Introduction
Neoplasms of the salivary glands constitute 2–10% of
all head and neck tumors (1,2).
Among these, pleomorphic adenoma (PA), also known as ‘mixed tumor’,
is one of the most common types of benign neoplasm of the salivary
gland (3). PA presents as a
slow-growing, painless and multi-nodular tumor that most frequently
occurs in the parotid gland and less often in the submandibular
gland, while it rarely occurs in the sublingual gland. The standard
treatment for PA is extra-capsular excision of the tumor with or
without partial excision of the affected gland. However, partial PA
may recur or transform into carcinoma ex PA following
surgery, which can lead to mortality in these patients (4).
Pleomorphic adenoma gene 1 (PLAG1), located on
chromosome 8q12, is best known for its involvement in the
development of various human tumors, such as lipoblastoma and
hepatoblastoma (5,6). Targeted disruption of PLAG1 may cause
fetal growth restriction, and reduce sperm output and motility
(7,8). In salivary gland tumors, PLAG1 was
initially identified as an oncogene associated with PA (9–11).
More recently, it was demonstrated that this gene is also
associated with carcinoma ex PA (12,13).
In our previous studies, PLAG1 transgenic mice were established,
whose submandibular PA resembles the salivary gland PA of humans
(14–16).
Long non-coding RNAs (lncRNAs) are a class of
regulatory molecules that consist of >200 nucleotides and are
not translated into proteins. A variety of lncRNAs have been
identified in human and mouse diseases using large-scale analyses
of RNA sequence data (17,18). However, the correlation between
lncRNAs and salivary gland diseases has not yet been widely
studied. The lncRNA LINC00473 is a downstream target and an
important mediator of the CRTC1-MAML2 fusion oncoprotein, which
represents a promising biomarker and therapeutic target for human
CRTC1-MAML2-positive mucoepidermoid carcinoma (19). Numerous lncRNA transcripts have
been reported to be dysregulated in the labial salivary glands of
patients with primary Sjögren's syndrome, which may serve important
roles in the pathogenesis of this disease (20). These previous studies have
indicated the crucial role of lncRNAs in salivary gland diseases.
However, the expression of lncRNAs in PA remains unclear.
To the best of our knowledge, the present study is
the first to use an lncRNA microarray to characterize the
expression profiles of lncRNAs and mRNAs in the PLAG1 transgenic
murine model of PA. The results indicated that dysregulated lncRNAs
may serve a vital role in the pathogenesis of PA.
Materials and methods
Animals and sample collection
All animal experiments were approved by the Ethics
Committee of the Faculty of Medicine, Shanghai Jiao Tong University
(Shanghai, China). The PLAG1 transgenic mice were generated as
described in our previous studies (14,15).
Briefly, mouse mammary tumor virus-PLAG1 transgenic mice were
maintained in C57BL/6 background. A total of three male transgenic
mice (age, 12 weeks; weight, 25–30 g) were used for subsequent
experiments. In addition, three male wild type mice (age, 12 weeks;
weight, 25–30 g) were used as control group. All animals ere
purchased from The Shanghai Slake Laboratory Animal Co., Ltd
(Shanghai, China). All mice were housed in cages under standard
conditions at 24°C under a 12-h light/dark cycle with a relative
humidity of 55%. All mice had free access to food and water.
Submandibular tumors (average diameter, 1.5 cm) from 3-month-old
transgenic mice and normal submandibular glands from wild-type mice
were collected. Three pairs of mouse tumors and control glands were
divided into two groups, the tumor group, including mouse tumor 1
(MT1), MT2 and MT3 and the control group (Con1, Con2 and Con3).
These samples were analyzed independently for the expression of
lncRNAs and mRNAs. Another six pairs of mouse tumors and control
glands were used for subsequent polymerase chain reaction (PCR)
detection.
RNA extraction
Total RNA was extracted from the mouse tumors and
control glands using TRIzol® reagent (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). RNA quantity and quality were
measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher
Scientific, Inc.). RNA integrity was assessed by standard
denaturing agarose gel electrophoresis.
Microarray analysis
Arraystar Mouse LncRNA Microarray V3.0 (Arraystar,
Inc., Rockville, MD, USA) is designed for the global profiling of
mouse lncRNAs and protein-coding transcripts. Following RNA
labeling and array hybridization, the microarray was scanned using
the Agilent Microarray Scanner (Agilent p/n G2565BA; Agilent
Technologies, Inc., Santa Clara, CA, USA). The Agilent Feature
Extraction Software was used to extract data and for subsequent
data processing. The lncRNA exhibiting a fold-change ≥2 and
P<0.05 compared with the control were considered as
differentially expressed.
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis
GO analysis was conducted to identify the functions
of differentially expressed genes, including biological processes,
molecular functions and cellular components. Furthermore, the
pathways associated with the differentially expressed mRNAs were
assessed using KEGG analysis. Significant GO terms and pathways
were selected on the basis of P<0.05 and false discovery rate of
<0.05. The unsupervised hierarchical clustering of the top 20
dysregulated lncRNAs and mRNAs was also performed.
Reverse transcription-quantitative PCR
(RT-qPCR) validation
Subsequent to extraction, total RNA was reverse
transcribed into cDNA using the PrimeScript RT Master Mix (Takara
Biotechnology Co., Ltd., Dalian, China), according to the
manufacturer's protocol. A total of four upregulated and two
downregulated lncRNAs were selected for PCR validation on the basis
of fold change (FC). The qPCR experiment was performed using SYBR
Premix Ex Taq II (Takara Biotechnology Co., Ltd.) on the 7500
Sequence Detection system (ABI; Thermo Fisher Scientific, Inc.).
The following thermocycling conditions were used: Initial
denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for
10 sec and 60°C for 60 sec. GAPDH (mouse) was used as an endogenous
control, and relative expression levels of each lncRNA was
calculated using the 2−ΔΔCq method. The primer sequences
of investigated lncRNAs are listed in Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer name | Sequence | Annealing temperature
(°C) | Product length
(bp) |
|---|
| GAPDH (mouse) | F:
5′-CACTGAGCAAGAGAGGCCCTAT-3′ | 60 | 144 |
|
| R:
5′-GCAGCGAACTTTATTGATGGTATT-3′ |
|
|
|
ENSMUST00000140495 | F:
5′-TCGTGGTGTTTTATCAAGCAAG-3′ | 60 | 203 |
|
| R:
5′-ATAGGCTCCGCACTGTTGT-3′ |
|
|
|
ENSMUST00000125649 | F:
5′-CGATGACAAGCTGAGGGACA-3′ | 60 | 118 |
|
| R:
5′-GCCTTCTTGCTGATCATATTTCTG-3′ |
|
|
| AK006017 | F:
5′-TGACACCGGGGTCTGAGTAT-3′ | 60 | 210 |
|
| R:
5′-CCGTAGGTCTGAAATATTCTGC-3′ |
|
|
| AK146794 | F:
5′-TGGATGGTAACAAAAACATGTG-3′ | 60 | 65 |
|
| R:
5′-TGAGAGAATGAGGCAAGAGAAG-3′ |
|
|
|
ENSMUST00000120145 | F:
5′-AGAAGCTAGTTCTATAGGAAGCAG-3′ | 60 | 137 |
|
| R:
5′-CATAGATGTGCTCCAGAAAAAG-3′ |
|
|
| AK089826 | F:
5′-GTGGTACAGTATCGCTAAAGGAC-3′ | 60 | 142 |
|
| R:
5′-GGGAAAACATAGTGATAGGAGC-3′ |
|
|
Construction of the lncRNA and mRNA
co-expression network
The aforementioned validated lncRNAs were used to
construct a co-expression network to explore specific mRNAs
involved in PA tumorigenesis. The Pearson correlation coefficient
(PCC) was calculated for correlations between the validated lncRNAs
and all the differentially expressed mRNAs. The lncRNAs and mRNAs
with an absolute value of Pearson correlation coefficients
>0.995 were selected to draw the gene network using the program
Cytoscape (version 3.0.1; http://cytoscape.org/).
Statistical analysis
Statistical analyses were performed using SPSS
software (version 19.0; IBM Corp., Armonk, NY, USA). For normally
distributed data, the differences between two groups were
determined using a two-tailed Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Overview of lncRNA and mRNA expression
profiles
To identify the differentially expressed lncRNAs
associated with PA tumorigenesis, the present study examined the
expression pattern of lncRNAs in three submandibular tumors from
PLAG1 transgenic mice and three normal submandibular samples from
control wild-type mice. The expression profiles of the lncRNAs and
mRNAs are listed in Table II. The
present data demonstrated that 25.36% of the lncRNAs (9,110/35,923)
and 31.15% of the mRNAs (7,750/24,881) were significantly
differentially expressed in the tumor group as compared with those
in the control group (FC>2, P<0.05). The unsupervised
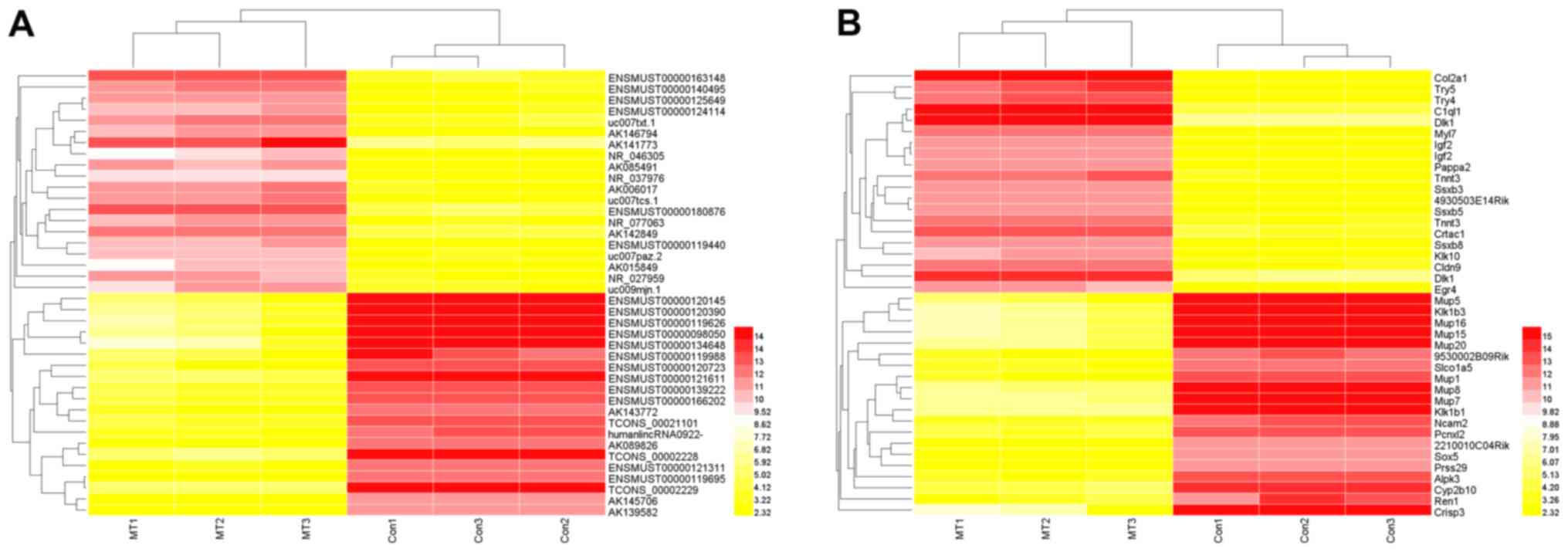
hierarchical clustering of the top 20 upregulated and downregulated
lncRNAs and mRNAs indicated that the expression patterns of the
three samples in each group were similar, as indicated by the heat
map (Fig. 1).
 | Table II.Summary of the results of lncRNA and
mRNA profile analyses. |
Table II.
Summary of the results of lncRNA and
mRNA profile analyses.
| Probe class | Total, n | Differentially
expressed, n (%) | Upregulated, n
(%) | Downregulated, n
(%) |
|---|
| lncRNAs | 35,923 | 9,110 (25.36) | 4,521 (12.59) | 4,589 (12.77) |
| mRNAs | 24,881 | 7,750 (31.15) | 4,201 (16.88) | 3,549 (14.26) |
| Combined | 60,804 | 16,860 (27.73) | 8,722 (14.34) | 8,138 (13.38) |
Differentially expressed lncRNAs in
PLAG1 transgenic mice and wild-type mice
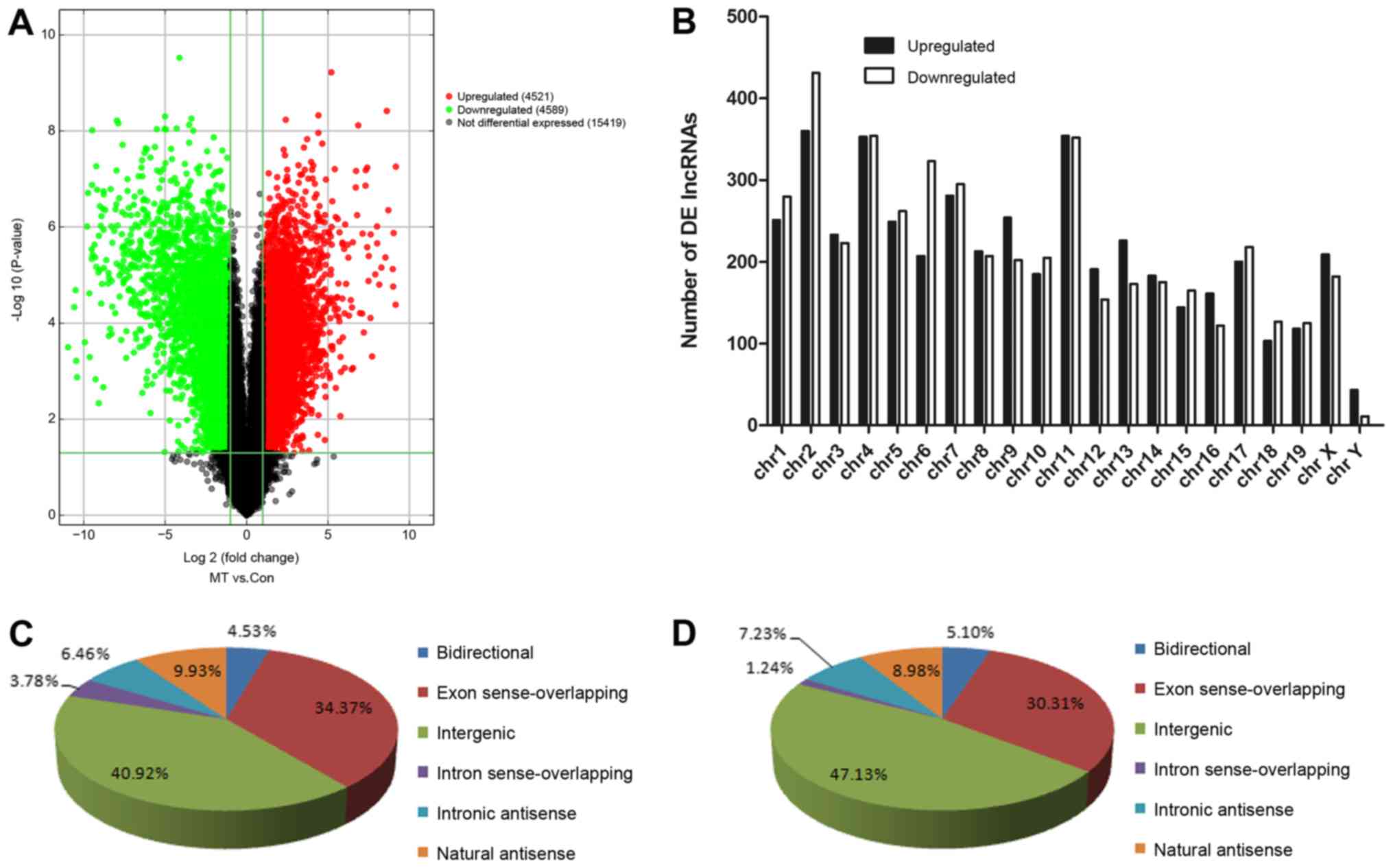
Volcano plot filtering was used to identify the
differences in the expression of lncRNAs between the PLAG1
transgenic mice and control mice (Fig.
2A). The plot demonstrated that a total of 9,110 lncRNAs (4,521
upregulated and 4,589 downregulated) were significantly
differentially expressed. The chromosome distribution of the
differentially expressed lncRNAs was then determined. As
illustrated in Fig. 2B, chromosome
2 exhibited the highest number of differentially expressed lncRNAs,
while chromosome Y exhibited the lowest number. On the basis of
different transcriptional forms, the differentially expressed
lncRNAs were classified into six different subgroups, including
bidirectional, exon sense-overlapping, intergenic, intron
sense-overlapping, intronic antisense and natural antisense
lncRNAs. The majority of the differentially expressed lncRNAs were
classified as intergenic and exon sense-overlapping lnRNAs, both in
the upregulated and downregulated groups (Fig. 2C and D).
GO and KEGG pathway analyses of
differentially expressed mRNAs
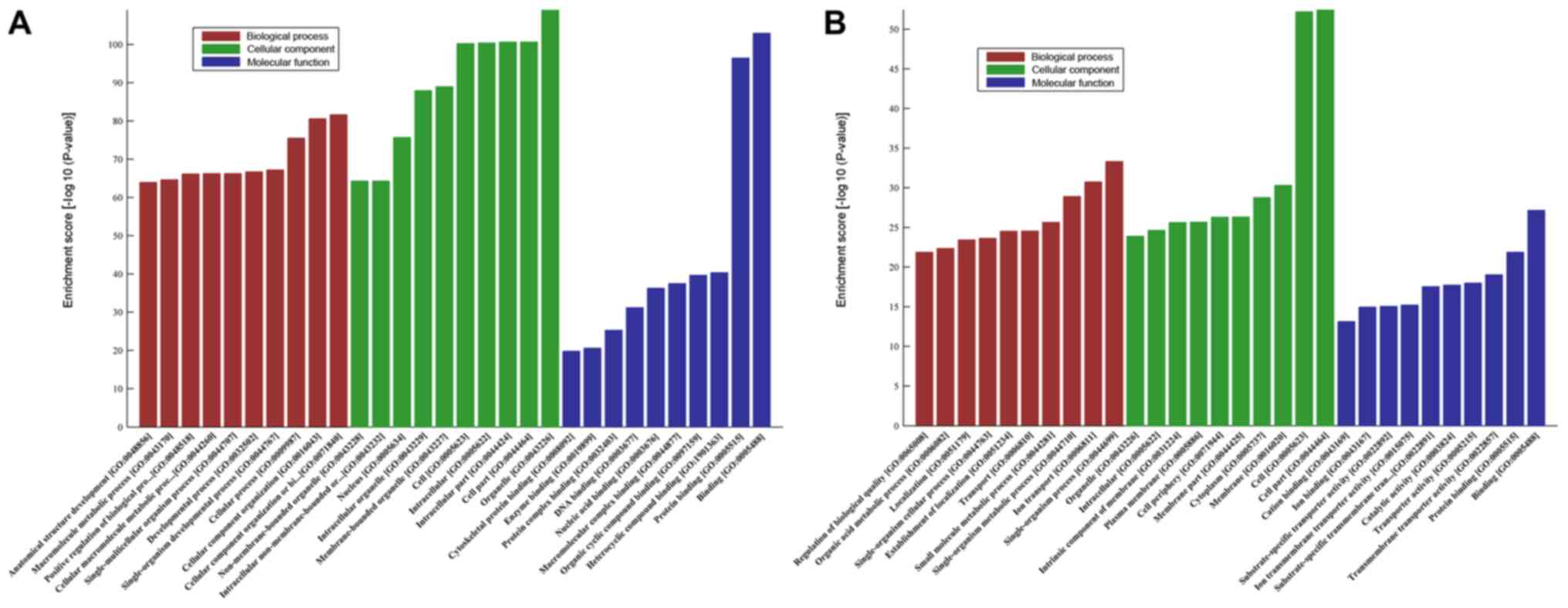
GO analysis was conducted to determine the
enrichment of the differentially expressed mRNAs in the biological
process, cellular component and molecular function categories. The
upregulated mRNAs were significantly enriched in a number of GO
terms, with the two most prominent molecular function terms being
‘binding’ and ‘protein binding’ (GO:0005488 and GO:0005515;
Fig. 3A), the most significant
biological process term was ‘anatomical structure development’
(GO:0048856; Fig. 3A) and the most
enriched cellular component was ‘non-membrane-bounded organelle’
(GO:0043228; Fig. 3A). The
downregulated mRNAs were also significantly enriched in a number of
GO terms, including ‘single-organism process’ (GO:0044699) in
biological processes and ‘cell part’ (GO:0044464) in cellular
components (Fig. 3B). In addition,
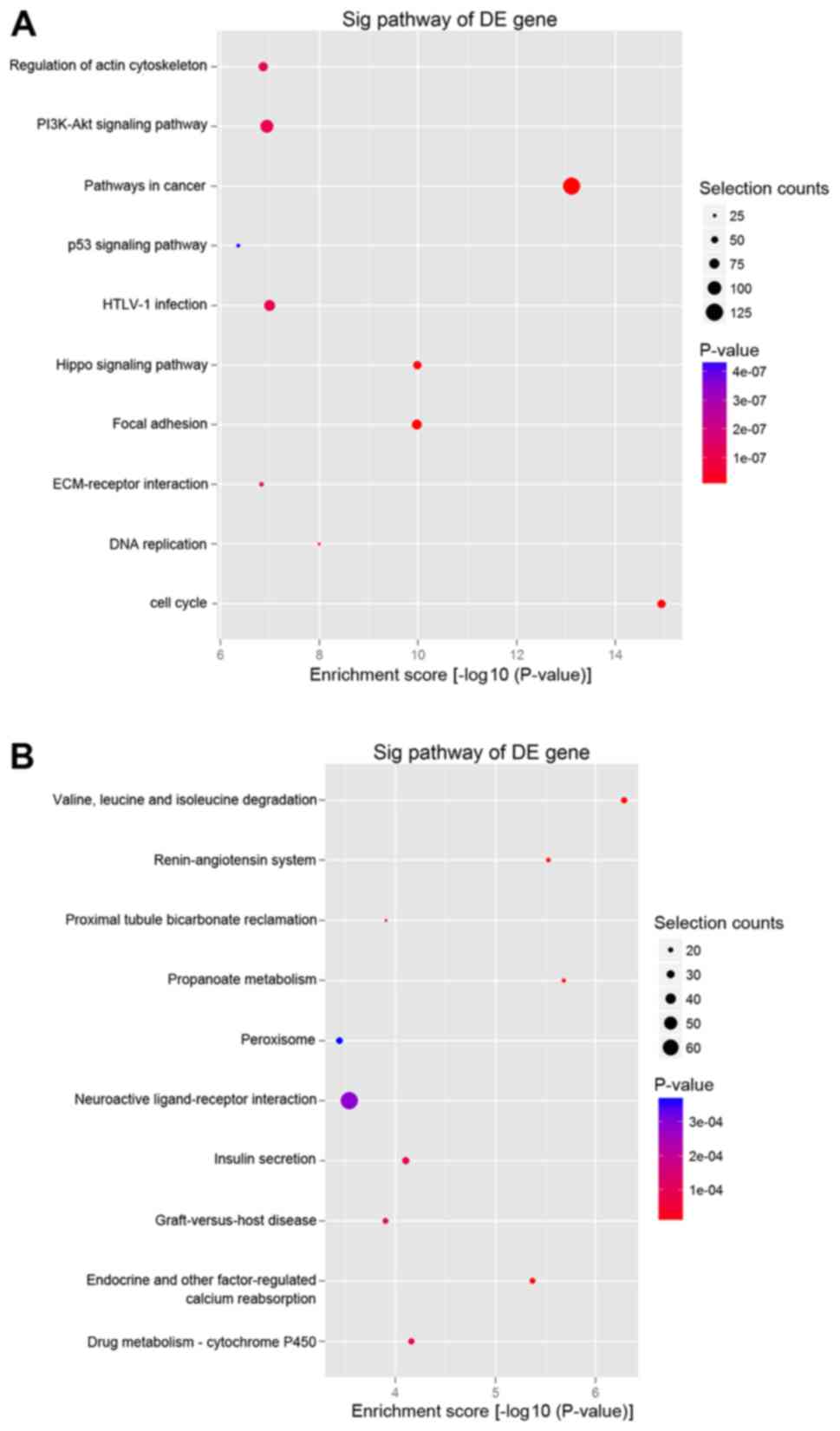
KEGG pathway analysis of the upregulated mRNAs indicated that the
two most enriched pathways were ‘pathways in cancer’ and ‘PI3K-Akt
signaling pathway’ (Fig. 4A),
while the downregulated mRNAs were significantly involved in
‘valine, leucine and isoleucine degradation’ (Fig. 4B).
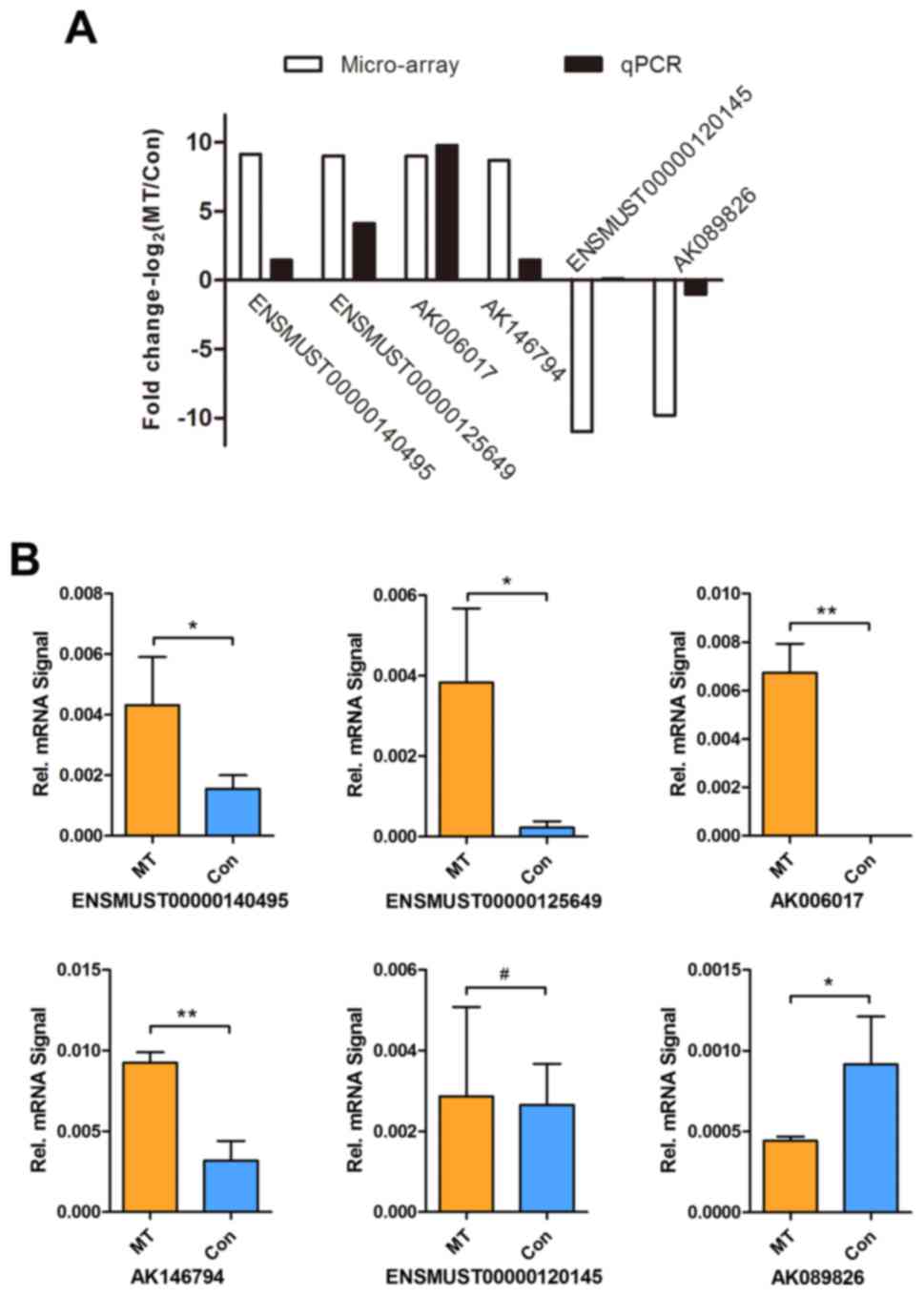
RT-qPCR validation
To validate the microarray data, RT-qPCR was
conducted to detect the expression levels of six most significantly
dysregulated lncRNAs (four upregulated and two downregulated) that
were selected from the differentially expressed lncRNAs. These
selected lncRNAs included ENSMUST00000140495, ENSMUST00000125649,
AK006017, AK146794 ENSMUST00000120145 and AK089826. The PCR data
were consistent with the microarray analysis results, with a
concordance rate of 83.33% (5/6), supporting the reliability of the
microarray data (Fig. 5A).
Furthermore, as illustrated in Fig.
5B, RT-qPCR revealed that the expression levels of
ENSMUST00000140495, ENSMUST00000125649, AK006017 and AK146794 were
significantly increased in the tumor group compared with those in
the control group, while AK089826 expression was significantly
decreased. By contrast, there was no significant difference in the
expression of ENSMUST00000120145 between the tumor and control
groups (P>0.05; Fig. 5B).
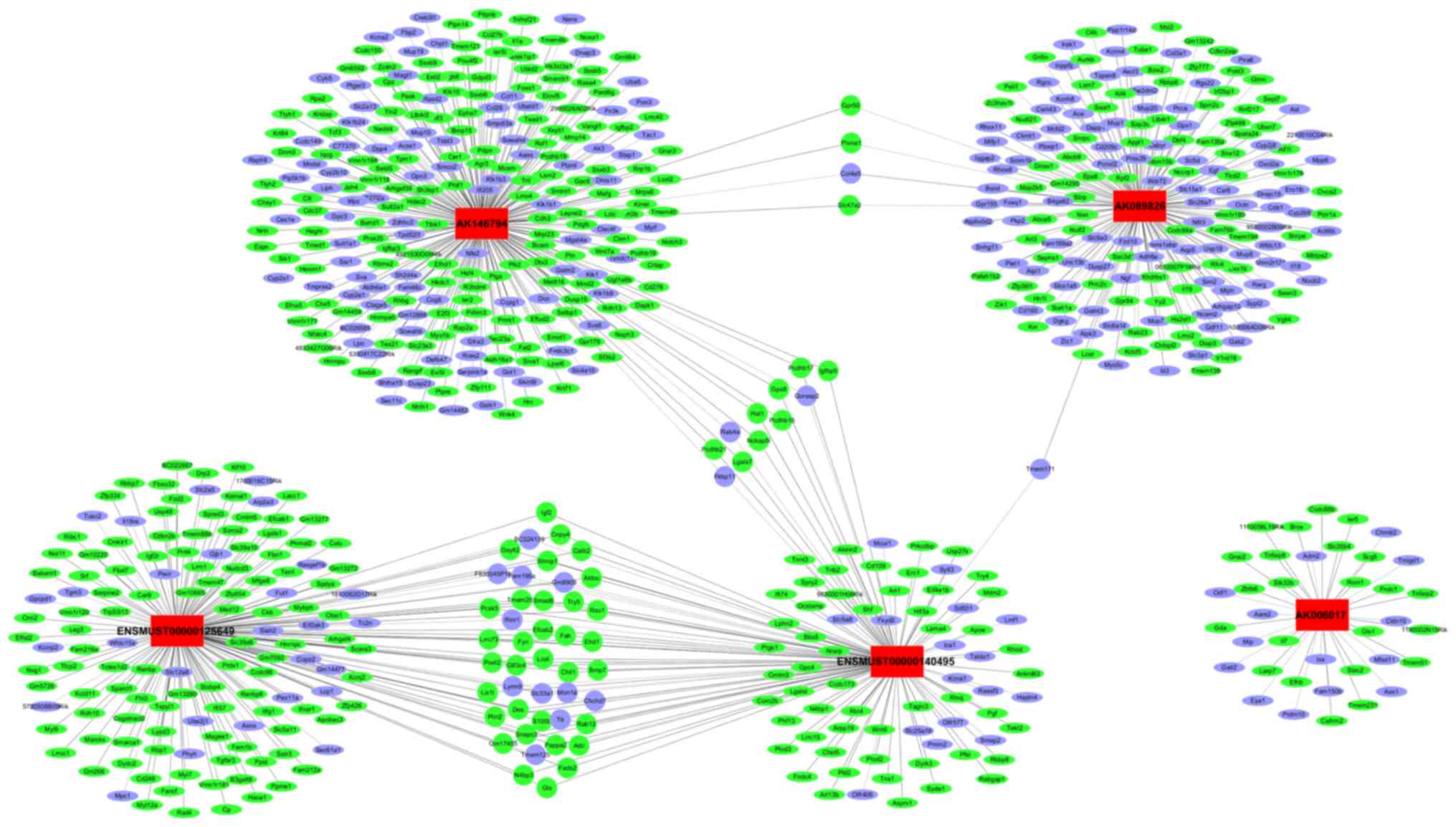
Establishment of the lncRNA and mRNA
co-expression network
Next, the study attempted to predict the target
genes of lncRNAs and to investigate the potential interactions
between the lncRNAs and mRNAs in PA in PLAG1 transgenic mice. The
associations between the five validated lncRNAs and corresponding
mRNAs were thus assessed by calculating the PCC values. The
co-expression network was constructed and visualized using
Cytoscape software based on the PCC value (PCC>0.99). The
network illustrated a number of interactions between the five
lncRNAs and a total of 725 mRNAs. The network also indicated that a
single lncRNA may regulate the mRNA expression of multiple genes,
and certain lncRNAs may co-regulate the expression of the same
gene. For instance, AK146794 was able to regulate the expression of
264 mRNAs, while IGF2 was simultaneously regulated by
ENSMUST00000140495 and ENSMUST00000125649 (Fig. 6).
Discussion
PA is the most common type of salivary gland tumor
with a complex pathogenesis. PLAG1 transgenic mice have been
utilized as a useful animal model to study PA. Although a number of
genes (including PLAG1, HMGA2 and IGF2) have been proven to be
involved in the pathogenesis of PA (7,21,22),
it remains unclear how those genes are specifically regulated. With
the rapid development of genomic technology, numerous non-coding
RNAs (including microRNAs and lncRNAs) have been identified, and
their roles in regulating mRNAs are currently researched.
In the present study, the lncRNA expression patterns
in submandibular PA tissues of PLAG1 transgenic mice and normal
tissues of wild-type C57BL/6 mice were compared using microarray
analysis. The data demonstrated that a total of 9,110 lncRNAs were
differentially expressed in the submandibular PA tissues obtained
from PLAG1 transgenic mice when compared with their counterparts
from the wild-type mice, which may facilitate the understanding of
the PA-associated global transcriptome. In addition, it was
observed that the differentially expressed lncRNAs and mRNAs in PA
were distributed unequally among all chromosomes, including the X
chromosome. Notably, all chromosomes were implicated in PA,
providing additional evidence that the pathogenesis of PA is
relatively complex. Among the differentially expressed lncRNAs,
intergenic lncRNAs comprised the largest category, with 1,850
upregulated and 2,163 downregulated members. Intergenic lncRNAs are
conserved across multiple species and may function via various
mechanisms, including transcriptional regulation, splicing
regulation and post-transcriptional regulation (23). Furthermore, the majority of the
validated lncRNAs in the present study belong to the category of
intergenic lncRNAs, including ENSMUST00000140495, AK006017,
AK146794 ENSMUST00000120145 and AK089826, which is consistent with
the universality of intergenic lncRNAs.
To further understand the biological functions and
molecular mechanisms of PA-associated mRNAs, GO and KEGG pathway
analyses were also performed to identify the biological functions
and signaling networks enriched among the differentially expressed
mRNAs in the current study. Notably, these mRNAs were involved in
multiple pathways associated with PA, including ‘PI3K-Akt signaling
pathway’ and ‘p53 signaling pathway’. The PI3K-Akt signaling
pathway, a key oncogenic pathway, has been reported to be
overactivated in numerous tumors (24,25).
The microarray results of the present study indicated that the
expression of a number of key molecules of the PI3K-Akt pathway,
including IRS1, PI3K, Ras and Akt, was increased. With regard to
the p53 signaling pathway, it was observed that p21, Bax, Fas and
CyclinD were also activated. However, with respect to intrinsic
complex regulatory mechanisms, the specific role of these pathways
in PA tumorigenesis warrants further investigation.
Unlike microRNAs, there are no reliable softwares
that may be used to predict the target genes of lncRNAs and their
functions via their sequence information or secondary structure.
Thus, the lncRNA-mRNA co-expression network has been developed to
predict the potential roles of lncRNAs. In the present study, it
was elucidated that the five validated lncRNAs may contribute to
the development and progression of PA by altering the expression of
target genes. Specifically, the lncRNA ENSMUST00000125649 was
observed to be correlated with the IGF2, IGF2R and IGFBP5 mRNAs. In
previous studies, it has been demonstrated that PLAG1
overexpression may be responsible for the frequent upregulation of
IGF2 (5,21,22,26).
Notably, thousands of dysregulated lncRNAs were
identified in the current study, which is possibly due to the
influence of PLAG1 overexpression in the transgenic mice. Future
studies, particularly functional characterization of these lncRNAs,
are thus required. Previous studies have demonstrated that lncRNAs
have a wide range of biological functions mediated via various
mechanisms, which include cis-targeting, trans-targeting and
allosteric modification, and function as enhancers, scaffolds, and
co-activators or co-repressors (23,27).
A literature search conducted in the current study provided no
research results on the aforementioned candidate lncRNAs, which
suggests that further research is needed. Additionally, how these
functional lncRNAs interact with the associated mRNAs in PA is
another issue that merits further study, such as gain- and
loss-of-function experiments in vitro, and tumor formation
assays in vivo.
In conclusion, the present results demonstrated that
a set of lncRNAs were differentially expressed in PA tissues
obtained from PLAG1 transgenic mice. These lncRNAs may offer novel
insights into the pathogenesis of PA and act as novel therapeutic
targets for this tumor.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81302359) and the Research
Grants from the Science and Technology Commission of Shanghai
Municipality (grant no. 16ZR1418800).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SS and WY conceived and designed the experiments. WX
conducted RNA sequencing and analysis. LL and HL performed the
experiments. WX and CZ wrote the manuscript. LL, JF and CZ
conducted data analysis.
Ethics approval and consent to
participate
All animal experiments were approved by The Ethics
Committee of the Faculty of Medicine, Shanghai Jiao Tong
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing of
interests.
References
|
1
|
Li LJ, Li Y, Wen YM, Liu H and Zhao HW:
Clinical analysis of salivary gland tumor cases in West China in
past 50 years. Oral Oncol. 44:187–192. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lima SS, Soares AF, de Amorim RF and
Freitas Rde A: Epidemiologic profile of salivary gland neoplasms:
Analysis of 245 cases. Braz J Otorhinolaryngol. 71:335–340.
2005.(In Portuguese). View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tian Z, Li L, Wang L, Hu Y and Li J:
Salivary gland neoplasms in oral and maxillofacial regions: A
23-year retrospective study of 6982 cases in an eastern Chinese
population. Int J Oral Maxillofac Surg. 39:235–242. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hu YH, Zhang CY, Xia RH, Tian Z, Wang LZ
and Li J: Prognostic factors of carcinoma ex pleomorphic adenoma of
the salivary glands, with emphasis on the widely invasive
carcinoma: A clinicopathologic analysis of 361 cases in a Chinese
population. Oral surg Oral Med Oral Pathol Oral Radiol.
122:598–608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zatkova A, Rouillard JM, Hartmann W, Lamb
BJ, Kuick R, Eckart M, von Schweinitz D, Koch A, Fonatsch C,
Pietsch T, et al: Amplification and overexpression of the IGF2
regulator PLAG1 in hepatoblastoma. Genes Chromosomes Cancer.
39:126–137. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Astrom A, D'Amore ES, Sainati L, Panarello
C, Morerio C, Mark J and Stenman G: Evidence of involvement of the
PLAG1 gene in lipoblastomas. Int J Oncol. 16:1107–1110.
2000.PubMed/NCBI
|
|
7
|
Abi Habib W, Brioude F, Edouard T, Bennett
JT, Lienhardt-Roussie A, Tixier F, Salem J, Yuen T, Azzi S, Le Bouc
Y, et al: Genetic disruption of the oncogenic HMGA2-PLAG1-IGF2
pathway causes fetal growth restriction. Genet Med. 20:250–258.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Juma AR, Grommen SVH, O'Bryan MK, O'Connor
AE, Merriner DJ, Hall NE, Doyle SR, Damdimopoulou PE, Barriga D,
Hart AH, et al: PLAG1 deficiency impairs spermatogenesis and sperm
motility in mice. Sci Rep. 7:53172017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Matsuyama A, Hisaoka M, Nagao Y and
Hashimoto H: Aberrant PLAG1 expression in pleomorphic adenomas of
the salivary gland: A molecular genetic and immunohistochemical
study. Virchows Arch. 458:583–592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Debiec-Rychter M, Van Valckenborgh I, Van
den Broeck C, Hagemeijer A, Van de Ven WJ, Kas K, Van Damme B and
Voz ML: Histologic localization of PLAG1 (pleomorphic adenoma gene
1) in pleomorphic adenoma of the salivary gland: Cytogenetic
evidence of common origin of phenotypically diverse cells. Lab
Invest. 81:1289–1297. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Avadhani V, Cohen C and Siddiqui MT:
PLAG1: An immunohistochemical marker with limited utility in
separating pleomorphic adenoma from other basaloid salivary gland
tumors. Acta Cytol. 60:240–245. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Katabi N, Xu B, Jungbluth AA, Zhang L,
Shao SY, Lane J, Ghossein R and Antonescu CR: PLAG1
immunohistochemistry is a sensitive marker for pleomorphic adenoma:
A comparative study with PLAG1 genetic abnormalities.
Histopathology. 72:285–293. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Brito BS, Giovanelli N, Egal ES,
Sánchez-Romero C, Nascimento JS, Martins AS, Tincani ÁJ, Del Negro
A, Gondak RO, Almeida OP, et al: Loss of expression of Plag1 in
malignant transformation from pleomorphic adenoma to carcinoma ex
pleomorphic adenoma. Hum Pathol. 57:152–159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao X, Ren W, Yang W, Wang Y, Kong H,
Wang L, Yan L, Xu G, Fei J, Fu J, et al: Wnt pathway is involved in
pleomorphic adenomas induced by overexpression of PLAG1 in
transgenic mice. Int J Cancer. 118:643–648. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen S, Yang W, Wang Z, Lei X, Xu L, Wang
Y, Wang L, Huang L, Yu Z, Zhang X, et al: Tumor-initiating cells
are enriched in CD44(hi) population in murine salivary gland tumor.
PLoS One. 6:e232822011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Shang W, Lei X, Shen S, Zhang H,
Wang Z, Huang L, Yu Z, Ong H, Yin X, et al: Opposing functions of
PLAG1 in pleomorphic adenoma: A microarray analysis of PLAG1
transgenic mice. Biotechnol Lett. 35:1377–1385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang W, Wei C, Li P, Wang L, Li W, Chen K,
Zhang J, Zhang W and Jiang G: Integrative analysis of mRNA and
lncRNA profiles identified pathogenetic lncRNAs in esophageal
squamous cell carcinoma. Gene. 661:169–175. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Jin L, Dong A, Zhou X and Yuan H:
Microarray expression profile analysis of long non-coding RNAs in
optineurin E50K mutant transgenic mice. Mol Med Rep. 16:1255–1261.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Z, Lin S, Li JL, Ni W, Guo R, Lu J,
Kaye FJ and Wu L: CRTC1-MAML2 fusion-induced lncRNA LINC00473
expression maintains the growth and survival of human
mucoepidermoid carcinoma cells. Oncogene. 37:1885–1895. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi H, Cao N, Pu Y, Xie L, Zheng L and Yu
C: Long non-coding RNA expression profile in minor salivary gland
of primary Sjögren's syndrome. Arthritis Res Ther. 18:1092016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Declercq J, Van Dyck F, Van Damme B and
Van de Ven WJ: Upregulation of Igf and Wnt signalling associated
genes in pleomorphic adenomas of the salivary glands in PLAG1
transgenic mice. Int J Oncol. 32:1041–1047. 2008.PubMed/NCBI
|
|
22
|
Voz ML, Agten NS, Van de Ven WJ and Kas K:
PLAG1, the main translocation target in pleomorphic adenoma of the
salivary glands, is a positive regulator of IGF-II. Cancer Res.
60:106–113. 2000.PubMed/NCBI
|
|
23
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fortin J and Mak TW: Targeting PI3K
signaling in cancer: A cautionary tale of two AKTs. Cancer Cell.
29:429–431. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Zhang X, Wang G, Wang L, Lin Y and
Sun F: Hsa-miR-513b-5p suppresses cell proliferation and promotes
P53 expression by targeting IRF2 in testicular embryonal carcinoma
cells. Gene. 626:344–353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Akhtar M, Holmgren C, Göndör A, Vesterlund
M, Kanduri C, Larsson C and Ekström TJ: Cell type and
context-specific function of PLAG1 for IGF2 P3 promoter activity.
Int J Oncol. 41:1959–1966. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|