Introduction
Papillary renal cell carcinoma (PRCC) is a complex
malignant neoplasm and the second most frequent renal cell
carcinoma (RCC) following renal clear cell carcinoma (CCRC),
comprising ~18.5% of the total cases of RCC (1). In 1997, Delahunt and Elbe (2) described PRCC as comprising two
subtypes, type 1 and type 2; however, Chevarie-Davis et al
(3) reported that the frequency of
‘overlapping’ PRCC, which possessed a certain overlapping features,
was ~47%. Furthermore, the survival of patients with PRCC varies,
particularly in cases of sporadic PRCC. Ha et al (4) reported that the subclassification of
PRCC did not affect the prognosis of patients with PRCC, whereas
Tsimafeyeu et al (5)
demonstrated that the expression of fibroblast growth factor
receptor 2 was a prognostic factor in the survival of patients with
metastatic PRCC. At present, there is no effective treatment for
patients with advanced PRCC (6).
A number of genes have been previously identified to
be frequently mutated in PRCC, including MET, SETD2, NF2, KDM6A,
SMARCB1, FAT1, BAP1, PBRM1, STAG2, NFE2L2 and TP53 (7,8). At
present, the research conducted regarding PRCC-associated
biomarkers is insufficient to meet clinical requirements for the
diagnosis and prognosis of patients, and there remains a lack of
knowledge regarding the use of PRCC-associated noncoding RNAs as
biomarkers and their internal interactions. A previous study
reported that the classification of different molecular subtypes
may aid the prognosis of PRCC (9).
The requirement for improved understanding of the molecular
mechanisms underlying the development and progression of PRCC is
increasing. A recent study analyzed the expression of long
noncoding RNAs (lncRNAs), which may serve as useful biomarkers for
tumor staging, in PRCC using The Cancer Genome Atlas (TCGA)
(10). At present, there has been
limited analysis of PRCC-associated competing endogenous RNA
(ceRNA) networks involving lncRNAs in an entire genome.
The ceRNA hypothesis involves a complex
post-transcriptional regulatory network in which lncRNAs, mRNAs and
other RNAs act as natural microRNA (miRNA) sponges to suppress
miRNA function by sharing one or more miRNA response elements
(11). At present, increasing
evidence indicates that regulatory networks serve important roles
in the occurrence, development and regulation of tumors, including
breast, ovarian, kidney, colon and liver cancers (12–17).
To investigate the regulatory mechanisms of ceRNA
networks in PRCC, and aid improvements in the diagnosis and
prognosis of PRCC, a comprehensive analysis of the genomic and
epigenomic landscape of PRCC was conducted to identify
statistically significant genetic alterations in tumors in the
present study. Following a series of analyses, an lncRNA-miRNA-mRNA
ceRNA network was constructed and important genes were identified,
which may aid the identification of the functions of noncoding RNAs
in PRCC, and the associations between miRNA, lncRNA and mRNA.
Reverse transcription-quantitative PCR (RT-qPCR) analysis revealed
that the expression of a central lncRNA in the ceRNA network,
lncRNA maternally expressed 3 (MEG3), was downregulated in PRCC
tumor tissues compared with adjacent non-tumor tissues.
Furthermore, Kaplan-Meier survival analyses identified 13 mRNAs, 15
lncRNAs and six miRNAs that significantly predicted the prognosis
of patients with PRCC. The results of the present study provide a
novel approach for the investigation of molecular mechanisms and
prognostic biomarkers in PRCC.
Materials and methods
Data source
mRNA and miRNA expression profiles and clinical
characteristics of patients with PRCC were obtained from TCGA using
the Data Transfer Tool (cancergenome.nih.gov/), using the search terms
‘Kidney’, ‘Kidney Renal Papillary Cell Carcinoma’ and
‘Transcriptome Profiling’, resulting in 289 PRCC sample tissues and
32 non-tumor tissues. mRNA and miRNA sequence (seq) data are
available open access, and the present research met the
requirements of TCGA publishing guidelines. Following the
acquisition of mRNA-seq and miRNA-seq data, lncRNA expression data
were obtained by relocating probes in mRNA expression profiles to
lncRNAs based on annotations from the GENCODE project (version 28;
gencodegenes.org/) (18,19).
Identification and analysis of
PRCC-associated mRNAs, lncRNAs and miRNAs
Differential expression analysis of mRNAs, lncRNAs
and miRNAs was conducted using data from tumor and adjacent
non-tumor tissues from patients with PRCC using edgeR, a
Bioconductor package (version 3.6) in R software (version 3.5.0)
(20), with thresholds of
|log2-fold change| ≥2.0 and adjusted P<0.05; the false discovery
rate (FDR) was adjusted using the Benjamini-Hochberg method
(21).
Functional annotation of
differentially expressed mRNAs (DEmRNAs) and construction of the
protein-protein interaction (PPI) network
To determine the biological significance of DEmRNAs,
Gene Ontology (GO) terms were assigned using the GO database
(geneontology.org/) (22) via the Database for Annotation
Visualization and Integrated Discovery (DAVID; version 6.8;
david.ncifcrf.gov/) (23,24).
Additionally, Kyoto Encyclopedia of Genes and Genomes (KEGG)
(25–27) pathway enrichment analysis of
DEmRNAs was performed using KOBAS 3.0 (http://kobas.cbi.pku.edu.cm/index.php) (28).
To further understand the protein-protein
interactions between DEmRNAs, a PPI network was generated using the
Search Tool for the Retrieval of Interacting Genes (version 10.5;
string-db.org/) database (29,30).
PPI networks were visualized using Cytoscape version 3.6.1 software
(31). Furthermore, the CytoHubba
(Version 0.1) plug-in for Cytoscape (32) was used to identify hub genes by
ranking the participation degree in PPI networks.
Construction of the ceRNA network
DElncRNAs were compared using the miRcode database
(version 11; mircode.org/) (33), then miRNAs in selected pairs were
compared with previously identified DEmiRNAs to obtain the final
integrated lncRNA-miRNA pairs. Prediction of target genes for
selected lncRNA-miRNA pairs of miRNAs was performed using three
databases, miRTarBase (version 7.0) (34), miRanda (version 3.3a) (35), and TargetScan (version 7.1)
(36). Candidate target mRNAs were
included in the three databases and intersected with previously
identified DEmRNAs. Then, the lncRNA-miRNA-mRNA ceRNA network was
reconstructed by assembling DElncRNA-DEmiRNA-DEmRNA associations
visualized using Cytoscape (version 3.6.1). The sub-network of the
ceRNA network was extracted using the Cytoscape plug-in MCODE
(version 1.5.1) (37).
RNA extraction and RT-qPCR
validation
lncRNA MEG3 was selected from the ceRNA network for
expression analysis in 12 PRCC tumor tissues and adjacent non-tumor
tissues. A total of 12 pairs of paraffin-embedded tissue samples
from PRCC patients were collected at the Affiliated Hospital of
Southwest Medical University (Luzhou, China) between January 2017
and February 2018. The patients ranged in age from 39 to 80 years,
with a median age of 60.5 years, including 8 males and 4 females.
All patients were confirmed with primary PRCC by pathological
examination of surgical specimens and were not subject to any
preoperative radiotherapy or chemotherapy, other malignant disease,
or acute injury. The study was approved by the Ethical Review
Committee of the Affiliated Hospital of Southwest Medical
University. Written informed consent was obtained from all patients
prior to sample collection. Total RNA was extracted from PRCC tumor
and adjacent normal tissues using a paraffin-embedded tissue RNA
extraction kit (Bioteke Corporation, Beijing, China). RT was
performed using a ReverTra Ace qPCR RT Master Mix with gDNA Remover
(Toyobo Life Science, Tokyo, Japan), according to the
manufacturer's protocols. Expression level of the lncRNA was
detected by qPCR using SYBR Green Realtime PCR Master Mix (Toyobo
Life Science) in an Applied Biosystems™ 7500 Fast
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). β-actin served as an internal control. The
following primer sequences were used for qPCR: lncRNA MEG3, forward
5′-GCCTGCTGCCCATCTACAC-3′, reverse 5′-CCTCTTCATCCTTTGCCATC-3′; and
β-actin, forward 5′-TCCTCTCCCAAGTCCACACA-3′ and reverse
5′-GCACGAAGGCTCATCATTCA-3′. qPCR was conducted as follows: 95°C for
30 sec; 40 cycles of 95°C for 5 sec and 60°C for 30 sec; and a
dissociation cycle of 95°C for 15 sec, 60°C for 60 sec, 95°C for 15
sec and 60°C for 15 sec. Each reaction was performed in triplicate,
and the relative expression levels (fold change) were calculated
using the 2−ΔΔCq method (38). A paired t-test was performed using
SPSS version 22.0 (IBM Corp., Armonk, NY, USA) to analyze
differences in the expression of lncRNA MEG3 between PRCC tumor
tissues and adjacent nontumor tissues (n=12/group). P<0.05 was
considered to indicate a statistically significant difference.
Survival analysis
To identify prognostic DERNAs for patients with PRCC
from TCGA, clinical data were obtained and mapped Kaplan-Meier
curves for various DElncRNAs, DEmiRNAs and DEmRNAs were calculated.
Patients with PRCC were divided into high expression and low
expression groups according to the median value of gene expression.
Significant differences in survival between groups were determined
using log-rank tests; P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of DEmRNAs, DElncRNAs
and DEmiRNAs in PRCC
Transcriptome sequencing data for mRNAs, lncRNAs and
miRNAs were analyzed separately using 289 PRCC tumor samples and 32
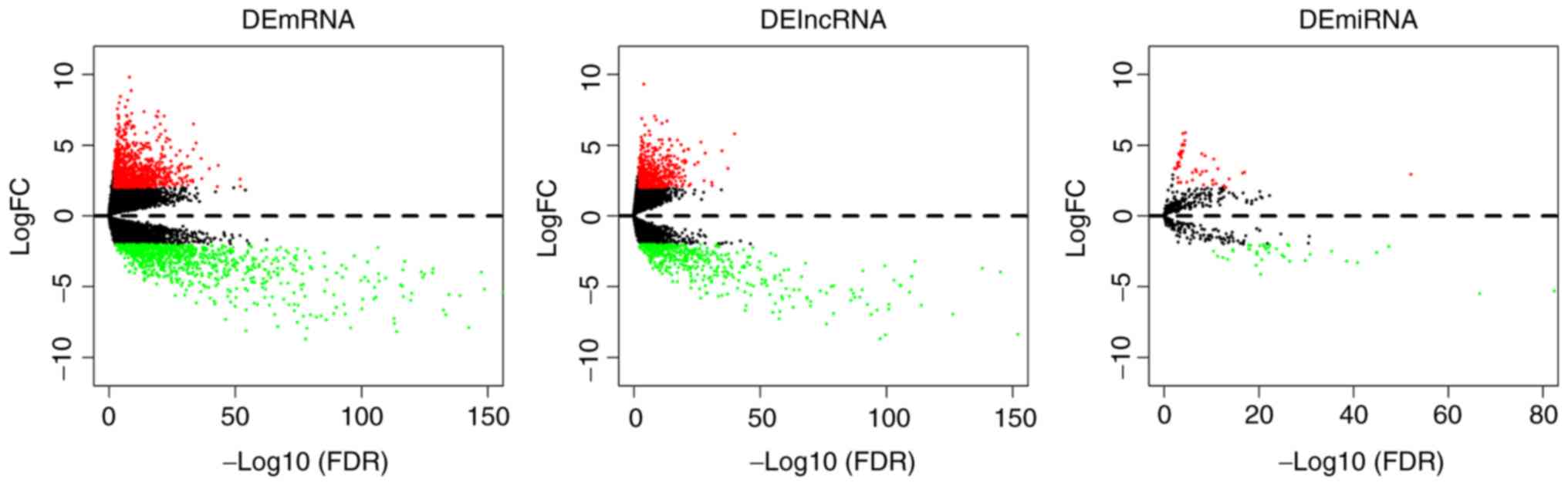
paracancerous tissue samples. A total of 1,970 DEmRNAs, 1,201
DElncRNAs and 96 DEmiRNAs were identified to have significantly
different expression (|log2-fold change| ≥2.0 and adjusted
P<0.05) in tumor tissues compared with the adjacent tissue.
Volcano plots were generated for the identified DEmRNAs, DElncRNAs
and DEmiRNAs (Fig. 1). The top 10
upregulated and downregulated DEmRNAs, DElncRNAs and DEmiRNAs are
listed in Table I.
 | Table I.Top 10 upregulated and downregulated
DEmRNAs, DElncRNAs and DEmiRNAs. |
Table I.
Top 10 upregulated and downregulated
DEmRNAs, DElncRNAs and DEmiRNAs.
| A, Upregulated |
|---|
|
|---|
| DEmRNA | DElncRNA | DEmiRNA |
|---|
|
|
|
|---|
| Name | log2FC | FDR P-value | Name | log2FC | FDR P-value | Name | log2FC | FDR P-value |
|---|
| DDB2 | 2.1 |
9.44×10−53 | AL365181.2 | 5.8 |
1.53×10−40 | hsa-mir-21 | 2.95 |
8.32×10−53 |
| BBC3 | 2.59 |
1.22×10−52 | AC019069.1 | 3.37 |
6.25×10−38 | hsa-mir-561 | 3.08 |
1.08×10−17 |
| HK2 | 3.56 |
5.82×10−44 | AL365181.3 | 4.61 |
1.46×10−35 | hsa-mir-592 | 3.03 |
2.78×10−17 |
| LRRC20 | 2.07 |
1.25×10−43 | AC083862.2 | 2.15 |
1.05×10−31 | hsa-mir-1254-1 | 2.62 |
2.02×10−14 |
| GPRIN1 | 3.34 |
1.62×10−40 | AC005041.3 | 2.37 |
1.52×10−31 | hsa-mir-3934 | 2.01 |
9.04×10−14 |
| TMSB10 | 2.63 |
1.97×10−37 | AC092535.4 | 4.43 |
4.79×10−29 | hsa-mir-4768 | 2.14 |
2.40×10−13 |
| TNFSF9 | 4.06 |
3.18×10−37 | LACTB2-AS1 | 2.49 |
1.73×10−28 | hsa-mir-1293 | 3.34 |
4.41×10−12 |
| HAMP | 5.16 |
2.97×10−35 | PAQR9-AS1 | 5.21 |
2.75×10−27 | hsa-mir-584 | 2.29 |
9.54×10−12 |
| APOC1 | 6.5 |
3.13×10−34 | GAS6-AS1 | 3.8 |
4.41×10−26 | hsa-mir-7156 | 4.01 |
3.23×10−11 |
| TREM2 | 4.7 |
4.32×10−34 | AL590666.2 | 3.64 |
8.35×10−25 | hsa-mir-4777 | 2.53 |
4.57×10−11 |
|
| B,
Downregulated |
|
| DEmRNA |
DElncRNA | DEmiRNA |
|
|
|
| Name | log2FC | FDR
P-value | Name | log2FC | FDR
P-value | Name | log2FC | FDR
P-value |
|
| MFSD4A | −4.72 |
1.48×10−274 | AC068631.1 | −5.58 |
1.62×10−177 | hsa-mir-184 | −5.3 |
4.38×10−83 |
| UMOD | −12.3 |
4.41×10−252 | AC002401.1 | −6.42 |
1.11×10−173 | hsa-mir-216b | −5.48 |
2.32×10−67 |
| CALB1 | −8.29 |
1.71×10−214 | AC079310.1 | −8.36 |
9.26×10−153 | hsa-mir-126 | −2.15 |
3.64×10−48 |
| EGF | −6.21 |
4.85×10−206 | AC008264.2 | −3.96 |
8.45×10−146 | hsa-mir-33a | −2.57 |
1.37×10−45 |
| GP2 | −8.23 |
4.61×10−199 | AC019080.1 | −3.69 |
1.11×10−138 | hsa-mir-129-2 | −3.3 |
1.45×10−41 |
| GGACT | −3.57 |
4.85×10−191 | AC106772.2 | −6.95 |
5.04×10−127 | hsa-mir-129-1 | −3.2 |
2.57×10−39 |
| DDN | −7.01 |
4.88×10−187 | AC027309.1 | −6.33 |
1.66×10−114 | hsa-mir-145 | −2.49 |
4.73×10−36 |
| CRHBP | −6.08 |
5.81×10−186 | AC010442.1 | −3.21 |
6.42×10−112 | hsa-mir-323a | −2.72 |
1.45×10−31 |
| PTGER1 | −5.4 |
1.20×10−156 | AC099684.2 | −4.27 |
9.72×10−111 | hsa-mir-206 | −3.16 |
1.70×10−30 |
| SLC26A4 | −5.18 |
3.04×10−149 | AC105384.1 | −5.39 |
7.77×10−110 | hsa-mir-489 | −3.19 |
3.16×10−27 |
Analysis of DEmRNAs
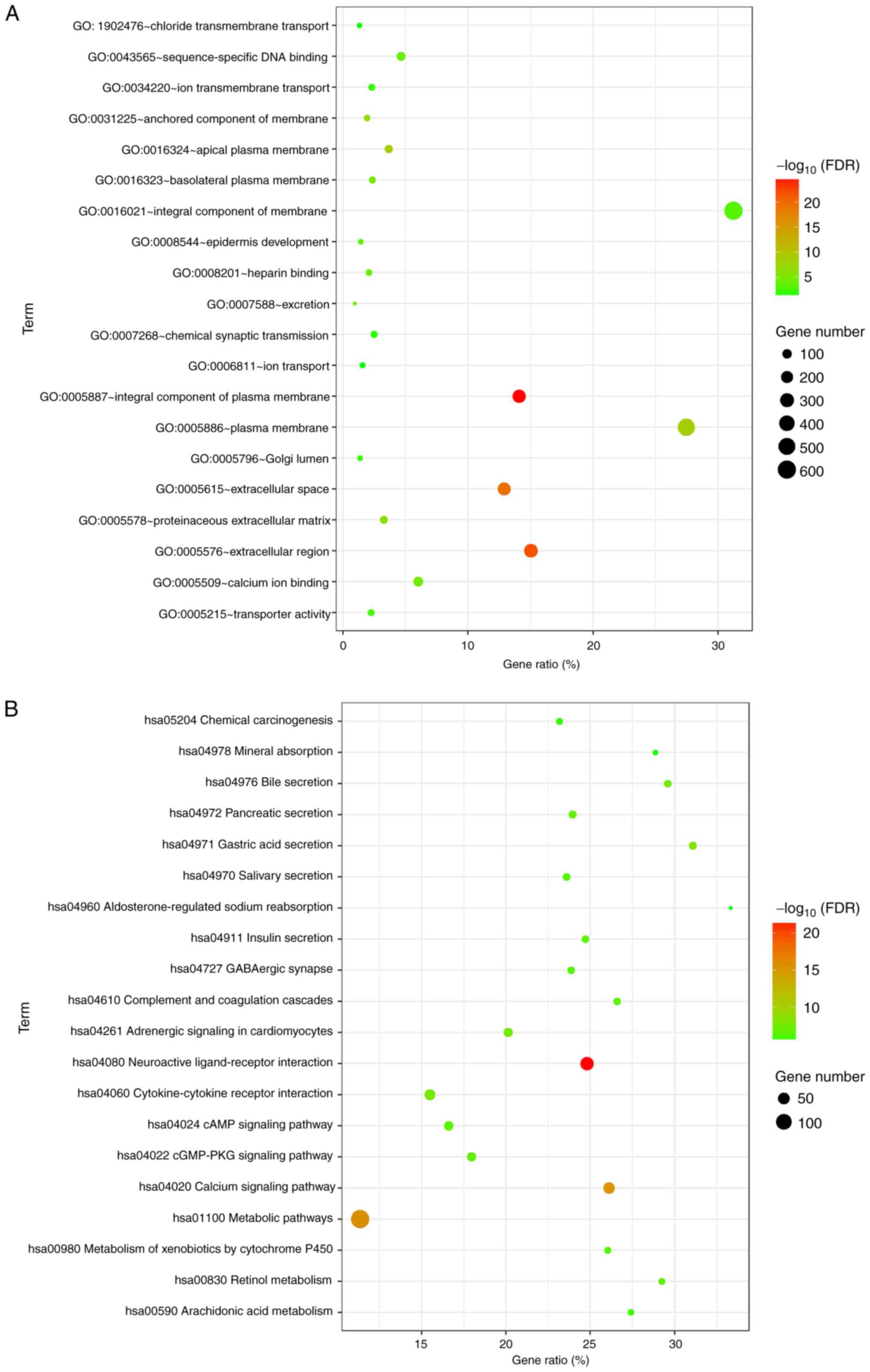
Of the 1,970 identified DEmRNAs, 1,092 mRNAs were
upregulated and 878 were downregulated. To understand the
biological significance of these DEmRNAs, GO and pathway enrichment
analyses were conducted. GO analysis provides three categories of
information: ‘Biological process’, ‘cellular component’ and
‘molecular function’. Identified DEmRNAs were primarily enriched in
‘metabolic pathways’, ‘neuroactive ligand-receptor interaction’,
‘calcium signaling pathways’, ‘pathways in cancer’ and
‘cytokine-cytokine receptor interactions’. GO and KEGG pathway
analysis results are presented in Fig.
2.
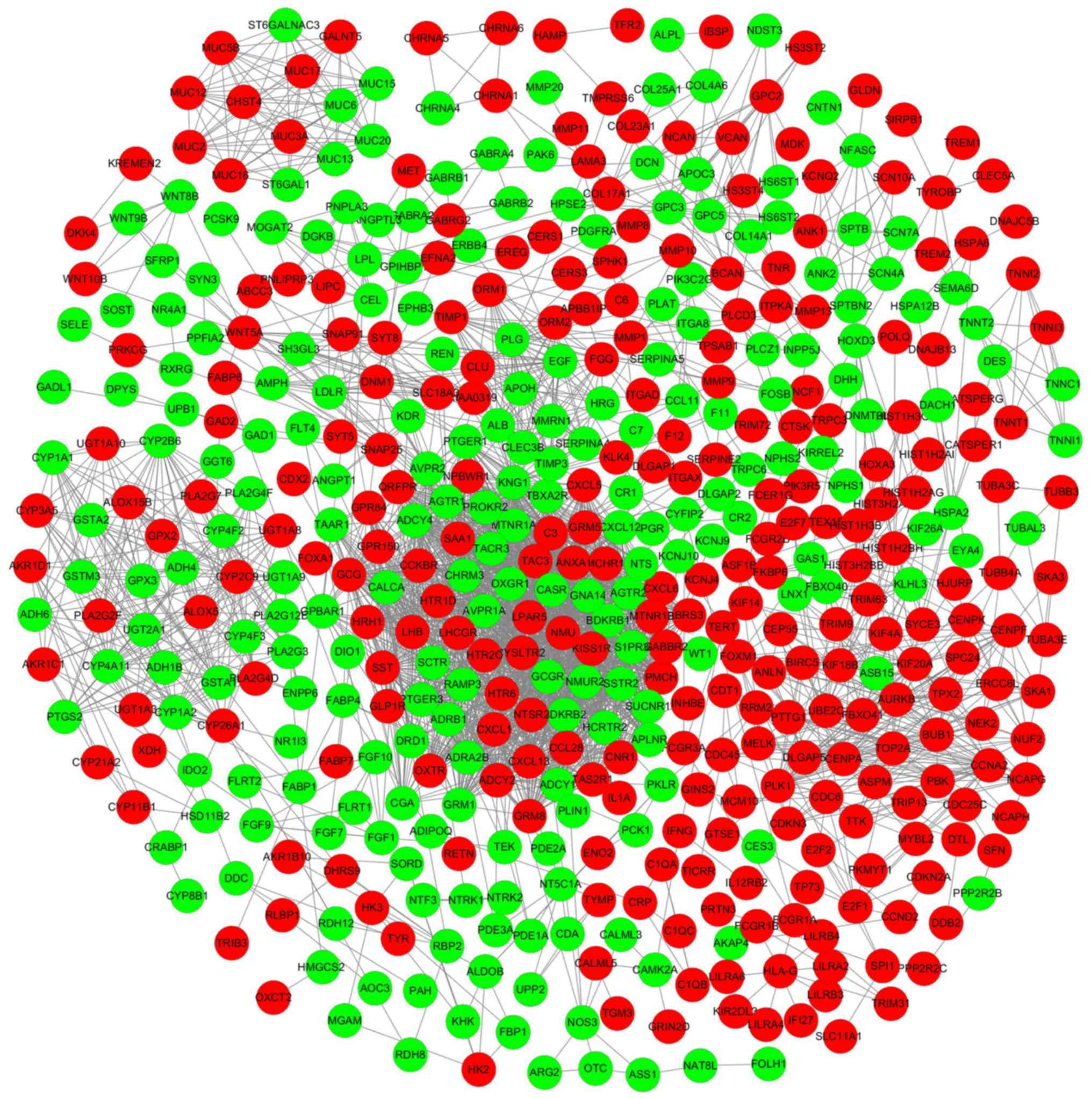
A PPI network was constructed to investigate the
interactions between the identified DEmRNAs. The PPI network was
visualized using Cytoscape (Fig.
3). In addition, the top 10 hub DEmRNAs were identified using
the CytoHubba plug-in according to degree levels: albumin (ALB),
DNA topoisomerase II α (TOP2A), epidermal growth factor (EGF),
kininogen 1 (KNG1), leucine rich repeat kinase 2 (LRRK2),
baculoviral IAP repeat containing 5 (BIRC5), polo like kinase 1
(PLK1), cyclin A2 (CCNA2), cyclin dependent kinase inhibitor 3
(CDKN3), and aurora kinase B (AURKB); the interaction network is
presented in Fig. 4.
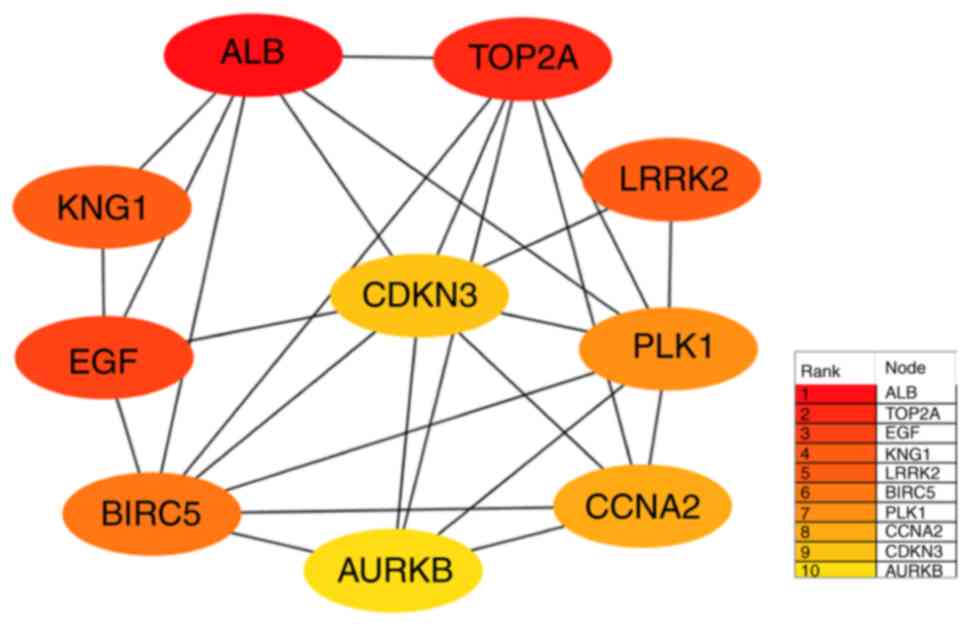 | Figure 4.Top 10 hub DEmRNAs extracted from the
protein-protein interaction network. Circles indicate
protein-coding genes; lines between DEmRNAs indicate direct
interactions. DE, differentially expressed; ALB, albumin; TOP2A,
DNA topoisomerase II α; KNG1, kininogen 1; LRRK2, leucine-rich
repeat kinase 2; CDKN3, cyclin dependent kinase inhibitor 3; EGF,
epidermal growth factor; PLK1, polo like kinase 1; BIRC5,
baculoviral IAP repeat containing 5; CCNA2, cyclin A2; AURKB,
aurora kinase B. |
Construction and analysis of the
lncRNA-miRNA-mRNA ceRNA network
To investigate how lncRNAs and miRNAs cooperate to
regulate mRNA expression in PRCC, miRNA-mRNA and lncRNA-miRNA
regulatory associations were used to construct an lncRNA-miRNA-mRNA
ceRNA network. The differential expression profile consisted of 26
DEmRNA nodes, 65 DElncRNA nodes, 15 DEmiRNA nodes (Table II) and 287 edges. This
reconstructed ceRNA network was visualized using Cytoscape
(Fig. 5). From the networks, it
was determined that DElncRNA MEG3 exhibited potential interactions
with 14 DEmiRNAs (hsa-miR-507, hsa-miR-145, hsa-miR-519d,
hsa-miR-184, hsa-miR-206, hsa-miR-211, hsa-miR-21, hsa-miR-214,
hsa-miR-216a, hsa-miR-216b, hsa-miR-217, hsa-miR-508, hsa-miR-31
and hsa-miR-506). Therefore, it was hypothesized that lncRNA MEG3
may have an important role in regulating the ceRNA network in PRCC.
Furthermore, miR-519d interacted with 18 DElncRNAs [testis-specific
transcript Y-linked 14 (TTTY14), AP002478.1, long intergenic
non-protein coding RNA (LINC)00221, T cell leukemia/lymphoma 6
(TCL6), LINC00173, AC009061.1, AC061975.6, AC025278.1, MEG3,
LINC00269, glutamate metabotropic receptor 7-antisense RNA 3
(GRM7-AS3), LINC00462, zinc finger RANBP2-type containing
2-antisense RNA 1 (ZRANB2-AS1), LINC00330, HOXA distal transcript
antisense RNA (HOTTIP), versican-antisense RNA 1 (VCAN-AS1), Pvt1
oncogene (PVT1) and glutamate metabotropic receptor 5-antisense RNA
1 (GRM5-AS1)] and possessed a targeted regulatory association with
eight DEmRNAs [Rap guanine nucleotide exchange factor 4 (RAPGEF4),
E2F transcription factor 2 (E2F2), hyaluronan synthase 2 (HAS2),
Sal-like protein 3 (SALL3), ribonucleotide reductase regulatory
subunit M2 (RRM2), low density lipoprotein (LDL), DNA polymerase θ
(POLQ) and E2F transcription factor 1 (E2F1)]. The ceRNA subnetwork
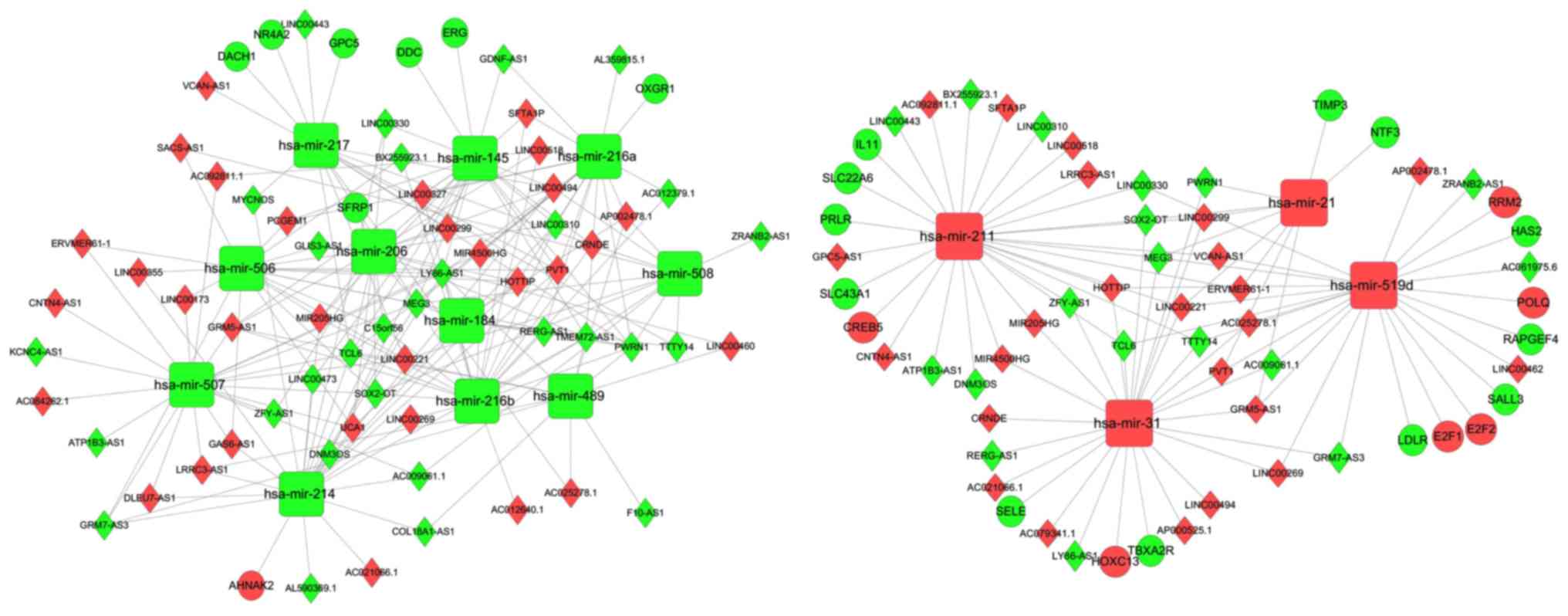
was extracted using the plug-in MCODE for Cytoscape (Fig. 6). This subnetwork consists of hub
genes, including lncRNA MEG3, lncRNA Prader-Willi region
non-protein coding RNA 1 (PWRN1), hsa-miR-508 and hsa-miR-21, plus
certain first neighbors, including neurotrophin 3 (NTF3), tissue
inhibitor of metalloproteinase 3 (TIMP3), GRM5-AS1, AP002478.1,
TTTY14 and hsa-miR-489.
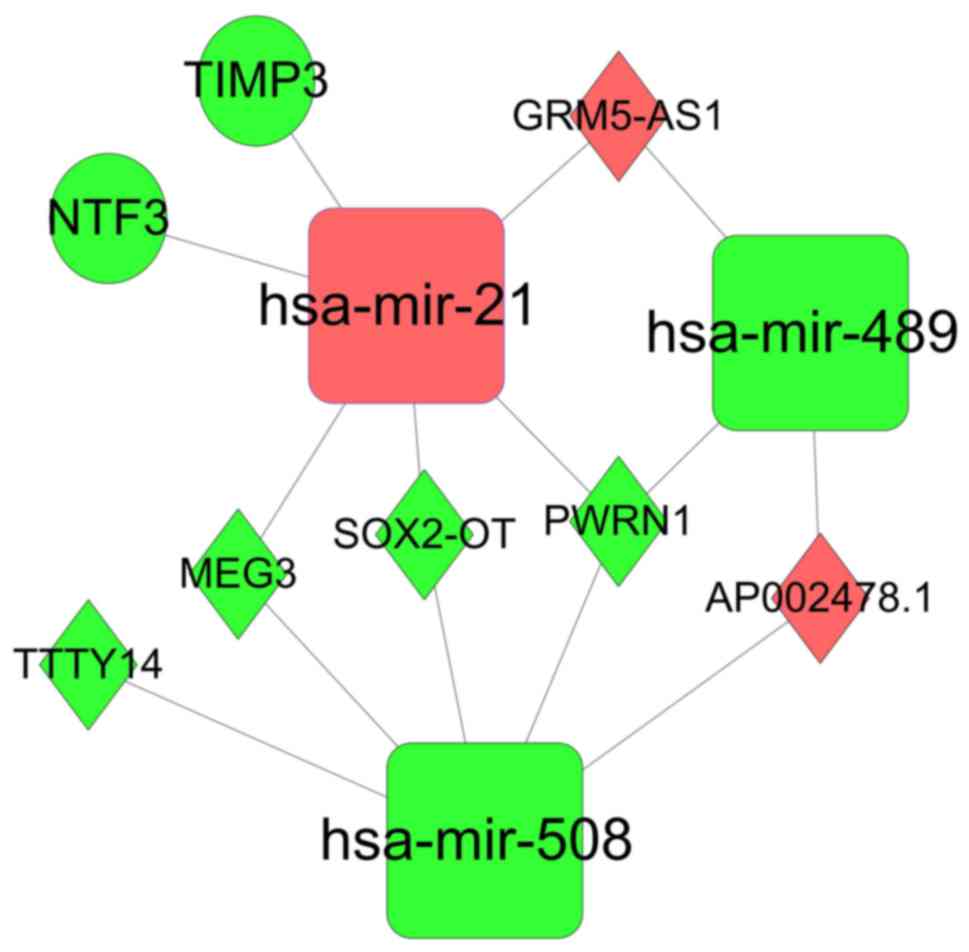 | Figure 6.Subnetwork of the lncRNA-miRNA-mRNA
ceRNA network. Diamonds indicate lncRNAs; circles indicate mRNA;
round rectangles indicate miRNAs; edges indicate interactions. Red
indicates upregulation; green indicates downregulation. lncRNA,
long noncoding RNA; miRNA/mir, microRNA; ceRNA, competing
endogenous RNA; TIMP3, tissue inhibitor of metalloproteinase 3;
GRM5-AS1, glutamate metabotropic receptor 5-antisense RNA 1; NTF,
neurotrophin 3; TTTY14, testis-specific transcript Y-linked 14;
MEG3, maternally expressed 3; SOX2-OT, SOX2 overlapping transcript;
PWRN1, Prader-Willi region non-protein coding RNA 1. |
 | Table II.Key DEmRNAs, DElncRNAs and DEmiRNAs
comprising the papillary renal cell carcinoma-associated competing
endogenous RNA network. |
Table II.
Key DEmRNAs, DElncRNAs and DEmiRNAs
comprising the papillary renal cell carcinoma-associated competing
endogenous RNA network.
| RNA type | Name |
|---|
| DEmRNA | SFRP1, NTF3, GPC5,
RAPGEF4, TIMP3, E2F2, LDLR, DACH1, ELE, PRLR, SLC43A1, POLQ, ERG,
RRM2, NR4A2, DDC, AHNAK2, IL11, CREB5, HAS2, E2F1, SALL3, TBXA2R,
HOXC13, OXGR1, SLC22A6 |
| DElncRNA | AC021066.1,
TMEM72-AS1, ZRANB2-AS1, AC084262.1, TTTY14, KCNC4-AS1, LINC00330,
UCA1, C15orf56, SFTA1P, LINC00494, MEG3, AP002478.1, GRM7-AS3,
AC012379.1, LINC00269, COL18A1-AS1, AC079341.1, GPC5-AS1, LY86-AS1,
LINC00518, PCGEM1, LINC00299, AL359815.1, LINC00221, LINC00310,
GLIS3-AS1, LINC00473, TCL6, LINC00355, DLEU7-AS1, F10-AS1,
BX255923.1, MIR4500HG, CNTN4-AS1, LINC00327, LINC00173, LRRC3-AS1,
AC012640.1, ZFY-AS1, AC009061.1, SACS-AS1, SOX2-OT, LINC00460,
AC061975.6, LINC00443, HOTTIP, LINC00462, AC092811.1, ERVMER61-1,
ATP1B3-AS1, GAS6-AS1, AC025278.1, DNM3OS, CRNDE, MYCNOS,
AP000525.1, MIR205HG, GDNF-AS1, PWRN1, RERG-AS1, AL590369.1,
VCAN-AS1, GRM5-AS1, PVT1 |
| DEmiRNA | hsa-mir-214,
hsa-mir-31, hsa-mir-519d, hsa-mir-184, hsa-mir-211, hsa-mir-217,
hsa-mir-508, hsa-mir-206, hsa-mir-216b, hsa-mir-506, hsa-mir-216a,
hsa-mir-489, hsa-mir-145, hsa-mir-507, hsa-mir-21 |
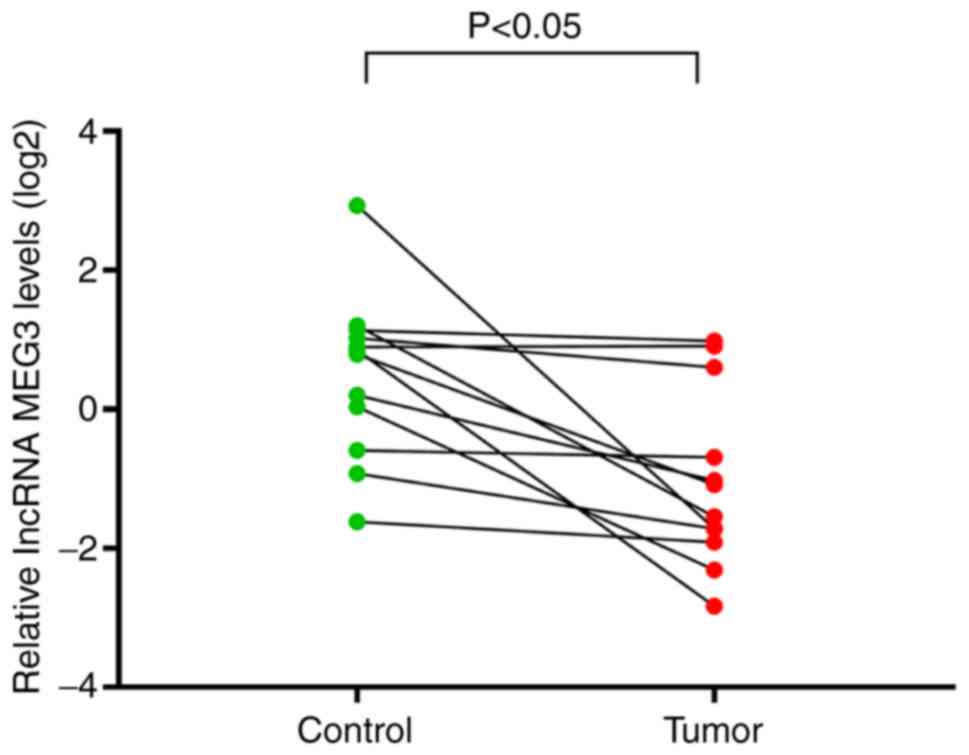
RT-qPCR validation
Of the 15 DEmiRNAs that formed the ceRNA network,
lncRNA MEG3 exhibited potential interactions with 14 DEmiRNAs, more
than any other DElnRNA, suggesting that this lncRNA may be most
likely to serve an important role in PRCC. To validate the
bioinformatics results, lncRNA MEG3 was selected for expression
analysis. LncRNA MEG3 was revealed to be significantly
downregulated in PRCC tumor tissue compared with adjacent non-tumor
tissues (Fig. 7), consistent with
the aforementioned bioinformatics analysis.
Survival analysis using lncRNAs,
miRNAs and mRNAs
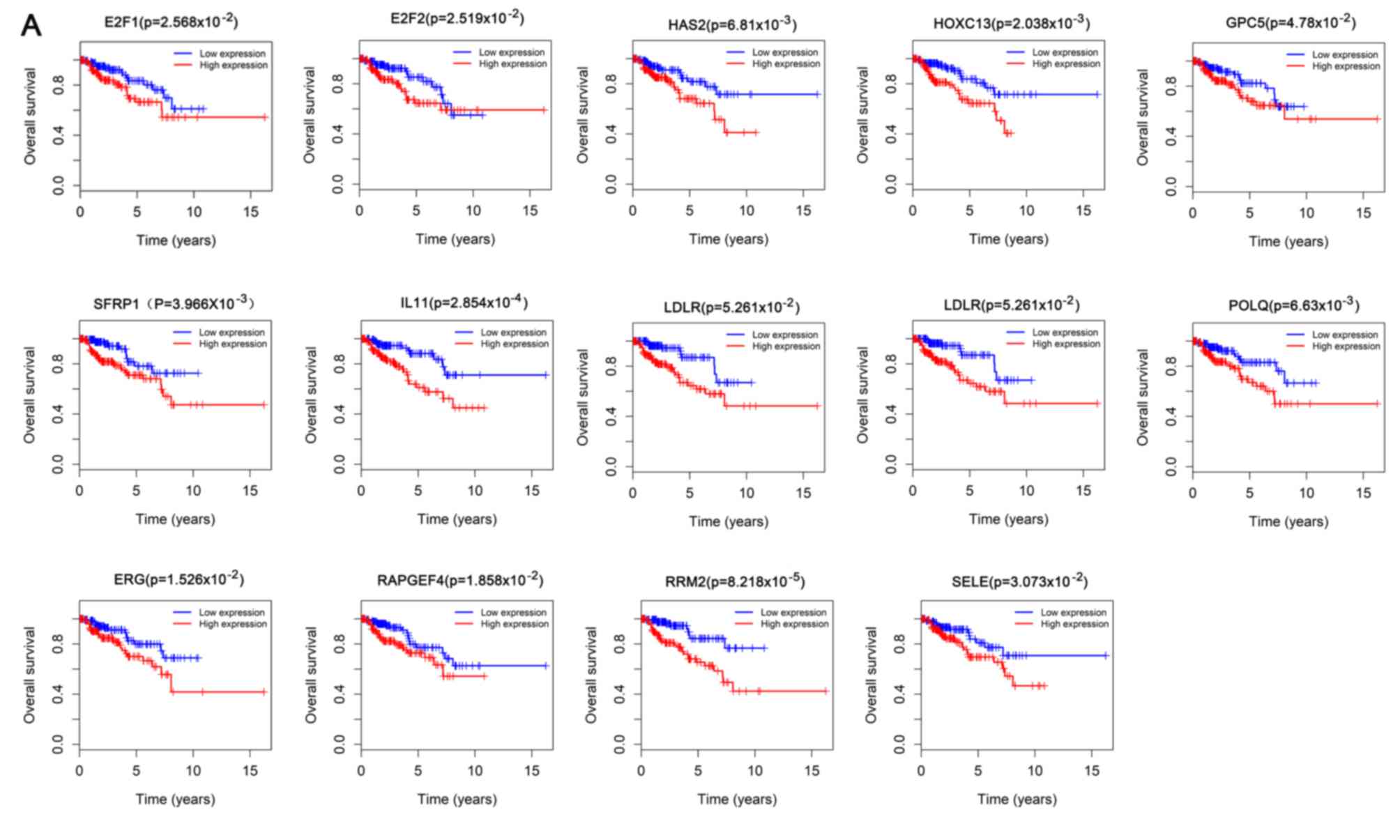
In the ceRNA network, 13 DEmRNAs were analyzed to
determine associations between expression levels and patient
survival, including E2F1, E2F2, ETS transcription factor, glypican
5 (GPC5), HAS2, homeobox C13 (HOXC13), interleukin 11 (IL11), LDL
receptor (LDLR), POLQ, RAPGEF4, RRM2, selectin E (SELE) and
secreted frizzled related protein 1 (SFRP1), all of which were
upregulated in patients with PRCC; expression levels of all these
genes were associated with overall survival (P<0.05; Fig. 8A). Similarly, the expression of
five DElncRNAs [colorectal neoplasia differentially expressed
(CRNDE), GAS6 antisense RNA 1 (GAS6-AS1), GPC5-antisense RNA 1
(GPC5-AS1), LINC00327 and SACS antisense RNA 1 (sacsin-AS1)] were
significantly positively associated with patient survival, whereas
10 [AP000525.1, glial cell-derived neurotrophic factor-antisense
RNA 1 (GDNF-AS1), GLIS family zinc finger 3-antisense RNA 1
(GLIS3-AS1), LINC00221, LINC00310, LINC00462, LINC00473, LRR
containing 3-antisense RNA 1 (LRRC3-AS1), surfactant associated 1,
pseudogene (SFTA1P) and DNM3 opposite strand (DNM3OS)] were
negatively associated (P<0.05; Fig.
8B). One (hsa-miR-211) and five DEmiRNAs (hsa-miR-145,
hsa-miR-184, hsa-mir-214, hsa-miR-216a, hsa-miR-217) were
significantly positively and negatively associated with overall
survival in PRCC, respectively (P<0.05; Fig. 8C).
Discussion
PRCC accounts for ~18.5% of total cases of RCC
(1), and it is generally
considered to exhibit an improved prognosis compared with CRCC
(3). Therefore, there is a notably
reduced level of research into PRCC. Prior to the TCGA report into
PRCC (8), no large-scale study
systematically investigated the pathogenesis of this disease or
aimed to identify prognosis-associated biomarkers. Using TCGA, an
in-depth analysis was conducted involving a comprehensive genomics
approach to characterize the pathology of 161 cases of PRCC in
subtypes 1 and 2 (8); however, due
to the heterogeneity of PRCC, the pathogenesis, development and
prognosis remain unclear, particularly concerning important ceRNA
network-associated mechanisms. PRCC is considered to be highly
heterogeneous; however, it was previously reported that ~50% of
cases exhibit a certain degree of overlap between type I and type
II (3). Therefore, in the present
study, sample data for PRCC in TCGA were analyzed to identify
factors frequently associated with the pathogenesis, development
and prognosis of PRCC.
A large number of samples were obtained from the
TCGA database, and gene pathways and hub genes associated with PRCC
were identified to determine the mechanisms underlying PRCC
incidence. A previous study involving TCGA data mining observed
that type 2 tumors were characterized by CDKN2A silencing (8). In the present study, it was reported
that CDKN3 was one of 10 hub genes in the ceRNA network. GO and
KEGG pathway analyses are frequently used to determine the
biological functions of DE coding genes. It was revealed via GO
analysis of DEmRNAs that there was significant enrichment of 170 GO
‘biological processes’ (P<0.01), including ‘excretion’,
‘epidermis development’, ‘integral components of plasma membrane’,
‘extracellular regions’, ‘calcium ion binding’ and ‘heparin
binding’. The biological functions of the aforementioned DE genes
are consistent with the formation and function of renal cells. It
has been established that calcium ions affect almost every aspect
of cellular life (39), and that
variations in cytosolic calcium concentrations induce important
cellular events (40).
Intracellular calcium overload can initiate mitochondrial-dependent
apoptosis (40), which may be a
strategy for inhibiting the proliferation of cancer cells (41,42).
Identified DEmRNAs were also significantly enriched in calcium
signaling pathways, as determined by KEGG pathway analysis. Raynal
et al (43) reported that
targeting calcium signaling can reverse the epigenetic silencing of
tumor suppressor genes. Additionally, renal cell carcinoma is
closely associated with abnormal alterations in metabolic pathways
involved in oxygen sensing, energy sensing and nutrient sensing
cascades (44–46). A previous study demonstrated that
metabolic pathways were altered in metastatic RCC, with
downregulation of citric acid cycle genes and upregulation of the
pentose phosphate pathway (47).
Furthermore, identified DEmRNAs were also enriched in ‘pathways in
cancer’, providing a theoretical basis for further research.
Therefore, investigation of these signaling pathways may have
notable implications for the identification of biological processes
and molecular functions involved in tumorigenesis, progression and
metastasis.
The roles of noncoding RNA have been identified
following advancements in genetic research, including their central
role in the diagnosis, treatment and prognosis of various tumors
(48–50). Therefore, a lncRNA-miRNA-mRNA ceRNA
network was reconstructed to investigate the roles of noncoding
RNAs associated with PRCC via a combination of differential gene
expression profile and target analyses. From the ceRNA network, it
was hypothesized that lncRNA MEG3 and miR-519d may serve important
roles in the regulation of ceRNA networks associated with PRCC. It
has been reported that lncRNA MEG3 acts as a lncRNA tumor
suppressor in numerous tumors (51–56)
via interactions with the tumor suppressor p53 and the regulation
of the expression of p53 target genes (57); however, the role of lncRNA MEG3 in
PRCC has not yet been investigated.
It was also revealed that miR-519d may occupy an
important position in the constructed ceRNA network. Downregulation
of miR-519d was reported in studies investigating the molecular
mechanisms underlying various tumors, including gastric, ovarian
and colorectal cancers (58–60).
Additionally, the extraction of a subnetwork identified potentially
important RNAs, including lncRNA MEG3, lncRNA PWRN1, hsa-miR-508
and hsa-miR-21. lncRNA PWRN1 has been reported to target miR-425-5p
and suppress the development of gastric cancer via p53 signaling
(61). miR-508 suppressed the
epithelial-mesenchymal transition, migration and invasion of
ovarian cancer cells via the mitogen-activated protein kinase 1/ERK
signaling pathway (62).
Similarly, miR-21 has been reported be involved in numerous
molecular mechanisms underlying tumorigenesis (63–65),
including ceRNA network regulatory mechanisms (66).
To validate the results of the bioinformatics
analyses, lncRNA MEG, a core lncRNA in the ceRNA network, was
selected for expression analysis in PRCC tumor tissues and adjacent
tissues. RT-qPCR analysis revealed that lncRNA MEG3 was
downregulated in tumor tissues compared with adjacent non-tumor
tissues. It was recently reported that lncRNA MEG3 expression was
decreased in CCRC tissues and cells, affecting the apoptosis,
proliferation, migration and invasion of CCRC cells by regulating
miR-7/RAS like family 11 member B (67).
Survival analysis revealed that the expression of 13
out of 26 DEmRNAs, 15 out of 65 DElncRNAs and 6 out of 15 DEmiRNAs
were significantly associated with survival, indicating that these
RNAs may be potential biomarkers for the prognosis of patients with
PRCC (P<0.05). It was observed that the expression of RRM2 was
the most significantly associated with survival out of all the
RNAs. Wang et al (68)
reported that low expression of RRM2 was associated with increased
time to progression and overall survival in patients with non-small
cell lung cancer. Similarly, Zhang et al (69) revealed that reduced expression of
GPC5 was an independent prognostic marker for the overall survival
of patients with prostate cancer. GPC5 protein expression exhibited
an association with tumorigenesis and tumor progression in prostate
cancer, suggesting a potential application as a novel biomarker for
the prediction of diagnosis and prognosis of prostate cancer
(70). Furthermore, decreased
expression of lncRNA GAS6-AS1 predicted poor prognosis in patients
with non-small cell lung cancer (71). These prognosis-associated genes may
be potential targets for future clinical treatments, and were
identified to be significantly associated with the prognosis of
PRCC in the present study.
In conclusion, an lncRNA-miRNA-mRNA ceRNA network
was constructed via differential expression and target analyses,
demonstrating that lncRNA MEG3 and miR-519d may serve important
roles in PRCC. The expression of lncRNA MEG3 was observed to be
downregulated in PRCC tumor tissues compared with adjacent
non-tumor tissues. The present findings improve understanding of
ceRNA network regulatory mechanisms associated with PRCC and may
aid future studies into the molecular mechanisms underlying PRCC
and the identification of prognostic biomarkers.
Acknowledgements
Not applicable.
Funding
The present study received financial supported by
the Sichuan Science and Technology Program (grant nos.
2018TJPT0011, 2017TJPT0003, 2017HH0105 and 17KJFWSF0059).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QL and JL conceived and designed the study. MC and
QL collected the data. QL and MC analyzed the database. QD and QL
performed the experiments. QL prepared the diagrams and drafted the
manuscript. JL and QD reviewed and edited the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
For the use of human tissue samples, the study was
approved by the Ethics Committee of the Affiliated Hospital of
Southwest Medical University (Luzhou, China) and all patients
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Humphrey PA, Moch H, Cubilla AL, Ulbright
TM and Reuter VE: The 2016 WHO classification of tumours of the
urinary system and male genital organs-part B: Prostate and bladder
tumours. Eur Urol. 70:106–119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Delahunt B and Eble JN: Papillary renal
cell carcinoma: A clinicopathologic and immunohistochemical study
of 105 tumors. Mod Pathol. 10:537–544. 1997.PubMed/NCBI
|
|
3
|
Chevarie-Davis M, Riazalhosseini Y,
Arseneault M, Aprikian A, Kassouf W, Tanguay S, Latour M and Brimo
F: The morphologic and immunohistochemical spectrum of papillary
renal cell carcinoma: Study including 132 cases with pure type 1
and type 2 morphology as well as tumors with overlapping features.
Am J Surg Pathol. 38:887–894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ha YS, Chung JW, Choi SH, Lee JN, Kim HT,
Kim TH, Chung SK, Byun SS, Hwang EC, Kang SH, et al: Clinical
significance of subclassification of papillary renal cell
carcinoma: Comparison of clinicopathologic parameters and oncologic
outcomes between papillary histologic subtypes 1 and 2 using the
Korean renal cell carcinoma database. Clin Genitourin Cancer.
15:e181–e186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsimafeyeu I, Khasanova A, Stepanova E,
Gordiev M, Khochenkov D, Naumova A, Varlamov I, Snegovoy A and
Demidov L: FGFR2 overexpression predicts survival outcome in
patients with metastatic papillary renal cell carcinoma. Clin
Transl Oncol. 19:265–268. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Motzer RJ, Bacik J, Mariani T, Russo P,
Mazumdar M and Reuter V: Treatment outcome and survival associated
with metastatic renal cell carcinoma of non-clear-cell histology. J
Clin Oncol. 20:2376–2381. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Durinck S, Stawiski EW, Pavía-Jiménez A,
Modrusan Z, Kapur P, Jaiswal BS, Zhang N, Toffessi-Tcheuyap V,
Nguyen TT, Pahuja KB, et al: Spectrum of diverse genomic
alterations define non-clear cell renal carcinoma subtypes. Nat
Genet. 47:13–21. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cancer Genome Atlas Research Network, ;
Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis
C, Wheeler DA, Murray BA, Schmidt L, et al: Comprehensive molecular
characterization of papillary renal-cell carcinoma. N Engl J Med.
374:135–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang XJ, Tan MH, Kim HL, Ditlev JA, Betten
MW, Png CE, Kort EJ, Futami K, Furge KA, Takahashi M, et al: A
molecular classification of papillary renal cell carcinoma. Cancer
Res. 65:5628–5637. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lan H, Zeng J, Chen G and Huang H:
Survival prediction of kidney renal papillary cell carcinoma by
comprehensive LncRNA characterization. Oncotarget. 8:110811–110829.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y,
Zhao L, Zhang Y, Huang B and Lu J: LincRNA-ROR induces
epithelial-to-mesenchymal transition and contributes to breast
cancer tumorigenesis and metastasis. Cell Death Dis. 5:e12872014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen P, Fang X, Xia B, Zhao Y, Li Q and Wu
X: Long noncoding RNA LINC00152 promotes cell proliferation through
competitively binding endogenous miR-125b with MCL-1 by regulating
mitochondrial apoptosis pathways in ovarian cancer. Cancer Med.
7:4530–4541. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu K, Yao H, Wen Y, Zhao H, Zhou N, Lei S
and Xiong L: Functional role of a long non-coding RNA
LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to
pohotodynamic therapy. Biochim Biophys Acta Mol Basis Dis.
1864:2871–2880. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qu Y, Xiao H, Xiao W, Xiong Z, Hu W, Gao
Y, Ru Z, Wang C, Bao L, Wang K, et al: Upregulation of MIAT
regulates LOXL2 expression by competitively binding MiR-29c in
clear cell renal cell carcinoma. Cell Physiol Biochem.
48:1075–1087. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bai N, Peng E, Qiu X, Lyu N, Zhang Z, Tao
Y, Li X and Wang Z: circFBLIM1 act as a ceRNA to promote
hepatocellular cancer progression by sponging miR-346. J Exp Clin
Cancer Res. 37:1722018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Frankish A, Diekhans M, Ferreira AM,
Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J,
Armstrong J, et al: GENCODE reference annotation for the human and
mouse genomes. Nucleic Acids Res. 47:D766–D773. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statisti Soc Series B: Methodol. 57:289–300.
1995.
|
|
22
|
Gene Ontology Consortium: The gene
ontology (GO) project in 2006. Nucleic Acids Res 34 (Database
Issue). D322–D326. 2006. View Article : Google Scholar
|
|
23
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47(D1): D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res 45(D1). D353–D361. 2017.
View Article : Google Scholar
|
|
27
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res 36 (Database Issue). D480–D484.
2008.
|
|
28
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:W316–W322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jensen LJ, Kuhn M, Stark M, Chaffron S,
Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et
al: STRING 8-a global view on proteins and their functional
interactions in 630 organisms. Nucleic Acids Res 37 (Database
Issue). D412–D416. 2009. View Article : Google Scholar
|
|
31
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res 42 (Database
Issue). D78–D85. 2014. View Article : Google Scholar
|
|
35
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 42015.(doi: 10.7554/eLife.05005).
|
|
37
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Boehning D, Patterson RL, Sedaghat L,
Glebova NO, Kurosaki T and Snyder SH: Cytochrome c binds to
inositol (1,4,5) trisphosphate receptors, amplifying
calcium-dependent apoptosis. Nat Cell Biol. 5:1051–1061. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pinton P, Giorgi C, Siviero R, Zecchini E
and Rizzuto R: Calcium and apoptosis: ER-mitochondria Ca2+ transfer
in the control of apoptosis. Oncogene. 27:6407–6418. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim KY, Cho HJ, Yu SN, Kim SH, Yu HS, Park
YM, Mirkheshti N, Kim SY, Song CS, Chatterjee B and Ahn SC:
Interplay of reactive oxygen species, intracellular Ca2+ and
mitochondrial homeostasis in the apoptosis of prostate cancer cells
by deoxypodophyllotoxin. J Cell Biochem. 114:1124–1134. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xue J, Li R, Zhao X, Ma C, Lv X, Liu L and
Liu P: Morusin induces paraptosis-like cell death through
mitochondrial calcium overload and dysfunction in epithelial
ovarian cancer. Chem Biol Interact. 283:59–74. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Raynal NJ, Lee JT, Wang Y, Beaudry A,
Madireddi P, Garriga J, Malouf GG, Dumont S, Dettman EJ, Gharibyan
V, et al: Targeting calcium signaling induces epigenetic
reactivation of tumor suppressor genes in cancer. Cancer Res.
76:1494–1505. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Massari F, Ciccarese C, Santoni M,
Brunelli M, Piva F, Modena A, Bimbatti D, Fantinel E, Santini D,
Cheng L, et al: Metabolic alterations in renal cell carcinoma.
Cancer Treat Rev. 41:767–776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wettersten HI, Aboud OA, Lara PN Jr and
Weiss RH: Metabolic reprogramming in clear cell renal cell
carcinoma. Nat Rev Nephrol. 13:410–419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lucarelli G, Galleggiante V, Rutigliano M,
Sanguedolce F, Cagiano S, Bufo P, Lastilla G, Maiorano E, Ribatti
D, Giglio A, et al: Metabolomic profile of glycolysis and the
pentose phosphate pathway identifies the central role of
glucose-6-phosphate dehydrogenase in clear cell-renal cell
carcinoma. Oncotarget. 6:13371–13386. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
White NM, Newsted DW, Masui O, Romaschin
AD, Siu KW and Yousef GM: Identification and validation of
dysregulated metabolic pathways in metastatic renal cell carcinoma.
Tumour Biol. 35:1833–1846. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Renganathan A and Felley-Bosco E: Long
noncoding RNAs in cancer and therapeutic potential. Adv Exp Med
Biol. 1008:199–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dan J, Wang J, Wang Y, Zhu M, Yang X, Peng
Z, Jiang H and Chen L: LncRNA-MEG3 inhibits proliferation and
metastasis by regulating miRNA-21 in gastric cancer. Biomed
Pharmacother. 99:931–938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Feng SQ, Zhang XY, Fan HT, Sun QJ and
Zhang M: Up-regulation of LncRNA MEG3 inhibits cell migration and
invasion and enhances cisplatin chemosensitivity in bladder cancer
cells. Neoplasma. 65:925–932. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li Z, Yang L, Liu X, Nie Z and Luo J: Long
noncoding RNA MEG3 inhibits proliferation of chronic myeloid
leukemia cells by sponging microRNA21. Biomed Pharmacother.
104:181–192. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Long J and Pi X: lncRNA-MEG3 suppresses
the proliferation and invasion of melanoma by regulating CYLD
expression mediated by sponging miR-499-5p. Biomed Res Int.
2018:20865642018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sun KX, Wu DD, Chen S, Zhao Y and Zong ZH:
LncRNA MEG3 inhibit endometrial carcinoma tumorigenesis and
progression through PI3K pathway. Apoptosis. 22:1543–1552. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang SZ, Cai L and Li B: MEG3 long
non-coding RNA prevents cell growth and metastasis of osteosarcoma.
Bratisl Lek Listy. 118:632–636. 2017.PubMed/NCBI
|
|
57
|
Wei GH and Wang X: lncRNA MEG3 inhibit
proliferation and metastasis of gastric cancer via p53 signaling
pathway. Eur Rev Med Pharmacol Sci. 21:3850–3856. 2017.PubMed/NCBI
|
|
58
|
Pang Y, Mao H, Shen L, Zhao Z, Liu R and
Liu P: MiR-519d represses ovarian cancer cell proliferation and
enhances cisplatin-mediated cytotoxicity in vitro by targeting
XIAP. Onco Targets Ther. 7:587–597. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ye X and Lv H: MicroRNA-519d-3p inhibits
cell proliferation and migration by targeting TROAP in colorectal
cancer. Biomed Pharmacother. 105:879–886. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yue H, Tang B, Zhao Y, Niu Y, Yin P, Yang
W, Zhang Z and Yu P: MIR-519d suppresses the gastric cancer
epithelial-mesenchymal transition via Twist1 and inhibits
Wnt/β-catenin signaling pathway. Am J Transl Res. 9:3654–3664.
2017.PubMed/NCBI
|
|
61
|
Chen Z, Ju H, Yu S, Zhao T, Jing X, Li P,
Jia J, Li N, Tan B and Li Y: Prader-Willi region non-protein coding
RNA 1 suppressed gastric cancer growth as a competing endogenous
RNA of miR-425-5p. Clin Sci (Lond). 132:1003–1019. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hong L, Wang Y, Chen W and Yang S:
MicroRNA-508 suppresses epithelial-mesenchymal transition,
migration, and invasion of ovarian cancer cells through the
MAPK1/ERK signaling pathway. J Cell Biochem. 119:7431–7440. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhou B, Wang D, Sun G, Mei F, Cui Y and Xu
H: Effect of miR-21 on apoptosis in lung cancer cell through
inhibiting the PI3K/Akt/NF-κB signaling pathway in vitro and in
vivo. Cell Physiol Biochem. 46:999–1008. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhao MY, Wang LM, Liu J, Huang X, Liu J
and Zhang YF: MiR-21 suppresses anoikis through targeting PDCD4 and
PTEN in human esophageal adenocarcinoma. Curr Med Sci. 38:245–251.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Naro Y, Ankenbruck N, Thomas M, Tivon Y,
Connelly CM, Gardner L and Deiters A: Small molecule inhibition of
MicroRNA miR-21 rescues chemosensitivity of renal-cell carcinoma to
topotecan. J Med Chem. 11–Jul;2018.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang R and Xia T: Long non-coding RNA
XIST regulates PDCD4 expression by interacting with miR-21-5p and
inhibits osteosarcoma cell growth and metastasis. Int J Oncol.
51:1460–1470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
He H, Dai J, Zhuo R, Zhao J, Wang H, Sun
F, Zhu Y and Xu D: Study on the mechanism behind lncRNA MEG3
affecting clear cell renal cell carcinoma by regulating
miR-7/RASL11B signaling. J Cell Physiol. 233:9503–9515. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang L, Meng L, Wang XW, Ma GY and Chen
JH: Expression of RRM1 and RRM2 as a novel prognostic marker in
advanced non-small cell lung cancer receiving chemotherapy. Tumour
Biol. 35:1899–1906. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang C, Liu Z, Wang L, Qiao B, Du E, Li
L, Xu Y and Zhang Z: Prognostic significance of GPC5 expression in
patients with prostate cancer. Tumour Biol. 37:6413–6418. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yuan S, Yu Z, Liu Q, Zhang M, Xiang Y, Wu
N, Wu L, Hu Z, Xu B, Cai T, et al: GPC5, a novel epigenetically
silenced tumor suppressor, inhibits tumor growth by suppressing
Wnt/β-catenin signaling in lung adenocarcinoma. Oncogene.
35:6120–6131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Han L, Kong R, Yin DD, Zhang EB, Xu TP, De
W and Shu YQ: Low expression of long noncoding RNA GAS6-AS1
predicts a poor prognosis in patients with NSCLC. Med Oncol.
30:6942013. View Article : Google Scholar : PubMed/NCBI
|