette tip and the heal
distance was recorded after 24 h to study cell migration ability.
Cell migration was also analyzed using a Transwell chamber without
Matrigel coating. Harvested cells (5,000 per well) were added into
the upper chamber with serum-free medium and the lower upper
chamber was filled with 10% FBS medium. Invaded cells in the lower
chamber were stained with 0.1% crystal violet for 15 min at room
temperature after 4% paraformaldehyde fixation for 10 min after 12
h to analyze cell migration.
Statistical analysis
Statistical analysis was performed using SPSS
software (version 22.0; IBM, Corp., Armonk, NY, USA), and the BBD
experiment was designed and analyzed using Design-Expert software
(version 8.0; Stat-Ease, Inc.). Data is presented as the mean ±
standard deviation. The difference between two experimental groups
and multiple comparisons were analyzed by Student's t-test
(two-tailed and equal variance) and Tukey's honestly significant
difference method following ANOVA, respectively. P<0.05 was
considered to indicate a statistically significant differences.
Results
Screen of miRNAs targeting BLM
According published researches (18–20)
and bioinformatics analysis, five candidate miRNAs, miR-27b-3p,
miR-607, miR-338-3p, miR-361-3p and miR-628-5p, were identified
using TargetScan, miRanda, DIANA and others. miRNA function
prediction was analyzed using the DAVID system (the technological
process is presented in Fig. 1A).
Furthermore, the expression levels of miRNAs in PC3 and RWPE-2 cell
lines were detected to determine the association between miRNAs and
BLM expression, which demonstrated that miR-338-3p and miR-607 had
a similar expression pattern to BLM gene in cancer cells and normal
cells, whereas miR-628-5p, miR-361-3p and miR-27b-3p the opposite
pattern of expression (Fig. 2).
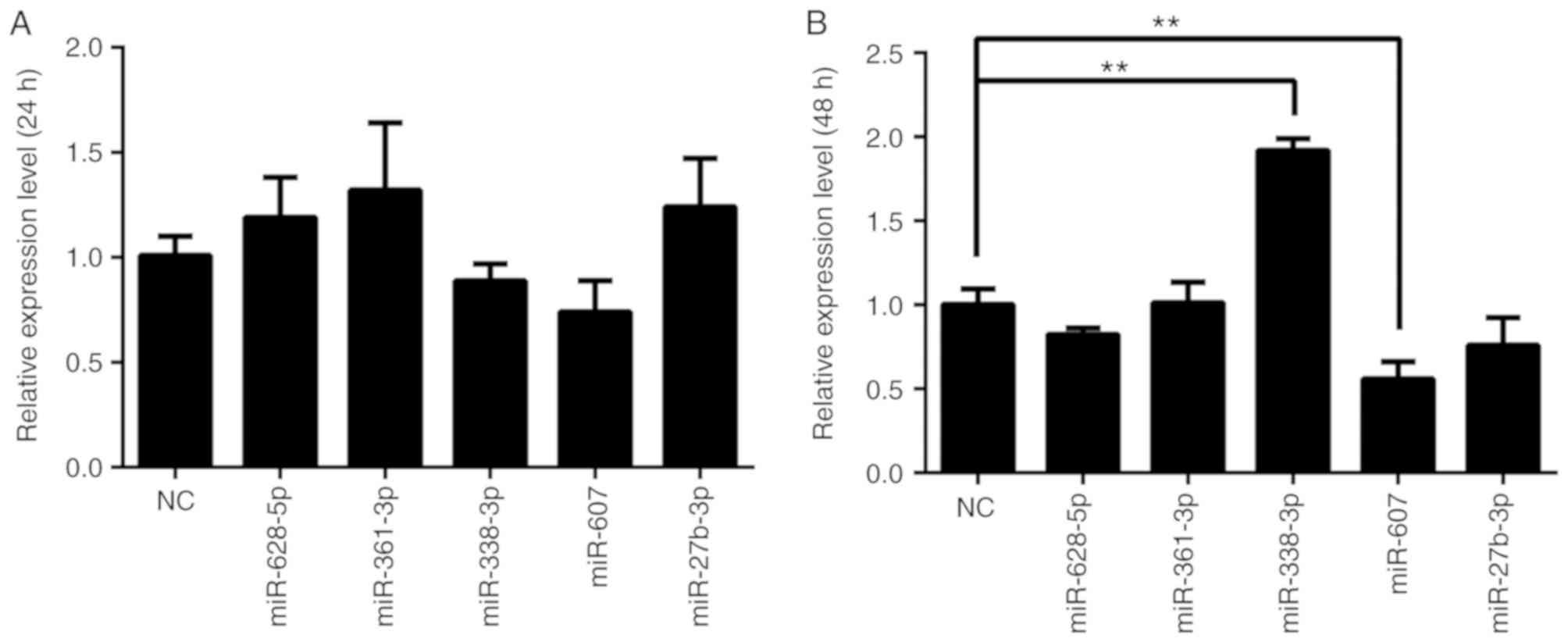
The effects of miRNA overexpression on BLM in PC3 cells were
analyzed at 24 and 48 h after transfection (Fig. 3). The results demonstrated that
miR-338-3p and miR-607 decreased BLM gene expression in mRNA levels
at 24 h, although the decrease was not statistically significant.
However, miR-338-3p transfection for 48 h significantly increased
BLM mRNA levels in PC3 cells (P<0.01). Additionally, miR-607
significantly decreased BLM mRNA expression at 48 h after
transfection (P<0.01). Other miRNAs had no significant effects
on BLM mRNA expression compared with the NC group (P>0.05). Dual
luciferase activity was determined following co-transfection of PC3
cells with miRNA and PGL-UTR-WT containing the wild type 3′-UTR of
BLM. miR-607 and miR-27b-3p modulated BLM gene relative luciferase
activity by directly targeting the BLM 3′-UTR (Fig. 4A).
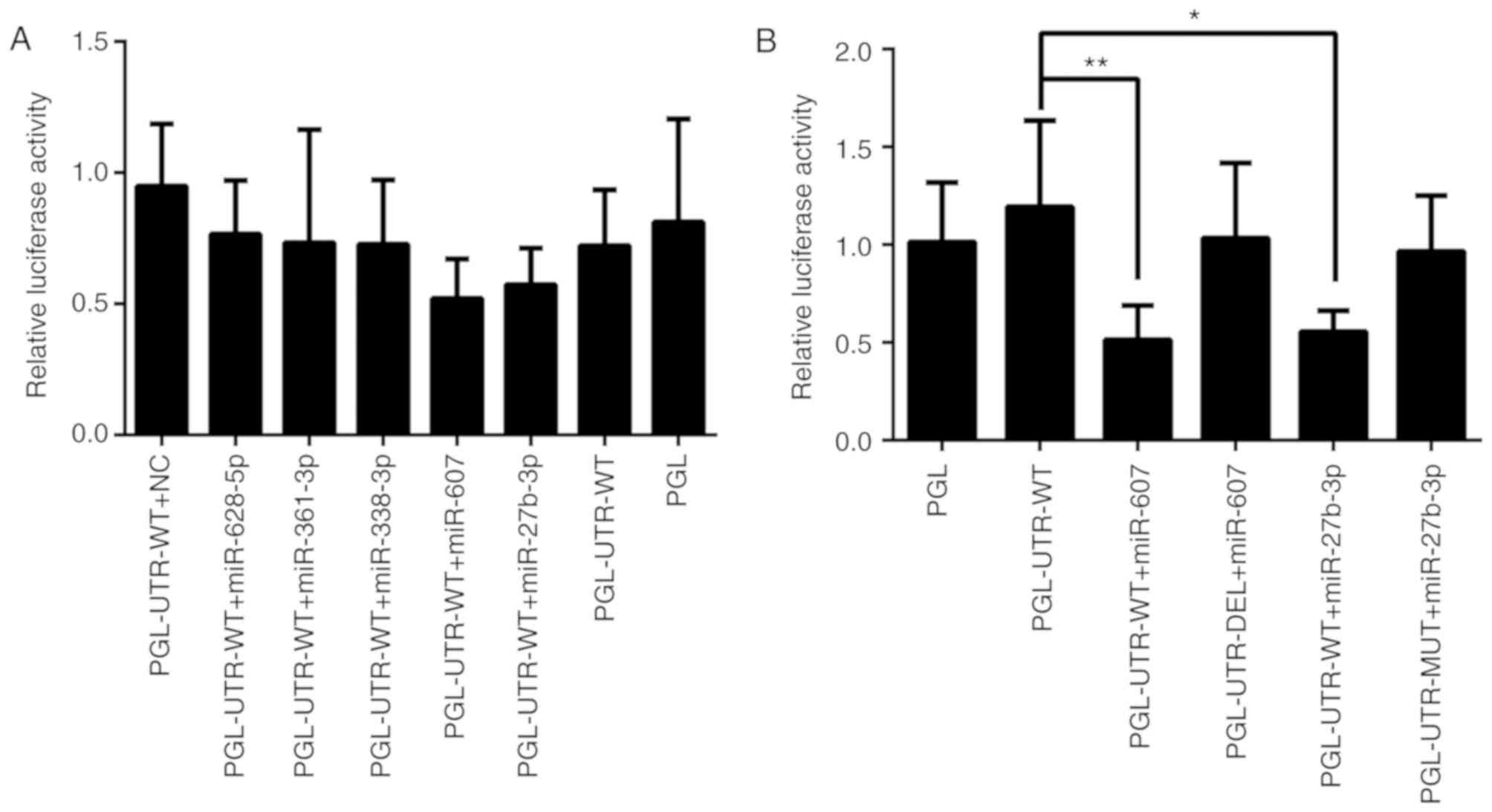 | Figure 4.Effects of miRs on relative
luciferase activity. PC3 cells were co-transfected with miR mimic
and luciferase reporter vectors. Relative luciferase activity was
detected at 24 h and the firefly luciferase activity of each sample
was normalized to the Renilla luciferase activity.
PGL-UTR-WT is a reporter vector containing the wild 3′-UTR sequence
BLM mRNA at the 3′-terminal of the firefly luciferase sequence in
the pmir-GLO vector. Similarly, PGL-UTR-MUT is a reporter vector
containing a mutated BLM 3′-UTR containing an miR-27b-3p
interaction region (changed at four bases). PGL-UTR-DEL is a
reporter vector with the BLM 3′-UTR sequence containing a miR-607
interaction region partially deleted. (A) Relative luciferase
activity results of PGL-UTR-WT + miR mimics. (B) Relative
luciferase activity of PGL-UTR-MUT or PGL-UTR-DEL + miR mimics.
n=6. *P<0.05, **P<0.01. BLM, BLM RecQ like helicase; PGL,
pmir-GLO; UTR, untranslated region; WT, wild-type; NC, negative
control; miR, microRNA; DEL, deletion; MUT, mutation. |
Validation of the effects of miR-607
and miR-27b-3p
Following the initial results, for investigation of
the regulatory roles of miR-607 and miR-27b-3p, PGL-UTR-MUT, in
which four key bases were mutated at the miR-27b-3p binding sites,
and PGL-UTR-DEL, in which miR-607 binding sites in the BLM 3′-UTR
was deleted, were used for further validation. The designed
mutation and deletion sites are presented in on Fig. 1B. PC3 cells were co-transfected
with PGL-UTR-MUT or PGL-UTR-DEL and miR-27b-3p or miR-607,
respectively, and relative luciferase activity was measured after
24 h (Fig. 4B). PGL-UTR-WT
co-transfection with miR-27b-3p or miR-607 decreased luciferase
activity compared with PGL-UTR-WT transfection alone (P<0.05 and
P<0.01, respectively), whereas PGL-UTR-MUT co-transfection with
miR-27b-3p, and PGL-UTR-DEL co-transfection with miR-607 produced
no significant difference compared the vector control (P>0.05).
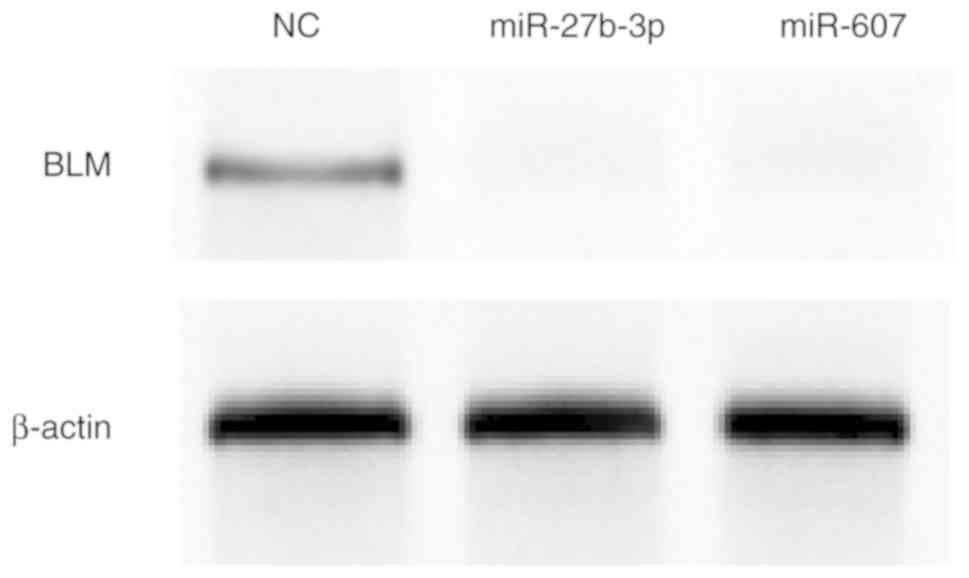
Finally, BLM helicase expression was determined by western blot
analysis after miR-27b-3p and miR-607 overexpression in PC3 cells.
miR-27b-3p and miR-607 reduced BLM helicase protein expression
compared with NC mimic transfection (Fig. 5).
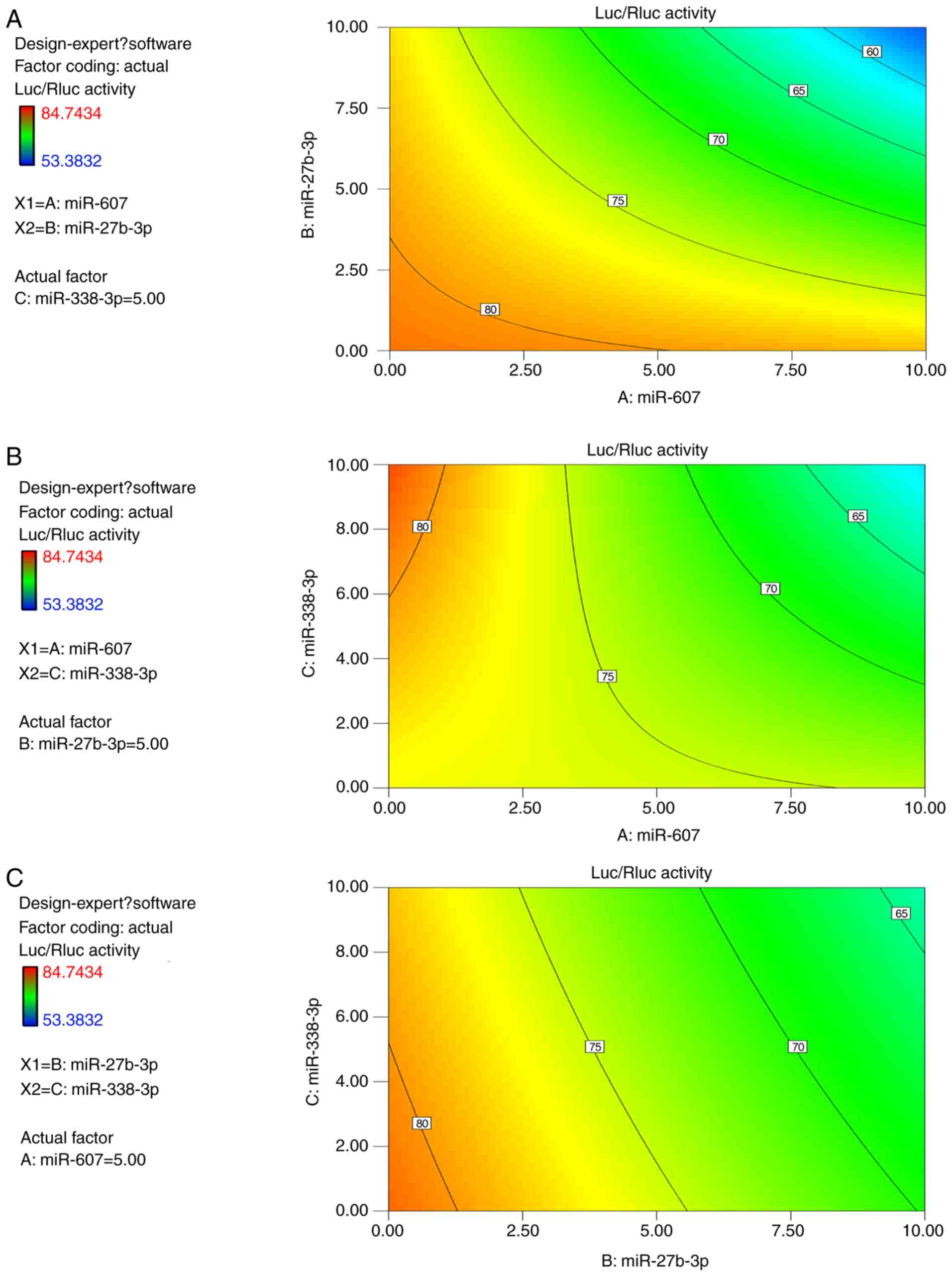
Interaction between miRNAs
BBD was applied to explore interactions between
miRNAs. In the previous assays, miR-338-3p had the activity to
impair relative luciferase and displayed a multiple effects in
various acting doses or times. Therefore, miR-338-3p was selected
as one factor to represent endogenous noise of miRNAs. The relative
luciferase activity of all runs including different miRNA dose
compositions is presented in Table
VII. The final results are presented in Table VIII, which demonstrated that the
calculated statistical model is statistically significant
(P<0.01) and the lack of fix is not significant (P>0.8). It
indicated that a proper formula mode had been built for prediction
of interaction effects between miRNAs on the luciferase activity of
PGL-UTR-WT containing the BLM 3′-UTR. miR-27b-3p and miR-607 were
significant factors (P<0.01) affecting relative luciferase
activity, but the effect of miR-338-3p on relative luciferase
activity was not significant (P>0.05). The results also
demonstrated that the interaction effects between miR-607 and
miR-27b-3p, and interaction effects between miR-607 and miR-338-3p
were statistically significant (P<0.05), but the interaction
effect of miR-27b-3p and miR-338-3p was not significant
(P>0.05). The calculated model coefficient is presented in
Table IX, and the negative values
of factors by code value calculation suggested a decrease in
relative luciferase activity. Thus, the calculated equation
indicated the synergetic inhibitory effects of miR-607, and
miR-27b-3p or miR-338-3p. The radian degree and direction of curves
on contours in Fig. 6 demonstrated
the intensity and manner of interactions between two factors in
different dose compositions. The interactional contour between
miR-607 and miR-27b-3p further demonstrated their synergistic
effects on the reduced BLM gene expression.
 | Table VII.Results of BBD for relative Luc/Rluc
activity. |
Table VII.
Results of BBD for relative Luc/Rluc
activity.
| Experiment
number | A, miR-607 | B, miR-27b-3p | C, miR-338-3p | Relative Luc/Rluc
activity (%) |
|---|
| 1 | −1 | −1 | 0 | 84.74 |
| 2 | 1 | −1 | 0 | 80.42 |
| 3 | −1 | 1 | 0 | 77.5 |
| 4 | 1 | 1 | 0 | 53.38 |
| 5 | −1 | 0 | −1 | 74.44 |
| 6 | 1 | 0 | −1 | 74.55 |
| 7 | −1 | 0 | 1 | 83.25 |
| 8 | 1 | 0 | 1 | 63.02 |
| 9 | 0 | −1 | −1 | 81.61 |
| 10 | 0 | 1 | −1 | 73.81 |
| 11 | 0 | −1 | 1 | 75.6 |
| 12 | 0 | 1 | 1 | 64.61 |
| 13 | 0 | 0 | 0 | 70.44 |
| 14 | 0 | 0 | 0 | 69.9 |
| 15 | 0 | 0 | 0 | 68.86 |
| 16 | 0 | 0 | 0 | 81.05 |
| 17 | 0 | 0 | 0 | 71.08 |
 | Table VIII.Results of statistics for Box-Behnken
design. |
Table VIII.
Results of statistics for Box-Behnken
design.
| Source | Sum of squares | df | Mean square | F-value | P-value (Prob
>F) |
|---|
| Model | 890.82 | 6 | 148.47 | 8.89 | 0.0016 |
| A (miR-607) | 294.76 | 1 | 294.76 | 17.65 | 0.0018 |
| B (miR-27b-3p) | 352.00 | 1 | 352.00 | 21.08 | 0.0010 |
| C (miR-338-3p) | 40.16 | 1 | 40.16 | 2.41 | 0.1520 |
| AB | 97.98 | 1 | 97.98 | 5.87 | 0.0359 |
| AC | 103.38 | 1 | 103.38 | 6.19 | 0.0321 |
| BC | 2.54 | 1 | 2.54 | 0.15 | 0.7048 |
| Residual | 166.96 | 10 | 16.70 |
|
|
| Lack of fit | 67.81 | 6 | 11.30 | 0.46 | 0.8137 |
| Pure error | 99.15 | 4 | 24.79 |
|
|
| Cor total | 1057.78 | 16 |
|
|
|
 | Table IX.Predicted equation by Box-Behnken
design in code value. |
Table IX.
Predicted equation by Box-Behnken
design in code value.
| Coefficient
factor | Standard
estimate | df | Error | 95% CI (low) | 95% CI (high) | VIF |
|---|
| Intercept | 73.43 | 1 | 0.99 | 71.22 | 75.64 |
|
| A (miR-607) | −6.07 | 1 | 1.44 | −9.29 | −2.85 | 1.00 |
| B (miR-27b-3p) | −6.63 | 1 | 1.44 | −9.85 | −3.41 | 1.00 |
| C (miR-338-3p) | −2.24 | 1 | 1.44 | −5.46 | 0.98 | 1.00 |
| AB | −4.95 | 1 | 2.04 | −9.50 | −0.40 | 1.00 |
| AC | −5.08 | 1 | 2.04 | −9.64 | −0.53 | 1.00 |
| BC | −0.80 | 1 | 2.04 | −5.35 | 3.76 | 1.00 |
Effects of miR-27b-3p and miR-607 on
PC3 proliferation, colony formation, migration and invasion
The effects of miR-27b-3p and miR-607 on PC3 cell
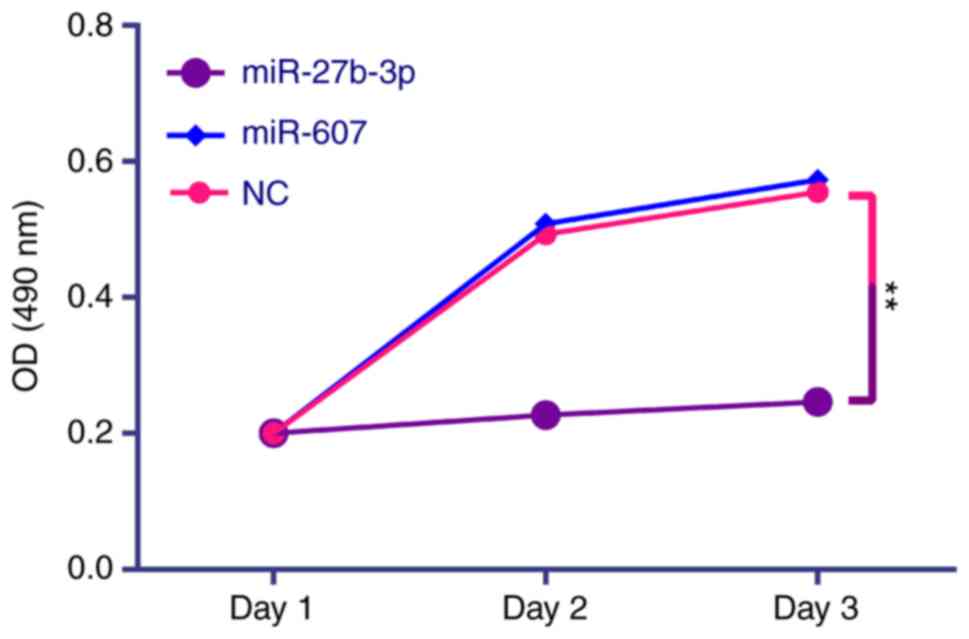
proliferation (Fig. 7), colony
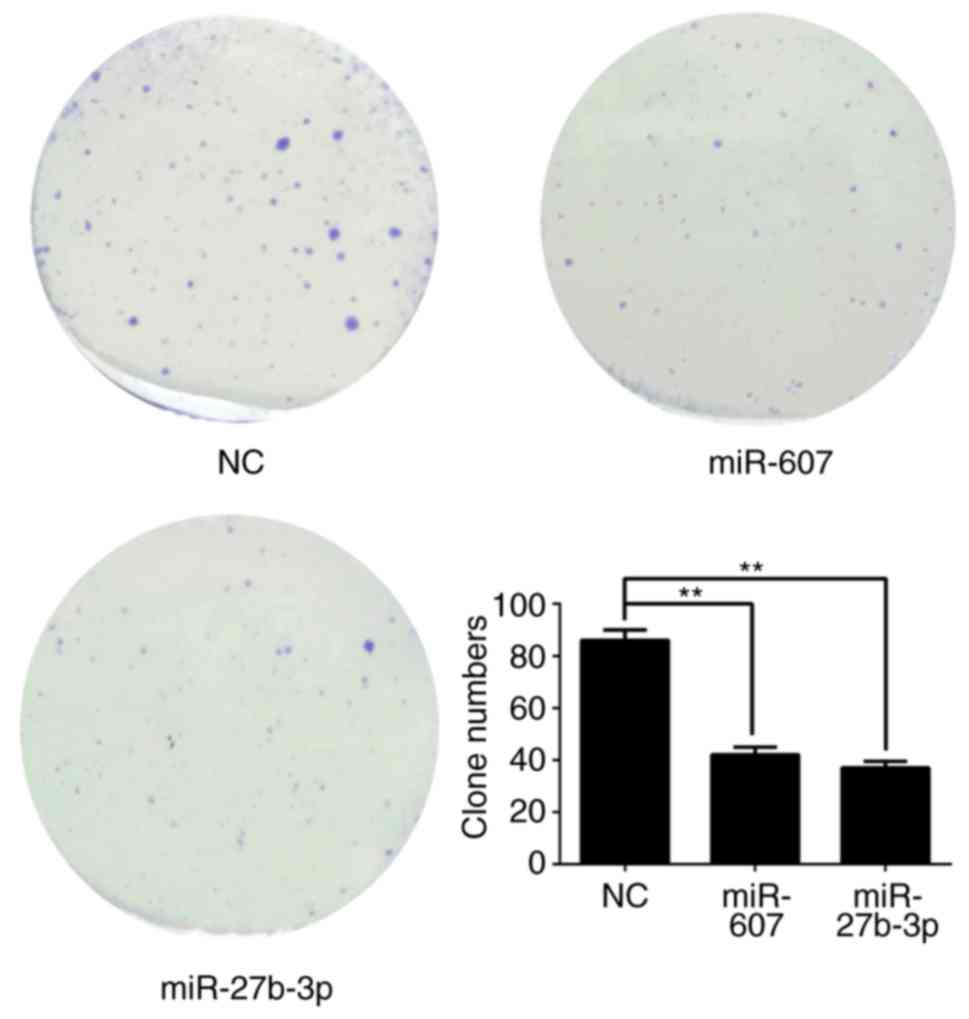
formation (Fig. 8), migration
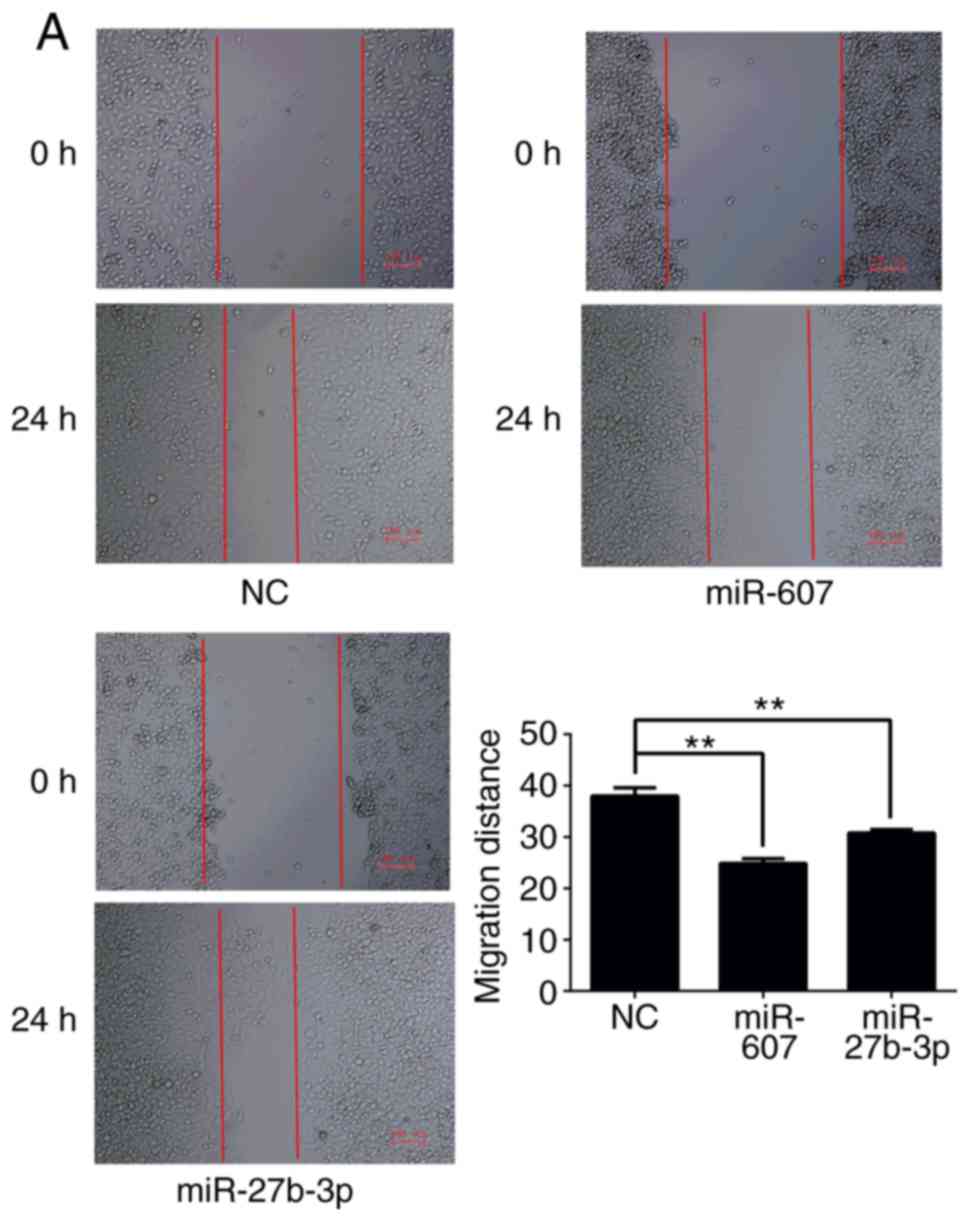
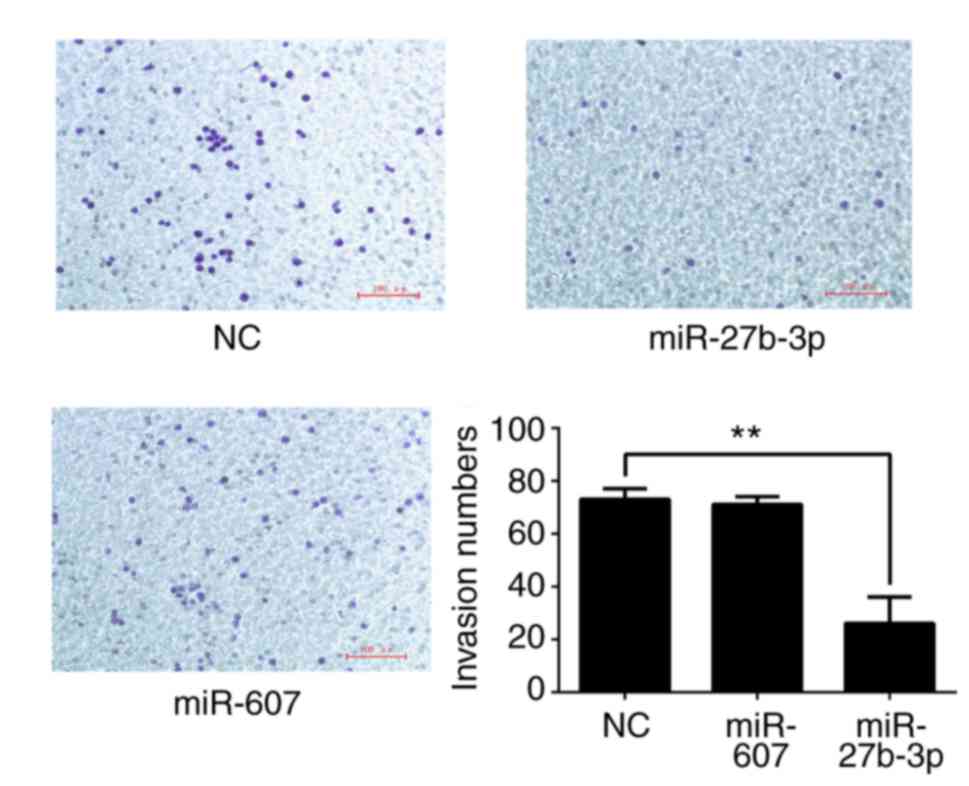
(Fig. 9) and invasion (Fig. 10) were analyzed. All results are
summarized in Table X, which
demonstrated that miR-27b-3p overexpression in PC3 cells reduces
proliferation, colony formation and invasion abilities compared
with the NC group (P<0.01). miR-607 overexpression significantly
reduced colony formation and migration of PC3 cells
(P<0.01).
 | Table X.Effect of miRs on PC3 cells. |
Table X.
Effect of miRs on PC3 cells.
| miR | Proliferation | Colony
formation | Migration (cell
scratch) | Migration
(transwell) | Invasion
(transwell) |
|---|
| miR-27b-3p | -a | -a |
|
| -a |
| miR-607 |
| -a | -a | -b |
|
Discussion
In the current study, five miRNAs were identified
using various online software tools, and miR-27b-3p and miR-607
were validated suppressors of BLM gene expression (19). The different online software
varying featured that were considered to identify the most
potential miRNAs for subsequent validation in vitro.
Bioinformatics selection (data not shown) identified miR-628-5p,
miR-361-3p, miR-338-3p, miR-607 and miR-27b-3p as potential
candidate miRs that may target BLM mRNA. Recent studies indicated
that miR-361-3p, miR-338-3p and miR-27b-3p have oncosuppressive
functions. There are fewer studies on miR-607 (24) and miR-628-5p (25) so further study is required.
Subsequently, the expression of the five miRNAs and BLM were
detected by RT-qPCR in normal cells (RWPE-2 cell line) and a cancer
cell line (PC3) to determine the expression correlation between
these miRNAs and BLM. The results indicated that miR-628-5p,
miR-361-3p and miR-27b-3p had lower expression in PC3 cells than in
RWPE-2 cells, whereas BLM, miR-338-3p and miR-607 had the opposite
pattern of expression. The negative association between BLM gene
expression, and miR-628-5p, miR-361-3p and miR-27b-3p levels
suggested that miR-628-5p, miR-361-3p and miR-27b-3p may be
potential miRNAs that regulate BLM gene expression at the
post-transcriptional level. The results of the dual luciferase
assay and western blot analysis revealed that miR-27b-3p and
miR-607 inhibit BLM gene expression. Additionally, previous studies
have indicated that miR-628-5p, miR-361-3p and miR-27b-3p may be
anti-cancer factors, as they have lower expression levels in cancer
compared with normal cells (18,26,27).
However, more tumor samples and cell lines are required to
determine whether these miRNAs are anti-cancer factors. By
contrast, the higher expression level of BLM, miR-338-3p and
miR-607 in PC3 cells indicate that they may have the potential to
be tumor markers for clinical diagnosis (21,28).
miRNA overexpression, dual luciferase assays and western blot
analysis were performed to explore the role of these miRNAs in the
regulation of BLM. The results indicated that miR-607 and
miR-338-3p decreased the level of BLM mRNA at 24 h
post-transfection, whereas miR-338-3p elevated the BLM mRNA level
at 48 h post-transfection. miR-607 significantly reduced the BLM
mRNA level at 48 h post-transfection, but miR-27b-3p had no evident
effect on BLM mRNA. However, the dual luciferase assay and western
blot results suggested that miR-27b-3p and miR-607 can target the
BLM 3′-UTR reduce the protein expression. This suggested that
miR-607 and miR-27b-3p act by decreasing BLM mRNA levels and
reducing the translation of BLM mRNA, respectively.
Multiple mechanisms of action of miRNAs allow for
positive or negative mutual effect between miRNAs. Mutual effect of
miRNAs may have many advantages for medicinal application, such as
higher drug efficacy and lower secondary actions (29,30).
Understanding the interaction mechanisms of miRNAs provides more
information on the regulation process of BLM gene expression at the
post-transcriptional level. BBD has been applied to determine
interactions between multiple factors in previous studies (31). Latin square design and the
equivalent line method (2,30–32)
are also able to calculate interaction effects, but the lower dose
application of factors, more reasonable and three-dimensional
arrangements are the main features of BBD which eliminates the dose
superposing effects from multiple factors (in the study of factor
interaction, multiple factors were used in one treatment group so
that the dose of multiple factors will be elevated to create effect
results). By contrast, Latin square design lacks consideration of
superposition of drug doses. Higher doses will lead to more errors
from random and toxic effects, but lower dose will lose effects,
which can be resolved using a BBD experiment. A parabolic
dose-effect curve is necessary for the equivalent line method and
two dose-effect curves of drugs must not cross. In brief, BBD was
the best choice for analyzing the interaction effects among the
miRNAs in the present study. miR-27b-3p and miR-607 were
demonstrated to reduce BLM protein expression, and understanding
their mutual effects has potential to assist future drug
development. miR-338-3p has been reported to influence the
proliferation and metabolism of cancer cells in recent reports
(19,33). According to our findings, action
time and dose altered the effects of miR-338-3p on BLM mRNA levels,
which implied miR-338-3p may be selected as a reference factor
which represents other functional miRNAs in PC3 cells to research
miRNA interactions in depth. Therefore, miR-338-3p is suitable to
be selected as a factor that represents endogenous miRNAs noise to
explore interaction effects between miR-27b-3p and miR-607.
miR-338-3p had been selected as auxiliary miRNA to analyze the
interaction between miR-27b-3p and miR-607 for a deeper
understating of the effects of multiple miRNA interactions on BLM
expression. It was demonstrated that the interaction between
miR-27b-3p with miR-607, and the interaction between miR-338-3p and
miR-607 decreased relative luciferase activity of the BLM reporter,
but there was no interaction between miR-27b-3p and miR-338-3p.
This indicated that miR-27b-3p and miR-338-3p have the same action
mode, so that they have no distinct interaction; whereas, the
different action modes between miR-607 and miR-27b-3p created a
synergistic effect between miR-27b-3p and miR-607 on the expression
suppression of BLM. miR-607 overexpression in PC3 resulted in
reduced BLM mRNA level, but miR-27b-3p did not reduce the mRNA
level, which also had supported the hypothesis of synergistic
effects between miR-27b-3p and miR-607 via different mechanisms of
action.
In previous studies, miR-27b-3p (22) and miR-607 (34) were to have anti-cancer functions.
It was indicated that miR-27b-3p and miR-607 alter normal
proliferation, invasion and migration of prostate cancer cells by
decreasing BLM protein expression. Additionally, the effects of
miR-27b-3p and miR-607 on the proliferation, migration, invasion
and colony formation of PC3 cells were determined. miRNAs regulate
important protein expression to alter various cell functions,
although the detail mechanisms of the roles of miR-27b-3p and
miR-607 in PC3 cells remain unknown as miRNAs commonly regulate
multiple target genes, and a gene is also regulated by multiple
miRNAs (17). The current study
demonstrated that miR-27b-3p and miR-607 cooperatively affected PC3
cells by directly targeting the BLM gene 3′-UTR to have
tumor-inhibiting functions. Previous research demonstrated that
ML216, which is a small molecule inhibitor of BLM, has
anti-proliferative activity in cells (9,35),
and BLM defects also increase the sensitivity of cancer cells to
cytotoxic drugs (36). Thus, the
effects of miRNAs on the proliferation, migration, invasion and
colony formation of PC3 cells was analyz ed to provide the evidence
and theoretical support for clinical treatment of prostate cancer
using miRNAs to inhibit BLM pathways. However, expression levels of
key protein factors, such as matrix metalloproteinases, cadherins,
vascular endothelial growth factor, catenin, need to be determined
for stronger evidence.
Finally, it was validated that miR-27b-3p and
miR-607 cooperatively reduce BLM gene expression by directly
targeting gene the BLM 3′-UTR in PC3 cells and provided information
on the post-transcriptional regulation of BLM gene expression. The
synergistic use of miR-27b-3p and miR-607 to target BLM gene was
more effective than their use alone. In conclusion, the presented
study provided evidence that miR-27b-3p and miR-607 have
anti-cancer activity, and cooperatively downregulate BLM expression
at the post transcriptional level in PC3 cells.
BLM helicase is crucial factor required cell DNA
metabolism and is involved in cell proliferation, migration,
invasion, apoptosis and the cell cycle in prostate cancer cells
(5). Prostate cancer is the second
leading cause of cancer-associated mortality in men (1). Targeting the key protein factors,
such as BLM, Her2/Neu, epidermal growth factor or tumor necrosis
factor-α, in cancer cells by anticancer drugs is main and valid
method for cancer remedy (3,5).
Targeting BLM helicase may be a useful future method to treat
patients with prostate cancer (5).
However, the post-transcription regulation of BLM gene has remained
unclear, which has limited the application and development of drugs
targeting BLM helicase. It is important to explore novel
anti-cancer drugs targeting BLM, to understand the BLM gene
regulation pathway and to identify other novel cancer targets and
tumor markers.
Acknowledgements
This work was supported by the Key Laboratory of
Animal Genetics, Breeding and Reproduction in Guizhou Province
(Guiyang, China). We thank Dr Chen Kun of Guizhou Provincial
People's Hospital (Guiyang, China) for providing the RWPE-2 cell
line and the expert technical assistance of Professor He Laping
(School of Liquor and Food Engineering, Guizhou University).
Funding
Funding was provided by the Natural Science
Foundation of China (grant no. 31361406).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YC designed the study, performed the research,
analyzed data and wrote the paper. HX and TG helped design the
study and checked the data. JZ and SW helped perform research and
analyzed data. ZD and WC helped designed the study and analyzed
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pandita A, Manvati S, Singh SK, Vaishnavi
S and Bamezai RN: Combined effect of microRNA, nutraceuticals and
drug on pancreatic cancer cell lines. Chem Biol Interact.
233:56–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi S, Han L, Deng L, Zhang Y, Shen H,
Gong T, Zhang Z and Sun X: Dual drugs (microRNA-34a and
paclitaxel)-loaded functional solid lipid nanoparticles for
synergistic cancer cell suppression. J Control Release.
194:228–237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Futami K, Ogasawara S, Goto H, Yano H and
Furuichi Y: RecQL1 DNA repair helicase: A potential tumor marker
and therapeutic target against hepatocellular carcinoma. Int J Mol
Med. 25:537–545. 2010.PubMed/NCBI
|
|
5
|
Qian X, Feng S, Xie D, Feng D, Jiang Y and
Zhang X: RecQ helicase BLM regulates prostate cancer cell
proliferation and apoptosis. Oncol Lett. 14:4206–4212. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bachrati CZ and Hickson ID: RecQ
helicases: Suppressors of tumorigenesis and premature aging.
Biochem J. 374:577–606. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sharma S, Doherty KM and Brosh RM Jr:
Mechanisms of RecQ helicases in pathways of DNA metabolism and
maintenance of genomic stability. Biochem J. 398:319–337. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu L: Role of the BLM helicase in
replication fork management. DNA Repair (Amst). 6:936–944. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nguyen GH, Dexheimer TS, Rosenthal AS, Chu
WK, Singh DK, Mosedale G, Bachrati CZ, Schultz L, Sakurai M,
Savitsky P, et al: A small molecule inhibitor of the BLM helicase
modulates chromosome stability in human cells. Chem Biol. 20:55–62.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Laitman Y, Boker-Keinan L, Berkenstadt M,
Liphsitz I, Weissglas-Volkov D, Ries-Levavi L, Sarouk I, Pras E and
Friedman E: The risk for developing cancer in Israeli ATM, BLM, and
FANCC heterozygous mutation carriers. Cancer Genet. 209:70–74.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Voer RM, Hahn MM, Mensenkamp AR,
Hoischen A, Gilissen C, Henkes A, Spruijt L, van Zelst-Stams WA,
Kets CM, Verwiel ET, et al: Deleterious germline BLM mutations and
the risk for early-onset colorectal cancer. Sci Rep. 5:140602015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Böhm S and Bernstein KA: The role of
post-translational modifications in fine-tuning BLM helicase
function during DNA repair. DNA Repair (Amst). 22:123–132. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Suspitsin EN, Yanus GA, Sokolenko AP,
Yatsuk OS, Zaitseva OA, Bessonov AA, Ivantsov AO, Heinstein VA,
Klimashevskiy VF, Togo AV and Imyanitov EN: Development of breast
tumors in CHEK2, NBN/NBS1 and BLM mutation carriers does not
commonly involve somatic inactivation of the wild-type allele. Med
Oncol. 31:8282014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sassi A, Popielarski M, Synowiec E,
Morawiec Z and Wozniak K: BLM and RAD51 genes polymorphism and
susceptibility to breast cancer. Pathol Oncol Res. 19:451–459.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matsuyama R, Okuzaki D, Okada M and
Oneyama C: MicroRNA-27b suppresses tumor progression by regulating
ARFGEF1 and focal adhesion signaling. Cancer Sci. 107:28–35. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wen C, Liu X, Ma H, Zhang W and Li H:
miR-338-3p suppresses tumor growth of ovarian epithelial carcinoma
by targeting Runx2. Int J Oncol. 46:2277–2285. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen D, Si W, Shen J, Du C, Lou W, Bao C,
Zheng H, Pan J, Zhong G, Xu L, et al: miR-27b-3p inhibits
proliferation and potentially reverses multi-chemoresistance by
targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis.
9:1882018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sui GQ, Fei D, Guo F, Zhen X, Luo Q, Yin S
and Wang H: MicroRNA-338-3p inhibits thyroid cancer progression
through targeting AKT3. Am J Cancer Res. 7:1177–1187.
2017.PubMed/NCBI
|
|
20
|
Hara ES, Ono M, Eguchi T, Kubota S, Pham
HT, Sonoyama W, Tajima S, Takigawa M, Calderwood SK and Kuboki T:
miRNA-720 controls stem cell phenotype, proliferation and
differentiation of human dental pulp cells. PLoS One. 8:e835452013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu S, Yi XM, Zhang ZY, Ge JP and Zhou WQ:
miR-129 predicts prognosis and inhibits cell growth in human
prostate carcinoma. Mol Med Rep. 14:5025–5032. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang G, Tian X, Li Y, Wang Z, Li X and
Zhu C: miR-27b and miR-34a enhance docetaxel sensitivity of
prostate cancer cells through inhibiting epithelial-to-mesenchymal
transition by targeting ZEB1. Biomed Pharmacother. 97:736–744.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Braoudaki M, Lambrou GI, Giannikou K,
Milionis V, Stefanaki K, Birks DK, Prodromou N, Kolialexi A,
Kattamis A, Spiliopoulou CA, et al: Microrna expression signatures
predict patient progression and disease outcome in pediatric
embryonal central nervous system neoplasms. J Hematol Oncol.
7:962014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Favreau AJ and Sathyanarayana P:
miR-590-5p, miR-219-5p, miR-15b and miR-628-5p are commonly
regulated by IL-3, GM-CSF and G-CSF in acute myeloid leukemia. Leuk
Res. 36:334–341. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu X, Zheng Y, Han B and Dong X: Long
noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin
resistance of gastric cancer via sponging miR-361. Biomed
Pharmacother. 99:832–838. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang S, Liu Z, Wu L and Wang Y: MiR-361
targets Yes-associated protein (YAP) mRNA to suppress cell
proliferation in lung cancer. Biochem Biophys Res Commun.
492:468–473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chiyomaru T, Seki N, Inoguchi S, Ishihara
T, Mataki H, Matsushita R, Goto Y, Nishikawa R, Tatarano S, Itesako
T, et al: Dual regulation of receptor tyrosine kinase genes EGFR
and c-Met by the tumor-suppressive microRNA-23b/27b cluster in
bladder cancer. Int J Oncol. 46:487–496. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ning S, Xu H, Al-Shyoukh I, Feng J and Sun
R: An application of a Hill-based response surface model for a drug
combination experiment on lung cancer. Stat Med. 33:4227–4236.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiao Q, Wang L and Xu H: Application of
kriging models for a drug combination experiment on lung cancer.
Stat Med. 38:236–246. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li C, Li L, Zhou HH, Xia C and He L:
Improving Yield of 1,3-diglyceride by whole-cell lipase fromA.
NigerGZUF36 catalyzed glycerolysis via medium optimization. J Braz
Chem Soc. 26:247–254. 2015.
|
|
32
|
Fang HB, Ross DD, Sausville E and Tan M:
Experimental design and interaction analysis of combination studies
of drugs with log-linear dose responses. Stat Med. 27:3071–3083.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Shi B, Chen J, Hu L and Zhao C:
MiR-338-3p targets pyruvate kinase M2 and affects cell
proliferation and metabolism of ovarian cancer. Am J Transl Res.
8:3266–3273. 2016.PubMed/NCBI
|
|
34
|
Mezlini AM, Wang B, Deshwar A, Morris Q
and Goldenberg A: Identifying cancer specific functionally relevant
miRNAs from gene expression and miRNA-to-gene networks using
regularized regression. PLoS One. 8:e731682013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rosenthal AS, Dexheimer TS, Nguyen G,
Gileadi O, Vindigni A, Simeonov A, Jadhav A, Hickson I and Maloney
DJ: Discovery of ML216, a small molecule inhibitor of bloom (BLM)
helicase. Probe Reports from the NIH Molecular Libraries Program;
Bethesda (MD): 2010
|
|
36
|
Gupta A, Ahmad A, Singh H, Kaur S, K M N,
Ansari MM, Jayamurugan G and Khan R: Nanocarrier composed of
magnetite core coated with three polymeric shells mediates LCS-1
delivery for synthetic lethal therapy of BLM-defective colorectal
cancer cells. Biomacromolecules. 19:803–815. 2018. View Article : Google Scholar : PubMed/NCBI
|