Introduction
Primary liver cancer, predominantly comprising
hepatocellular carcinoma (HCC), is reported to be the fifth most
common cancer globally and the third most common cause of
cancer-associated mortality (1).
There is an increasing understanding of the molecular mechanisms
that induce hepatocarcinogenesis, including chronic infections such
as hepatitis B or C (HBV/HCV), alcohol abuse and metabolic
syndromes (2). Surgical resection
followed by adjuvant drug therapy is the most common treatment in
clinical settings (3); however,
the metastasis and chemoresistance of tumor cells results in poor
outcomes for patients with advanced HCC (4). As the development of HCC is
accompanied with unique and continuous genomic and epigenetic
alterations, combining personalized approaches, including molecular
analysis-guided targeted therapy and immunotherapy may be a
potential strategy to improve the treatment of cancer (5,6).
Existing preclinical models comprise genetically engineered mouse
models and human tumor-derived cell lines; however, a reproducible
human model is required to accurately reproduce the important
characteristics of tumors in vivo and determine the
effectiveness of candidate therapeutics. Commercial tumor cell
lines have been extensively used in the investigation of
therapeutic targets; however, the establishment of in vitro
models that use tumor cells from individual patients may serve to
improve the clinical relevance of in vitro studies (3).
Tumor cells have been associated with strong
proliferative ability. This property is detrimental for the rapid
expansion of cells derived from adult tumor tissues while retaining
stable lineage commitment, particularly from liver tumors (7). Conditional reprogramming (CR) systems
have previously been used to establish patient-derived cell lines
from normal and tumor tissues that possess the ability to grow
indefinitely in vitro without genetic manipulation (8,9).
Potential applications for the CR system in clinical settings have
been investigated for breast (10,11),
lung (12) and prostate cancers
(13,14); however, it has been hypothesized
that the CR system cannot be used to expand patient-derived
metastatic lung cancer cells (15). In an in vitro study of
cultured liver cancer cells, Broutier et al (16) successfully constructed a primary
HCC organoid based on the CR system using a three-dimensional (3D)
culture method. On the contrary, whether CR may serve as a reliable
in vitro culture method to obtain matched tumor cells from
patients with HCC remains unclear.
The aim of the present study was to establish a
culture system with potential clinical applications that enabled
the amplification of genetically stable cells. Primary tumor cells
were isolated from tissue specimens from 20 patients with HCC and
were cultured using the CR system. The proliferative potential and
capacity of cells to undergo continuous regeneration, and the
expression of tumor-specific markers were evaluated to determine
the prospects for use in clinical settings. The study provided a
primary investigation into culture systems for HCC cells in
vitro, in preparation for future studies involving the
establishment of a conditionally reprogrammed culture model for
drug screening in the treatment of liver cancer drug screening.
Materials and methods
Cell isolation
A total of 20 samples of liver tumor tissue were
obtained from patients (10 males and 10 females, aged 38–67 years
old) undergoing orthotopic liver transplantation (OLT) or
hepatectomy at Tianjin First Center Hospital (Tianjin, China)
between January 2015 and December 2017. Tumors were graded using
the American Joint Commission on Cancer 8th edition staging system
for patients with HCC. The study was approved by the Tianjin First
Central Hospital Clinical Research Ethics Committee (review no.
2016N057KY). Informed written consent was obtained from all
patients. Procedures were conducted in accordance with the
Declaration of Helsinki (17).
Surgically resected liver tumor tissue was obtained from
individuals with HCC who had no history of viral-mediated
hepatitis. Half of the tissue was de-identified and supplied to lab
personnel for tissue culture; remaining tissue was used for
histological analysis. A 1 cm3 section of tumor tissue
was collected and transferred into a sterile tube containing 1×
cell protective fluid (Beijing Percans Oncology Research Co., Ltd.,
Beijing, China). The tissues were qualitatively divided into two
types: Hard tissues (brittle and non-viscous tissues) and soft
tissues (tissues with loose texture consisting of lumpy masses).
Following five washes with PBS, the samples were minced into ~2
mm3 sections and treated using a gentleMACS™ Tissue
Dissociator (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). The
samples were digested into single cells by 1×
collagenase/hyaluronidase solution (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) followed by 0.25% trypsin-EDTA (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) at 37°C for 2 h. Cell suspensions
were filtered using a MACS® SmartStrainer (Miltenyi
Biotec GmbH). The primary cells were suspended in Dulbecco's
modified Eagle's medium/Nutrient Mixture F-12 medium (DMEM/F12;
Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and then
centrifuged at 300 × g for 5 min at 37°C. Primary HCC cells were
further purified from stromal cells using a magnetic bead isolate
system (Beijing Percans Oncology Research Co., Ltd.) according to
the manufacturer's protocols (18).
H&E staining
The tumor tissues were fixed with 4%
paraformaldehyde overnight at 4°C and embedded in paraffin. The
samples in paraffin were cut into 3-µm sections, dewaxed in xylene
and rehydrated in a graded alcohol series. Sections were stained
with hematoxylin for 10 min at room temperature, and rinsed with
tap water for 10 min. Differentiation was performed with
differentiation buffer in the H&E staining kit (Beyotime
Institute of Technology, Shanghai, China) for 30 sec at room
temperature to the obtain clear structure. Following further
washing in running tap water for 10 min, the sections were stained
with eosin for 2 min at room temperature, then dehydrated and
mounted. Then, sections were imaged using an optical microscope
(magnification, ×40; Eclipse 80i; Nikon Corporation, Tokyo, Japan);
three images/sample were acquired for analysis.
CR culture
In the CR system, mouse embryonic fibroblast cells
(NIH-3T3; China Center for Type Culture Collection, Wuhan, China)
were irradiated at 30 Gy with gamma radiation to provide an
irradiated fibroblast feeder layer. HCC cells were passaged in
DMEM/F12 containing the following: 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.); 10 µg/ml Y-27632 (Enzo Life
Sciences, Inc., Farmingdale, NY, USA); 0.4 µg/ml hydrocortisone
(Sigma-Aldrich; Merck KGaA); 5 µg/ml insulin (Thermo Fisher
Scientific, Inc.); 8.6 ng/ml cholera toxin (Sigma-Aldrich; Merck
KGaA); 20 ng/ml epidermal growth factor (Sigma-Aldrich; Merck
KGaA), 20 ng/ml hepatocyte growth factor (Sigma-Aldrich; Merck
KGaA), 24 µg/ml adenine (Sigma-Aldrich; Merck KGaA) and 3 nM
dexamethasone (Sigma-Aldrich; Merck KGaA). The feeder cells were
removed following incubation for 1 min in 0.05% Trypsin-EDTA
(Sigma-Aldrich; Merck KGaA) at 37°C and replaced with fresh cells
(4×104 cells/cm2) during the passage of
HCC-CR cells. A 6-day culture period was required to generate
larger cell clones prior to the initial passage of P0 cells, then
the cells were passaged every 3–4 days. The HCC-CR cells were
sub-cultured at a 1:3 ratio upon reaching 60–80% confluence, and
these cells were cultured for a total of 6 generations in a 5%
CO2 incubator at 37°C. Subsequent passages of the HCC-CR
cells were referred to in a sequential numerical order starting at
P1. HCC-CR cells were observed using an optical microscope
(magnification, ×10; Olympus Corporation, Tokyo, Japan) following
culture for 50 and 100 h.
Cell colony formation and viability
assay
Following culturing under CR conditions for six
passages, the cell colony formation per passage was observed using
an inverted phase-contrast microscope (magnification, ×10; Olympus
Corporation). The HCC-CR cells were stained using a Live/Dead
Molecular Probes staining kit (Invitrogen; Thermo Fisher
Scientific, Inc.) for 30 min at 37°C and then analyzed using an
inverted fluorescence microscope (magnification, ×10; Olympus
Corporation). Three images/sample were acquired for analysis.
Cell proliferation assay
The proliferative ability of the P0 HCC-CR cells
following culturing for 10 days was quantified via
5-ethynyl-2′-deoxyuridine (EdU)/Hoechst 33342 staining using a
Cell-Light™ EdU in vitro imaging kit (Guangzhou RiboBio Co.,
Ltd., Guangzhou, China). Briefly, HCC-CR cells (4×104
cells/cm2) were seeded into a 24-well plate and
incubated with 50 mM EdU labeling solution (200 ml) at 37°C under
5% CO2 for 3 h. The HCC-CR cells were then sequentially
treated with 4% paraformaldehyde (PFA; pH 7.4) for 30 min, 2 mg/ml
glycine for 5 min, 0.5% Triton X-100 for 10 min, anti-EdU working
solution for 30 min and 5 mg/ml Hoechst 33342 dye for 30 min (all
at room temperature). The cells were imaged under a fluorescence
microscope (magnification, ×10; Leica Microsystems GmbH, Wetzlar,
Germany). Three images/sample were acquired for analysis. The
numbers of HCC-CR cells were counted for each passage, and a plot
of accumulated population doublings versus growth days was
constructed following culturing for 10, 14, 22 and 30 days as
previously described (19).
Western blotting
HCC-CR cells were separated from feeder cells by
differential trypsinization. Briefly, the cells were washed by PBS,
and then incubated by 0.05% trypsinization for 1 min at 37°C under
5% CO2. The feeder cells were separated by tapping the
bottom of the plates. Then, total protein was extracted from HCC-CR
cells using lysis buffer (10 mM Tris, 150 mM NaCl, 1% Triton X-100,
1% sodium deoxycholate, 0.1% SDS, 10 mM EDTA and protease inhibitor
cocktail, pH 7.4) on ice. The lysates were centrifuged at 14,000 ×
g for 10 min at 4°C. The supernatants were then collected and the
concentration of total protein was determined using a bicinchoninic
acid assay kit (Beyotime Institute of Technology) according to the
manufacturer's protocols. Equal amount of protein (30 µg/lane) of
the samples were boiled in water with SDS-PAGE sample loading
buffer (Beyotime Institute of Technology) for 10 min prior to
separation via 10% SDS-PAGE. The proteins were transferred onto
polyvinylidene difluoride membranes (Roche Diagnostics, Basel,
Switzerland) and blocked with 5% non-fat milk at room temperature
for 1 h. Then the proteins. Then the membranes were incubated with
primary antibodies against α-fetoprotein (AFP; 1:1,000; cat. no.
4448; Cell Signaling Technology, Inc., Danvers, USA) and β-actin
(1:1,000; cat. no. AF0003; Beyotime Institute of Technology). The
membranes were then incubated with horseradish peroxide-conjugated
secondary antibodies (1:1,000; cat. nos. A0208 and A0216; Beyotime
Institute of Technology) for 1 h at room temperature and washed
with TBS containing 1% Tween and PBS. Protein bands were visualized
using the enhanced chemiluminescence (ECL) detection system
(Pierce; Thermo Fisher Scientific, Inc.) with an ECL kit (EMD
Millipore, Billerica, MA, USA). Protein expression was quantified
using Quantity One 4.62 software (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Immunofluorescence staining
HCC-CR cells (4×104 cells/cm2)
were seeded into a 24-well plate and incubated with 50 mM EdU
labeling solution (200 ml) at 37°C under 5% CO2 for 3 h.
Then the cells were fixed with 4% (v/v) PFA for 10 min at room
temperature, permeabilized with 0.3% Triton X-100 and then blocked
with 1% (w/v) bovine serum albumin (cat. no. P0023B; Beyotime
Institute of Technology) at 37°C for 30 min. Following incubation
overnight at 4°C with a primary antibody (mouse anti-human AFP;
1:200; cat. no. ab3980; Abcam, Cambridge, UK), the cells were
washed with PBS and incubated with an Alexa Fluor®
488-conjugated secondary antibody (1:1,000; cat. no. R37120;
Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature for
1 h. The cells were then incubated with anti-EdU working solution
for 30 min and 100 ng/ml DAPI (Beyotime Institute of Technology,
Shanghai, China) for 5 min (all at room temperature). and the
samples were analyzed using a confocal laser scanning microscope
(magnification, ×10; SP5; Leica Microsystems GmbH, Wetzlar,
Germany).
Statistical analysis
Data were analyzed using using SPSS software 22.0
(SPSS, Inc., Chicago, IL, USA). AFP protein expression data were
analyzed by one-way analysis of variance, whereas cell
proliferation data at different time points were analyzed using
Student's t-tests. Data were presented as the mean ± standard
deviation of at least three independent experiments. Associations
between the success rate of HCC-CR culture and patients'
background, including gender and surgical method, were analyzed
with χ2 tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
Establishment of patient-derived
primary HCC-CR cell cultures
Tumor tissue samples from 20 patients with HCC that
had not been previously treated with radiotherapy or chemotherapy
were collected, harvested and used for cell culture in the present
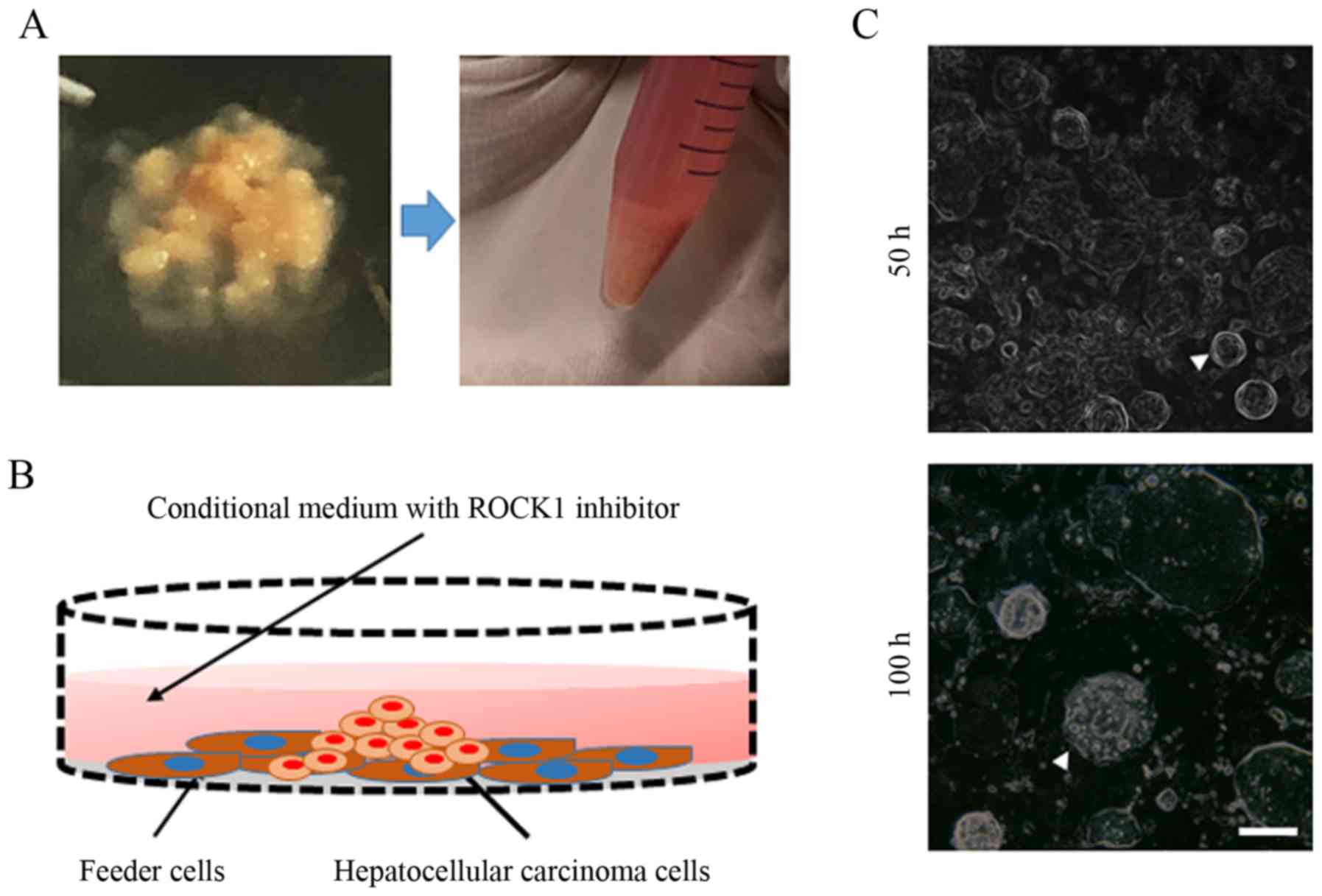
study (Fig. 1A). Cell cultures
were established by seeding HCC cells onto an NIH-3T3 fibroblast
feeder layer in CR medium (Fig.
1B). As presented in Fig. 1C,
adherent HCC cells exhibited small colonies at 50 h and progressed
into cell islands by 100 h.
The clinicopathological data of the patients are
presented in Table I. The median
age of the patients was 54.7 years; all patients suffered from HBV
and/or HCV. Tissue samples were obtained during OLT or hepatectomy.
A total of 35% (7/20) of patients exhibited T3/T4 grade tumors.
Additionally, 35% (7/20) of patients had tumor metastasis. HCC
cells from 55% of tumor tissues underwent successful continuous
passaging under CR conditions; the ability to establish continuous
passaging was markedly associated with the source and composition
of tumor tissues. With the exception of tissues from 100% of T1
patients and 54.5% of T2 patients, HCC cells were obtained via CR
culture from tissues from all other patients. HCC cells from
metastatic patients were more likely to be expanded in
vitro. The success rate of establishing CR cultures was
independent of the surgical method used (P=0.9510). HCC-CR cells
were not successfully generated from tissues with a necrosis rate
>30% and a stromal ratio >35%.
 | Table I.Clinicopathological data of
patients. |
Table I.
Clinicopathological data of
patients.
| Subject No. | Age | Sex | Grading | Metastasis |
Differentiation | Surgical
method | Necrosis (%) | Fibrosis (%) | CR culture passage
no. |
|---|
| 1 | 46 | Female | T2 | N1M1 | III | OLT | 0 | 10 | P6 |
| 2 | 59 | Male | T3 | N1M0 | II | Hepatectomy | 10 | 0 | P6 |
| 3 | 52 | Female | T1 | N0M0 | I | OLT | 30 | 30 | P2 |
| 4 | 47 | Male | T2 | N0M0 | II | OLT | 10 | 10 | P6 |
| 5 | 53 | Female | T3 | N1M1 | III | Hepatectomy | 10 | 30 | P6 |
| 6 | 44 | Female | T2 | N0M0 | I | OLT | 10 | 15 | P6 |
| 7 | 58 | Male | T1 | N0M0 | I | OLT | 20 | 30 | P2 |
| 8 | 54 | Male | T3 | N0M0 | II | Hepatectomy | 15 | 25 | P4 |
| 9 | 62 | Female | T2 | N0M0 | II | OLT | 20 | 25 | P3 |
| 10 | 41 | Male | T2 | N1M0 | III | OLT | 0 | 5 | P6 |
| 11 | 59 | Female | T3 | N1M0 | III | Hepatectomy | 0 | 15 | P6 |
| 12 | 67 | Male | T2 | N0M0 | II | OLT | 60 | 0 | N/A |
| 13 | 43 | Female | T2 | N0M0 | II | OLT | 20 | 0 | P6 |
| 14 | 53 | Male | T2 | N0M0 | II | OLT | 15 | 5 | P6 |
| 15 | 65 | Male | T1 | N0M0 | II | OLT | 30 | 35 | N/A |
| 16 | 62 | Female | T2 | N2M1 | III | OLT | 20 | 0 | P6 |
| 17 | 51 | Female | T3 | N0M0 | II | Hepatectomy | 60 | 0 | N/A |
| 18 | 38 | Female | T4 | N1M1 | III | Hepatectomy | 10 | 5 | P6 |
| 19 | 61 | Male | T2 | N0M0 | II | OLT | 30 | 15 | P4 |
| 20 | 51 | Male | T3 | N0M0 | II | Hepatectomy | 20 | 5 | P6 |
Propagation potential of primary
HCC-CR cells
Complete hepatic nodule or blood vessel-rich tissues
in primary HCC lesions were selected for further CR culture. A
6-day culture period was required to generate larger cell clones
prior to the initial passage of P0 cells, after which HCC cells
were generated every 3–4 days. CR cultures were determined to
possess the potential for continuous generation providing cells
were generated on a 3–4-day cycle, and colonies demonstrated a
continuous increase in quantity and volume. The morphologies of
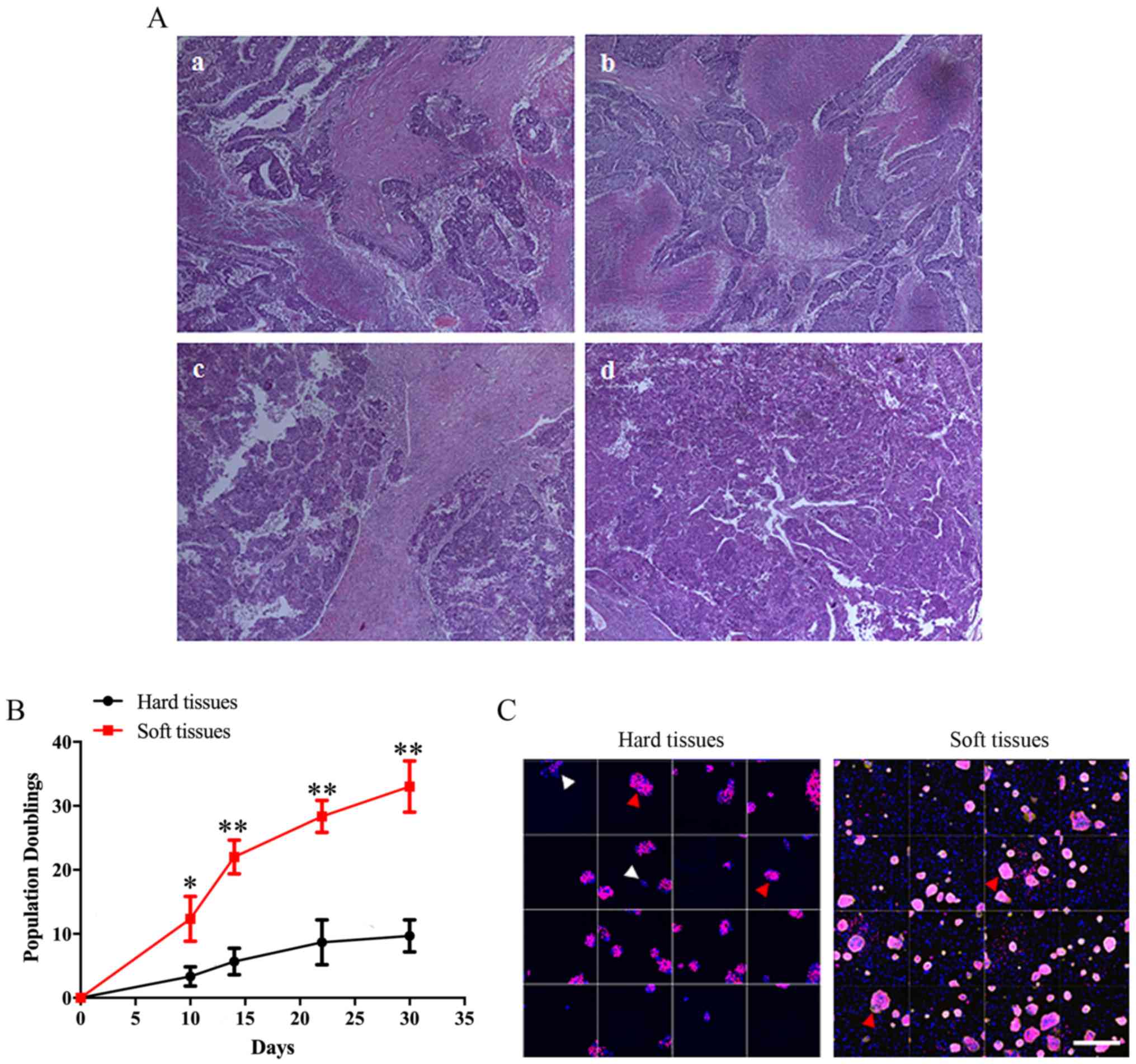
various tumor tissues were observed by H&E staining (Fig. 2A). Harder tissues exhibited larger
fibrotic areas and smaller necrotic regions. As presented in
Fig. 2B, the proliferation of
cells obtained from brittle, non-viscous tissues (hard tissues)
during continuous cultivation was significantly decreased compared
with cells from soft tissues with lumpy masses. Additionally,
following culture for 6 days, a markedly increased number of
colonies was observed in the soft tissues group compared with in
the hard tissues group (Fig. 2C).
Furthermore, EdU staining revealed that the number of cells with
proliferative potential in hard tissues cell colonies was notably
reduced compared with in soft tissue cell colonies.
Continuous generation capacity of
HCC-CR cells
HCC-CR cells that possessed proliferative ability in
the CR system were subjected to cryopreservation and resuscitation
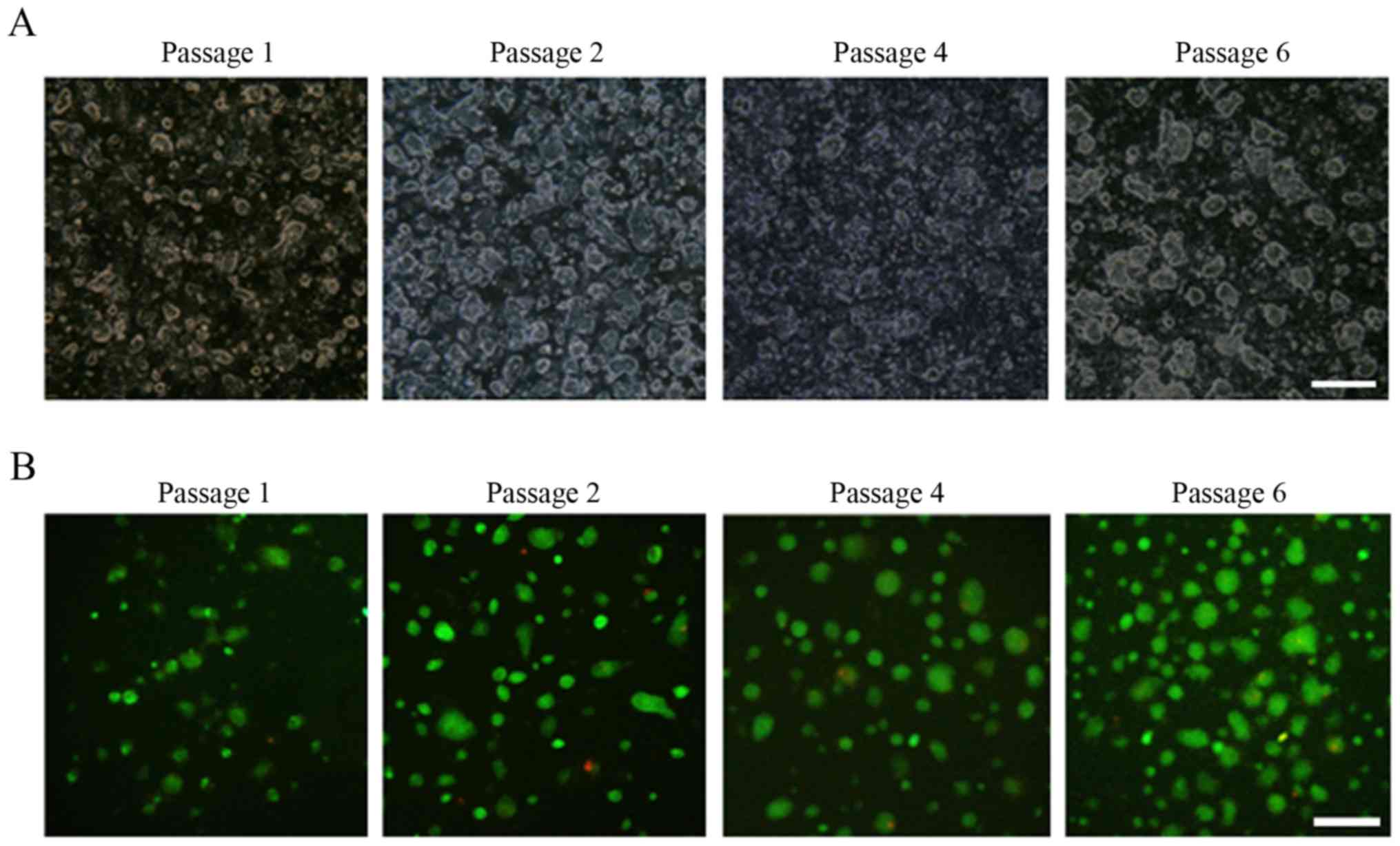
during every passage. As presented in Fig. 3A, HCC-CR cell colonies exhibited
continuous increases in quantity and volume across generations.
Additionally, live/dead cell staining revealed that HCC-CR cells
retained high viability following repeated cryopreservation and
resuscitation (Fig. 3B).
Expression of AFP in HCC-CR cells
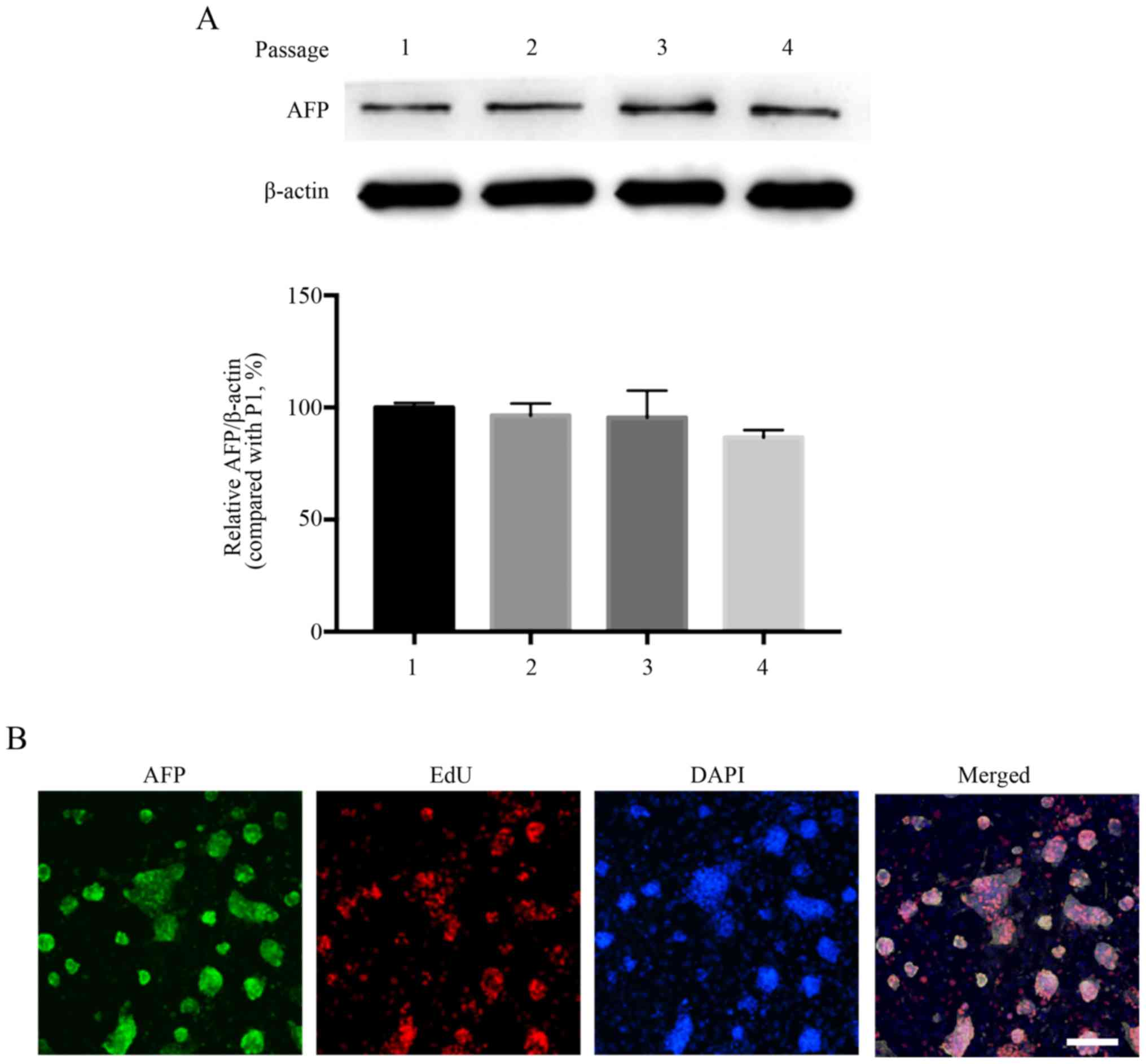
The identity of the cultured cells was demonstrated
by the stable expression of the HCC marker AFP during continuous
generation (Fig. 4A). AFP
expression was markedly downregulated with increasing passages;
however, there was no significant difference. Immunofluorescence
and EdU staining revealed that the cell purity was >90% and the
majority of AFP-positive cells possessed high proliferative
potential (Fig. 4B).
Discussion
It was previously hypothesized that only a small
proportion of tumor cells were able to form colonies in
vitro (20). In particular,
certain human epithelial cells, including those from the prostate,
lung and liver, possess very short lifespans in vitro and
can only undergo a limited number of passages, reducing their
potential use in cell biology studies (9). A novel culture system has been
developed for the indefinite propagation of various primary human
cells (normal and tumor cells) in vitro via co-culture with
irradiated fibroblast feeder cells and the rho-associated
coiled-coil containing protein kinase 1 inhibitor Y-27632 (21). This culture system is termed CR due
to the conditional induction of cell proliferation. As self-renewal
is a reported property of stem cells, it was hypothesized that CR
culture induced adult cells to exhibit adult stem cell-associated
properties (22).
The aim of the present study was to establish a
culture system in vitro for human hepatoma cells. Recent
studies have reported the ability of cancer cells to exhibit
sustained stable amplification under CR conditions (10–14).
Numerous studies have made advancements in the field of liver
cancer cell amplification; however, sufficient rapid amplification
of cells was not established for clinical use (23). A previous study reported the
successful construction of a primary HCC organoid; however, the
expansion time required to generate the 3D organoid was
impractically long (16). In the
present study, HCC cells were passaged every 3–4 days. The numbers
of harvested cells were counted following each passage, and a
constant growth of HCC-CR cells during each passage was observed
for 30 days (Fig. 2B). These
findings were consistent with previous studies and meet clinical
requirements (14,24). The identification of additional
biomarkers is required to further verify the reliability of the
in vitro CR culture method in obtaining matched tumor cells
from tissues from patients with HCC.
The ability to expand cells isolated from tissue
samples in vitro was associated with the quality of the
original tissue. In the present study, the HCC cells that were
continuously passaged under CR conditions were obtained from ~55%
of tumor samples; this ability was affected by the origin and
volume of actively proliferating tissue, as HCC cells from necrotic
or fibrotic tissues struggled to be continuously passaged. As
presented in Table I, the success
rate of HCC-CR culture was independent of the age of patients and
the surgical method used, which was consistent with a study
investigating nasopharyngeal carcinoma (25). Additionally, the proliferation and
cloning abilities of cells were notably increased in tissue samples
from patients with metastasis compared with in other samples. Cells
for amplification could not be isolated from calcified and necrotic
lesions. Furthermore, reduced proliferation and activation was
observed in tumor cells isolated from fibrotic samples. Based upon
these findings, to obtain HCC-CR cells that can be used for
subsequent experiments, original samples should be extracted from
patients with tumors in the T2/T3 stage. Furthermore, combined with
the sample information and the cell culture effect, it was
suggested that the tumor tissue should be >1 cm3 in
size, with a necrosis rate of <50% and a stromal ratio of
<40% to achieve acceptable expansion.
Increases in the number and area of cell colonies
are regarded as manifestations of the proliferation of tumor cells
in vitro (26). In the
present study, successfully amplified HCC-CR cells formed more and
larger cell colonies, thereby exhibiting adaptability to the CR
medium. Furthermore, the proliferation of cells, formation of
colonies and expression of AFP were markedly unaffected by repeated
cycles of cryopreservation and resuscitation, indicating that the
CR system met the requirements for successful hepatoma cell
culture. On the contrary, due to the apparent complexity of the
HCC-CR medium and the diversity of clinical samples, further
investigation is required to develop a simpler, more versatile CR
medium. Compared with tumor cells cultured under CR conditions,
commercial tumor cell lines fail to fully reproduce important
features of tumors in vivo. The heterogeneity of tumors
results in susceptibility to drug resistance, leading to tumor
recurrence and metastasis. In clinical settings, cells cultured
under CR conditions may be useful for drug screening. In the
present study, tumor cells were divided into soft and hard tissue
groups. In addition, stable expression of the HCC marker AFP during
continuous passaging was determined via western blotting. AFP is
the most extensively studied serological biomarker in the
surveillance of HCC and the only marker that has undergone all five
phases of biomarker development (27,28).
The findings of the present study are promising; however, the CR
system varies from physiological conditions. Therefore, analysis at
the transcriptional and translation levels should be conducted, and
in vivo transplants of CR-derived tumors should be
performed.
In conclusion, 20 tumor specimens from patients with
HCC were collected. HCC cells isolated from 55% of samples
exhibited continuous expansion under CR conditions; the ability to
undergo continuous passaging was associated with the source and
composition of the original tumor tissues. The expression of AFP in
HCC-CR cells was stable during passaging, and cells demonstrated
adaptability to CR culture conditions. These findings indicated
that the CR system may be a useful source for passaging HCC cells
required for clinical trials; however, the specific media
components require further optimization.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Tianjin
Clinical Research Center for Organ Transplantation Project (grant
no. 15ZXLCSY00070), the International S&T Cooperation Program
of China (grant no. 2015DFG31850), the Tianjin Science and
Technology Plan Project (grant no. 14RCGFSY00147) and the National
Human Genetic Resources Sharing Service Platform (grant no.
2005DKA21300).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
ZW made substantial contributions to the conception
and design of the present study, and the acquisition, analysis and
interpretation of data. BB and HS performed the experiments. LL
collected and analyzed patient information, and was involved in
drafting the manuscript and revising it critically for important
intellectual content. HZ performed some of the experiments and
provided final approval for the present version to be published. SW
was involved in the analysis and interpretation of data. ZS made
substantial contributions to the design of the study and agreed to
be accountable for all aspects of the study in ensuring that
questions regarding the accuracy or integrity of any part of the
work were appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Tianjin First
Central Hospital Clinical Research Ethics Committee (review no.
2016N057KY).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
CR
|
conditional reprogramming
|
|
DMEM/F12
|
Dulbecco's modified Eagle's
medium/Nutrient Mixture F-12 medium
|
|
AFP
|
α-fetoproteins
|
References
|
1
|
Bertuccio P, Turati F, Carioli G,
Rodriguez T, La Vecchia C, Malvezzi M and Negri E: Global trends
and predictions in hepatocellular carcinoma mortality. J Hepatol.
67:302–309. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ghouri YA, Mian I and Rowe JH: Review of
hepatocellular carcinoma: Epidemiology, etiology, and
carcinogenesis. J Carcinog. 16:12017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chan SL and Yeo W: Development of systemic
therapy for hepatocellular carcinoma at 2013: Updates and insights.
World J Gastroenterol. 20:3135–3145. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tovoli F, Granito A, De Lorenzo S and
Bolondi L: Regorafenib for the treatment of hepatocellular
carcinoma. Drugs Today (Barc). 54:5–13. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thorgeirsson SS and Grisham JW: Molecular
pathogenesis of human hepatocellular carcinoma. Nat Genet.
31:339–346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lohitesh K, Chowdhury R and Mukherjee S:
Resistance a major hindrance to chemotherapy in hepatocellular
carcinoma: An insight. Cancer Cell Int. 18:442018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huch M, Gehart H, van Boxtel R, Hamer K,
Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt
J, et al: Long-term culture of genome-stable bipotent stem cells
from adult human liver. Cell. 160:299–312. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu X, Ory V, Chapman S, Yuan H, Albanese
C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, et al:
ROCK inhibitor and feeder cells induce the conditional
reprogramming of epithelial cells. Am J Pathol. 180:599–607. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu X, Krawczyk E, Suprynowicz FA,
Palechor-Ceron N, Yuan H, Dakic A, Simic V, Zheng YL, Sripadhan P,
Chen C, et al: Conditional reprogramming and long-term expansion of
normal and tumor cells from human biospecimens. Nat Protoc.
12:439–451. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mahajan AS, Sugita BM, Duttargi AN, Saenz
F, Krawczyk E, McCutcheon JN, Fonseca AS, Kallakury B, Pohlmann P,
Gusev Y, et al: Genomic comparison of early-passage conditionally
reprogrammed breast cancer cells to their corresponding primary
tumors. PLoS One. 12:e01861902017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alamri AM, Kang K, Groeneveld S, Wang W,
Zhong X, Kallakury B, Hennighausen L, Liu X and Furth PA: Primary
cancer cell culture: Mammary-optimized vs conditional
reprogramming. Endocr Relat Cancer. 23:535–554. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao B, Huang C, Kernstine K, Pelekanou V,
Kluger Y, Jiang T, Peters-Hall JR, Coquelin M, Girard L, Zhang W,
et al: Non-malignant respiratory epithelial cells preferentially
proliferate from resected non-small cell lung cancer specimens
cultured under conditionally reprogrammed conditions. Oncotarget.
8:11114–11126. 2017.PubMed/NCBI
|
|
13
|
Saeed K, Rahkama V, Eldfors S, Bychkov D,
Mpindi JP, Yadav B, Paavolainen L, Aittokallio T, Heckman C,
Wennerberg K, et al: Comprehensive Drug Testing of Patient-derived
Conditionally Reprogrammed Cells from Castration-resistant Prostate
Cancer. Eur Urol. 71:319–327. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Timofeeva OA, Palechor-Ceron N, Li G, Yuan
H, Krawczyk E, Zhong X, Liu G, Upadhyay G, Dakic A, Yu S, et al:
Conditionally reprogrammed normal and primary tumor prostate
epithelial cells: A novel patient-derived cell model for studies of
human prostate cancer. Oncotarget. 8:22741–22758. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sette G, Salvati V, Giordani I, Pilozzi E,
Quacquarini D, Duranti E, De Nicola F, Pallocca M, Fanciulli M,
Falchi M, et al: Conditionally reprogrammed cells (CRC) methodology
does not allow the in vitro expansion of patient-derived primary
and metastatic lung cancer cells. Int J Cancer. 143:88–99. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Broutier L, Mastrogiovanni G, Verstegen
MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R,
Sidorova O, Gaspersz MP, et al: Human primary liver cancer-derived
organoid cultures for disease modeling and drug screening. Nat Med.
23:1424–1435. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
World Medical A; World Medical
Association, : World Medical Association Declaration of Helsinki:
Ethical principles for medical research involving human subjects.
JAMA. 310:2191–2194. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thery C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol.
30:3.22.1–3.22.29. 2006. View Article : Google Scholar
|
|
19
|
Zhu Y, Yang Y, Guo J, Dai Y, Ye L, Qiu J,
Zeng Z, Wu X, Xing Y, Long X, et al: Ex vivo 2D and 3D HSV-2
infection model using human normal vaginal epithelial cells.
Oncotarget. 8:15267–15282. 2017.PubMed/NCBI
|
|
20
|
Plaks V, Kong N and Werb Z: The cancer
stem cell niche: How essential is the niche in regulating stemness
of tumor cells? Cell Stem Cell. 16:225–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Agarwal S and Rimm DL: Making every cell
like HeLa a giant step for cell culture. Am J Pathol. 180:443–445.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Suprynowicz FA, Upadhyay G, Krawczyk E,
Kramer SC, Hebert JD, Liu X, Yuan H, Cheluvaraju C, Clapp PW,
Boucher RC Jr, et al: Conditionally reprogrammed cells represent a
stem-like state of adult epithelial cells. Proc Natl Acad Sci USA.
109:20035–20040. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gillet JP, Varma S and Gottesman MM: The
clinical relevance of cancer cell lines. J Natl Cancer Inst.
105:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hensley CT, Wasti AT and DeBerardinis RJ:
Glutamine and cancer: Cell biology, physiology, and clinical
opportunities. J Clin Invest. 123:3678–3684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu F, Lu Y, Tao L, Jiang YY, Lin DC, Wang
L, Petersson F, Yoshiyama H, Koeffler PH, Goh BC, et al:
Non-malignant epithelial cells preferentially proliferate from
nasopharyngeal carcinoma biopsy cultured under conditionally
reprogrammed conditions. Sci Rep. 7:173592017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
McMahon BJ, Alberts SR, Wainwright RB,
Bulkow L and Lanier AP: Hepatitis B-related sequelae. Prospective
study in 1400 hepatitis B surface antigen-positive Alaska native
carriers. Arch Intern Med. 150:1051–1054. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang BH, Yang BH and Tang ZY: Randomized
controlled trial of screening for hepatocellular carcinoma. J
Cancer Res Clin Oncol. 130:417–422. 2004. View Article : Google Scholar : PubMed/NCBI
|