Introduction
Lung cancers are the most commonly diagnosed and
most fatal type of cancers worldwide, of which non-small cell lung
cancer (NSCLC) accounts for ~80% of all primary lung cancer cases
(1–4). Since it has been reported that the
majority of patients with NSCLC are diagnosed at advanced stages,
chemotherapeutics are widely applied as the main first-line agents
for the treatment of these advanced stage NSCLC patients in
addition to surgical resection. Despite efforts to improve the
therapeutic efficacy of patients with NSCLC, the total 5-year
survival rate is <15% (5).
Therefore, it is urgent to uncover the molecular mechanisms
involved in NSCLS, which may help to provide new prognostic
biomarkers and therapeutic targets for patients with NSCLC.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
which downregulate specific mRNA targets through binding to
sequences located in the 3′-untranslated regions (UTRs), leading to
reduced gene transcription (6). It
has been estimated that miRNAs directly regulate ≥30% of human
genes. Therefore, miRNAs are involved in several physiological and
pathological processes (7–10). In addition, they serve important
roles in RNA silencing, and the majority of miRNAs are located at
fragile sites, which are frequently dysregulated in human cancers
(11,12). In recent decades, the dysregulation
of miRNA expression has been identified in numerous human diseases,
including cancer (13–15). A previous report revealed that
miR-539 was significantly downregulated in cisplatin
(DDP)-resistant NSCLC tissues and cells when compared with
DDP-sensitive NSCLC tissues and parental NSCLC cells. miR-539
inhibited DDP-resistant NSCLC cell invasion and migration through
targeting DCLK1 (16). miR-362 had
a greater expression in NSCLC tissues than in adjacent normal
tissues. In addition, miR-362 promoted NSCLC cell invasion,
migration and colony formation in vitro by targeting
Semaphorin-3A, which is significantly associated with metastasis
(17). In addition, some other
miRNAs were involved in non NSCLC, including miR-421 (18), miR-873 (19), miR-21 (20) and miR-486 (21).
Among all known miRNAs, miR-214 has been extensively
studied in cancer. It has been reported as a tumor suppressor in
gastric, cervical and colorectal cancer (22–24).
However, studies have also identified miR-214 as a promoter of
growth and metastases in lung cancer (25,26).
In addition, miR-214 may mediate hypoxia-induced cell proliferation
and apoptosis inhibition in pulmonary artery smooth muscle cells
(27). Hypoxia-inducible factor-1α
(HIF-1α) is overexpressed in several human cancers, and its
overexpression promotes tumor growth and metastasis by initiating
angiogenesis and regulating metabolism to overcome hypoxia.
Additionally, HIF-1α induces the expression of the angiogenic
protein vascular endothelial growth factor (VEGF) (28,29).
In the present study, the association between miR-214 with HIF-1α
and VEGF in A549 and H1299 lung cancer cells was explored. The
underlying mechanisms were also investigated, which may help to
clarify the associations between miR-214 and inhibitor of growth
family member 4 (ING4), HIF-1α, VEGF. Future studies should be
performed to test the therapeutic applications of miR-214.
Materials and methods
Clinical specimens
Fresh samples from lung cancer and corresponding
normal adjacent tissues were obtained from patients at The First
Affiliated and Shengjing Hospitals of China Medical University
(Shenyang, China) between May 2013 and December 2015. The present
study was conducted with the approval of the Ethics Committee of
China Medical University (Shenyang, China). Informed consent was
obtained from all patients. The mean patient age was 56.5 years
(range, 41 to 77 years). All patients underwent surgical resection
without prior chemotherapy or radiation therapy. There were 15 male
patients and five female patients. The tumor, node, metastasis
(TNM) staging system was used to classify specimens as stage I
(n=7), II (n=9) and III (n=4). There were 12 cases of
adenocarcinoma and eight cases of squamous cell carcinoma.
Cell culture and transfection
A549, H1299, H2228, H292, H3255 and H358 cell lines
were obtained from the American Type Culture Collection (Manassas,
VA, USA). Cells were cultured in Dulbecco's modified Eagle medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C in 5% CO2.
miR-214 mimics (cat. no. miR10000271; 100 nM)/mimic
control (cat. no. miR 01101; 100 nM) and miR-214 inhibitor (cat.
no. miR20000271; 150 nM)/inhibitor mimic (cat. no. miR 021011; 50
nM) were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou,
China; all sequences are not commercially available). ING4 small
interfering (si)RNA (cat. no. SR309575; 50 nM) and non-targeting
siRNA (cat. no. SR30004; 50 nM) were obtained from OriGene
Technologies, Inc. (Rockville, MD, USA). Cells were transfected
with miR-214 mimics, inhibitor or ING4 siRNA using DharmaFECT 1
transfection reagent (GE Healthcare Dharmacon, Inc., Lafayette, CO,
USA) according to the manufacturer's protocol; cells were used for
subsequent experimentation at 48 h post-transfection. pCMV6-ING4
plasmid and pCMV6 empty plasmid were purchased from OriGene
Technologies, Inc. Lipofectamine® 3000 transfection
reagent was used for plasmid transfection (Invitrogen; Thermo
Fisher Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from fresh tissue samples
and cells with TRIzol® reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Total RNA was reversed transcribed to cDNA using PrimeScript RT
Master mix (Takara Biotechnology Co., Ltd., Dalian, China). In
brief, reverse-transcription reaction solution (10 µl) was
prepared, which contained 2 µl 5X RT Master mix, 500 ng RNA and
DEPC H2O. Reverse transcription was performed at 37°C
for 15 min and 85°C for 5 sec using the PrimerScript RT Reagent Mix
kit (cat. no. RR037A; Takara Biotechnology Co., Ltd.). Bulge-Loop™
miRNA RT-qPCR primer sets for miR-214 and U6 were purchased from
Guangzhou RiboBio Co., Ltd. qPCR analyses were performed in an ABI
7500 Real-time PCR system (Thermo Fisher Scientific, Inc.) using
the SYBR Green Master mix (cat. no. a25778; Applied Biosystems;
Thermo Fisher Scientific, Inc.). The thermocycling conditions were
50°C for 2 min, 95°C for 2 min, followed by 45 cycles of 95°C for
15 sec and 60°C for 40 sec. Relative gene expression was determined
with the following formula: ΔCq=Cq gene-Cq reference. Fold changes
in gene expression were calculated using the 2−ΔΔCq
method (30). Primer sequences for
mRNA PCR were as follows: ING4 forward, 5′-GCCCGTTTTGAGGCTGAT-3′
and reverse, 5′-CACGAGCAGCTTTCTTCTCCT-3′; β-actin forward,
5′-ATAGCACAGCCTGGATAGCAACGTAC-3′ and reverse,
5′-CACCTTCTACAATGAGCTGCGTGTG-3′; miR-214 forward,
5′-TATACATCAAACAGCAGGCACA-3′, and reverse,
5′-CATTCGATCTTCTCCACAGTCTC-3′; U6 forward, 5′-CTGGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The ratio (>1.5) of
miR-214 in cancer tissues and the corresponding normal tissue was
defined as miR-214 upregulation.
Western blot analysis
Total proteins were extracted using
Radioimmunoprecipitation Assay lysis and extraction buffer (cat.
no. 89900; Thermo Fisher Scientific, Inc.). Proteins concentration
was determined using the Bradford method. Proteins (50 µg/lane)
were separated by 10% SDS-PAGE and transferred to polyvinylidene
difluoride membranes. The membranes were incubated overnight at 4°C
with primary antibodies against ING4 (1:800; cat. no. 10617-1-AP;
ProteinTech Group, Inc., Chicago, IL, USA), adenylate kinase 3
(AK3; 1:800; cat. no. 12562-1-AP; ProteinTech Group, Inc.), matrix
metalloproteinase 2 (MMP2; 1:800; cat. no. 10373-2-AP; ProteinTech
Group, Inc.), HIF-1α (1:1,000; cat. no. ab51608; Abcam, Cambridge,
MA, USA), VEGF (1:1,000; cat. no. ab53465ame; Abcam) and GAPDH
(1:2,000; cat. no. 2118; Cell Signaling Technology, Inc., Danvers,
MA, USA). Following incubation with horseradish
peroxidase-conjugated anti-mouse/rabbit IgG (cat. nos. #7076 and
#7074, respectively; 1:2,000; Cell Signaling Technology, Inc.) at
37°C for 2 h, target proteins were visualized using enhanced
chemiluminescence reagent (Pierce; Thermo Fisher Scientific, Inc.).
Images were captured using a MicroChemi (DNR Bio-Imagining Systems,
Ltd., Neve Yamin, Israel). The relative intensity of blotted
proteins was determined using ImageJ 1.8.0 software (National
Institutes of Health, Bethesda, MD, USA).
Matrigel invasion assay
The cell invasion assay was performed in a 24-well
Transwell chamber (pore size, 8 µm), and the inserts were coated
with 20 µl Matrigel (1:6 dilution; dilution with serum-free medium;
BD Biosciences; Becton, Dickinson and Company, Franklin, Lakes, NJ,
USA). The transfected cells were cultured for 48 h following
transfection at 37°C with 5% CO2. Then the cells were
trypsinized, transferred to the upper Matrigel chamber in 100 µl
serum-free medium (1×105 cells/ml) and incubated at 37°C
for 18 h. Medium supplemented with 10% fetal bovine serum (FBS;
Invitrogen; Thermo Fisher Scientific, Inc.) was added to the lower
chamber. Following this, non-invaded cells on the upper membrane
surface were removed and cells that invaded through the filter were
fixed in 4% paraformaldehyde at room temperature for 20 min and
stained with hematoxylin (Fuzhou Maixin Biotech Co, Ltd., Fuzhou
China) at room temperature for 5 min. The cells were counted under
a light microscope (magnification, ×200; Olympus CX22LED).
MTT assay
Cells (105/well) were plated in 96-well
plates and cultured overnight at 37°C. MTT (20 µl; 5 mg/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) solution was added
to each well. Following a 4 h incubation, the supernatant was
removed and 150 µl dimethyl sulfoxide was added to dissolve the
formazan crystals. Absorbance was measured at 490 nm and the data
were obtained from triplicate wells.
Colony formation assay
Cells (102/dish) were seeded into three 6
cm cell culture dishes and incubated for ~2 weeks. Following this,
plates were washed with PBS and stained with hematoxylin (Fuzhou
Maixin Biotech Co, Ltd.) at room temperature for 10 min. Colony
numbers were manually counted.
Wound healing assay
After 24 h of culture, cells were seeded into 6-well
plates until 70–90% confluence was reached. The monolayer was
gently scratched using a 1 ml pipette tip. Detached cells were
washed away with PBS and the plates were incubated at 37°C for 24
h. Photos of the stained monolayer were taken using a light
microscope (magnification, ×200; Olympus CX22LED).
Validation of target gene
A reporter vector (pmiR-RB) (Promega Corporation,
Madison, Wisconsin, USA) was used for the luciferase assay.
TargetScan 7.1 software was used to predict potential binding sites
(31). The wild-type miR-214
target site in ING43′-UTR was CCUGCUG. The mutant miR-214 target
site was CCCCCUG. Transfection of reporter plasmid was performed
using Attractene reagent (Qiagen, Inc., Valencia, CA, USA).
Luciferase activity was measured in cellular extracts using a dual
luciferase reporter gene assay kit (Promega Corporation) at 36 h
post-transfection. The relative activity of the reporter gene was
normalized to Renilla luciferase activity.
Statistical analysis
SPSS version 16 (SPSS, Inc., Chicago, IL, USA) was
used for all statistical analyses. Data was presented as the mean ±
standard deviation. Student's t-test or one way analysis of
variance with Tukey's post hoc test was used to compare differences
between control and treatment groups. Linear regression was used to
estimate the correlation between miR-214 and ING4 expression in
tissues. All P-values were based on a two-sided statistical
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-214 expression is upregulated in
lung cancer tissues
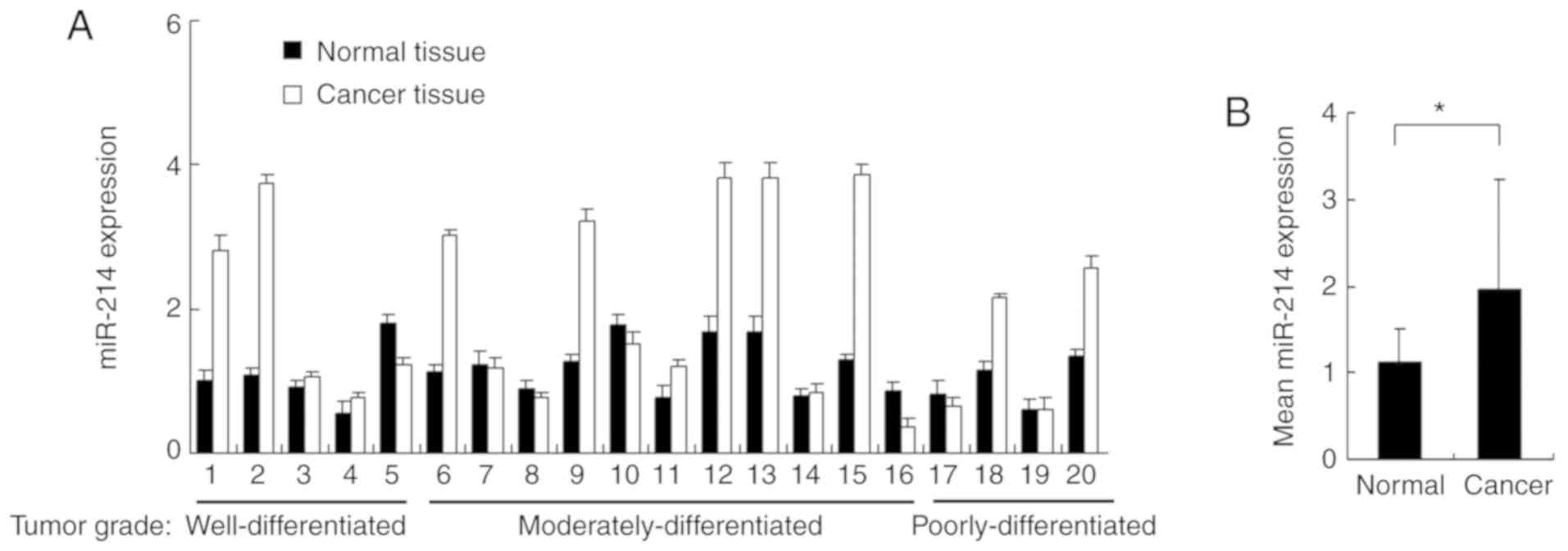
The expression of miR-214 was examined in 20 cases
of lung carcinoma tissues and paired adjacent normal tissues using
RT-qPCR (Fig. 1A). The mean value
of miR-214 expression was higher in cancer tissues compared with
normal tissues (Fig. 1B). A
cancer/normal ratio of >1.5 was defined as miR-214 upregulation,
which was observed in 10 out of 20 paired tissues (Fig. 1A). The percentage of miR-214
upregulation in tissues with different tumor grades was also
examined. miR-214 upregulation in well/moderately/poorly
differentiated tumors was present in 40% (2/5), 36.3% (5/11), and
50% (2/4) of cases, respectively. There was no obvious correlation
between miR-214 overexpression and tumor grade.
miR-214 promotes lung cancer cell
proliferation, invasion and migration
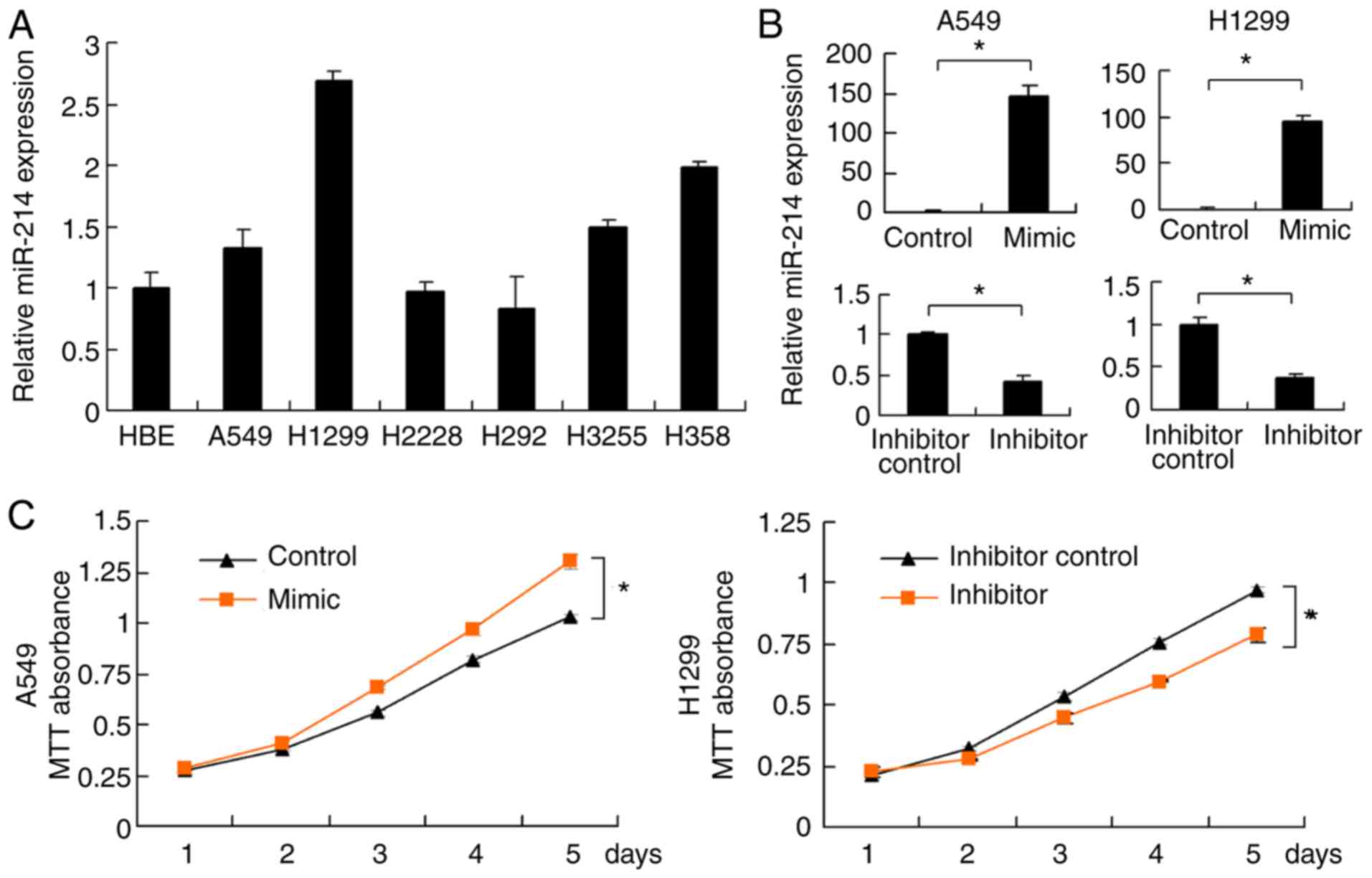
miR-214 expression levels were examined in several
lung cancer cell lines (A549, H1299, H2228, H292, H3255 and H358)
and normal bronchial epithelium cell line HBE (Fig. 2A). High miR-214 expression was
detected in H1299, H3255 and H358 cell lines. Relatively low
miR-214 expression was detected in HBE, A549, H2228 and H292 cell
lines. The A549 and H1299 cell lines were subsequently selected for
miR-214 mimic and inhibitor transfection; the A549 cell line was
used as it has high transfection efficiency and is widely used in
literature (32). Transfection
efficiency was confirmed by RT-qPCR. miR-214 mimics significantly
upregulated miR-214 expression, whereas miR-214 inhibitor
downregulated its expression in both cell lines (Fig. 2B). A MTT assay was performed for 5
days to examine the cell growth curves. As presented in Fig. 2C, miR-214 mimics promoted cell
growth rate, whereas miR-214 inhibitor decreased the cell growth
rate. The colony formation assay demonstrated that miR-214 mimics
promoted cell colony formation ability, whereas miR-214 inhibitor
decreased this ability (Fig. 2D).
Similarly, the Matrigel invasion assay demonstrated that miR-214
mimics promoted A549 cell invasion, whereas miR-214 inhibitor
downregulated H1299 invasion (Fig.
2E). To assess cell migration alterations, a wound healing
assay was performed. The results revealed that miR-214 mimics
facilitated A549 migration, while the miR-214 inhibitor prevented
H1299 cell invasion (Fig. 2F).
miR-214 regulates HIF-1α, VEGF, MMP2
and AK3 expression
To investigate the mechanisms underlying cell
proliferation and invasion regulation by miR-214, and to explore
the potential association between miR-214 and the hypoxia induced
effects, A549 and H1299 cells were transfected with miR-214 mimics
and inhibitors, and the expression of relevant proteins was
detected by western blotting (Fig.
3A). It was demonstrated that miR-214 upregulated the
expression of MMP-2, HIF-1α, as well as its target proteins VEGF
and AK3, compared with the corresponding control group.
Transfection of miR-214 inhibitor exhibited the opposite effects by
downregulating MMP2, HIF-1α, VEGF and AK3 expression, compared with
the corresponding inhibitor control (Fig. 3B).
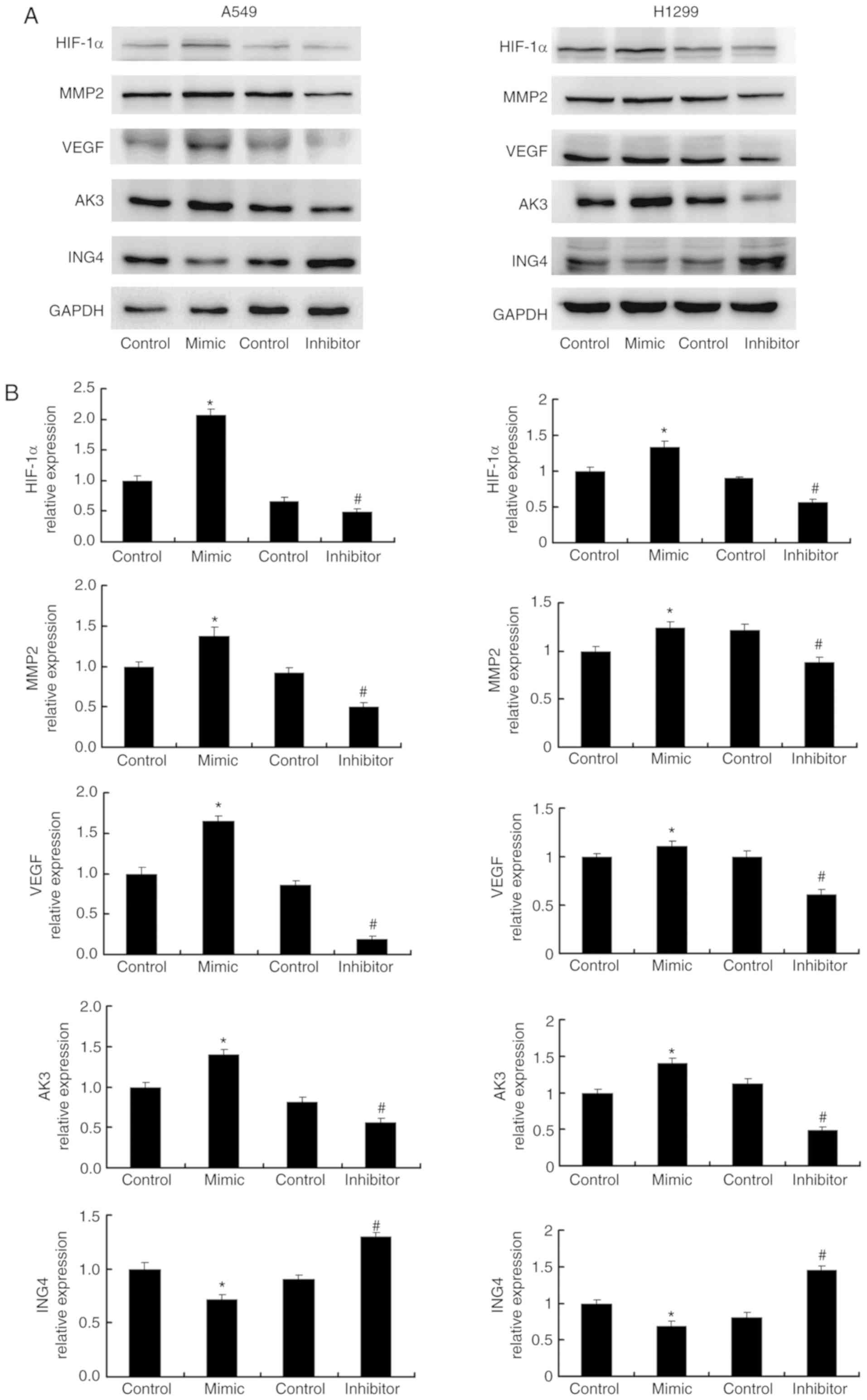 | Figure 3.miR-214 regulates ING4, HIF-1α, VEGF,
AK3 and MMP2 protein expression. (A) Western blotting showed that
miR-214 mimic transfection significantly increased HIF-1α, VEGF,
AK3 and MMP2 protein expression levels, and significantly decreased
ING4 expression in A549 and H1299 cell lines. HIF-1α, MMP2, AK3 and
VEGF expressions levels decreased, and ING4 expression increased in
A549 and H1299 cells transfected with miR-214 inhibitor. (B)
Quantification of the western blotting results. *P<0.05 vs.
mimic control; #P<0.05 vs. inhibitor control. ING4, inhibitor of
growth family member 4; HIF-1α, hypoxia-inducible factor 1α; AK3,
adenylate kinase 3; VEGF, vascular endothelial growth factor; MMP2,
matrix metalloproteinase 2; miR-214, microRNA-214. |
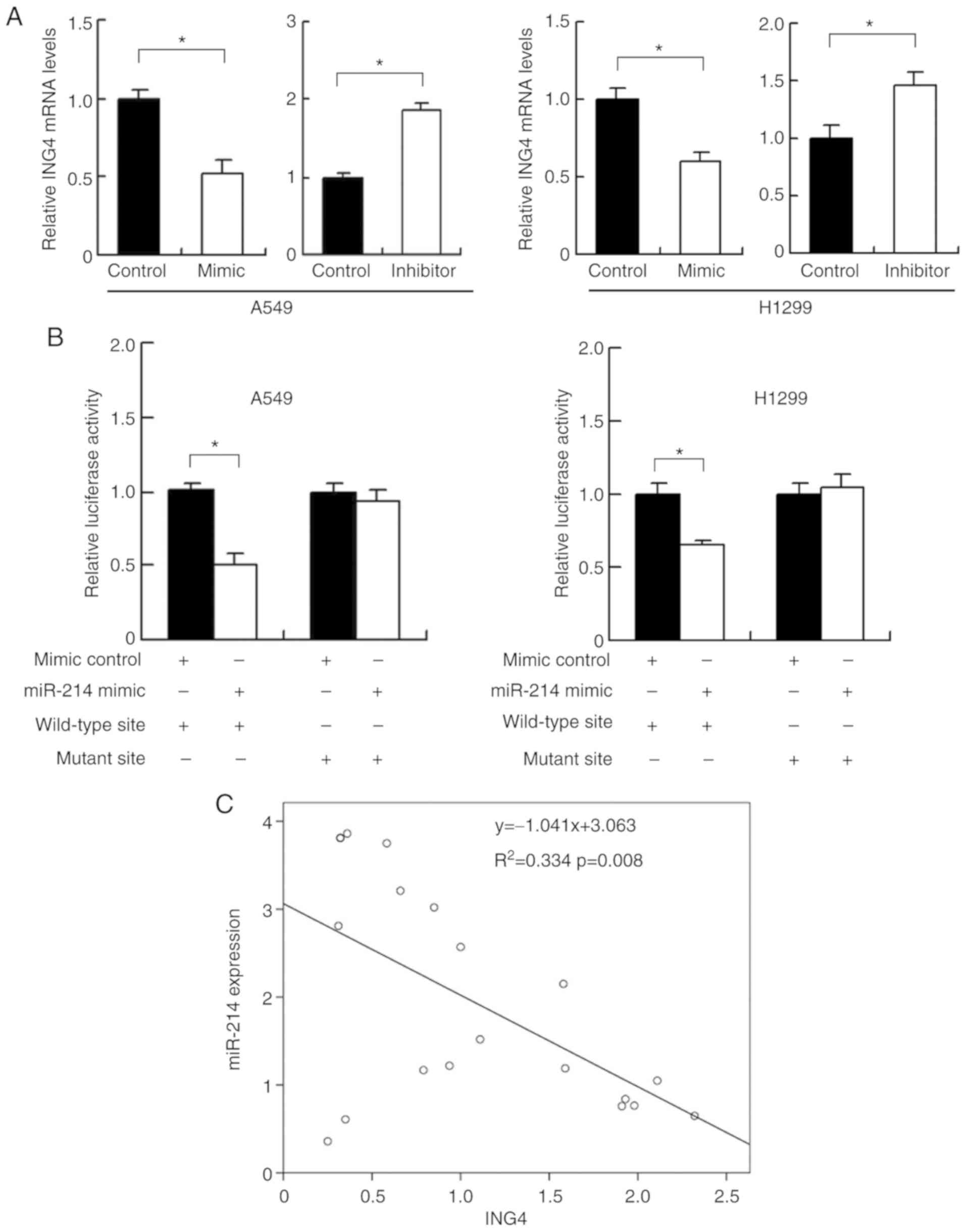
miR-214 targets and downregulates ING4
in A549 and H1299 cell lines
Using the online target prediction database
TargetScan 7.1, it was determined that miR-214 directly bound to
the 3′UTR of ING4, which has previously been reported to be a tumor
suppressor and inhibitor of HIF-1α. To determine their exact
relationship, ING4 expression was examined following transfection
with miR-214 mimics or inhibitor. miR-214 mimics significantly
downregulated ING4 expression, whereas miR-214 inhibitor
transfection upregulated ING4 expression at the mRNA and protein
level (Figs. 3A and 4A). To determine if ING4 was a direct
target of miR-214, luciferase reporter assays were performed.
Reporter plasmids with wild-type (CCUGCUG) or mutant (CCCCCUG)
3′-UTR binding sites for ING4 were transfected in A549 and H1299
cells together with miR-214 mimic. miR-214 mimics significantly
suppressed luciferase activity in cells transfected with the
wild-type vector (Fig. 4B). No
marked alteration was detected in cells with the mutant site
plasmid, suggesting that miR-214 bound to the ING4 3′-UTR and thus
downregulated ING4 expression. To validate their association in
lung cancer tissues, the mRNA expression of both miR-214 and ING4
was examined. Linear regression analyses demonstrated that there
was an inverse correlation between miR-214 and ING4 mRNA expression
(Fig. 4C; P=0.008).
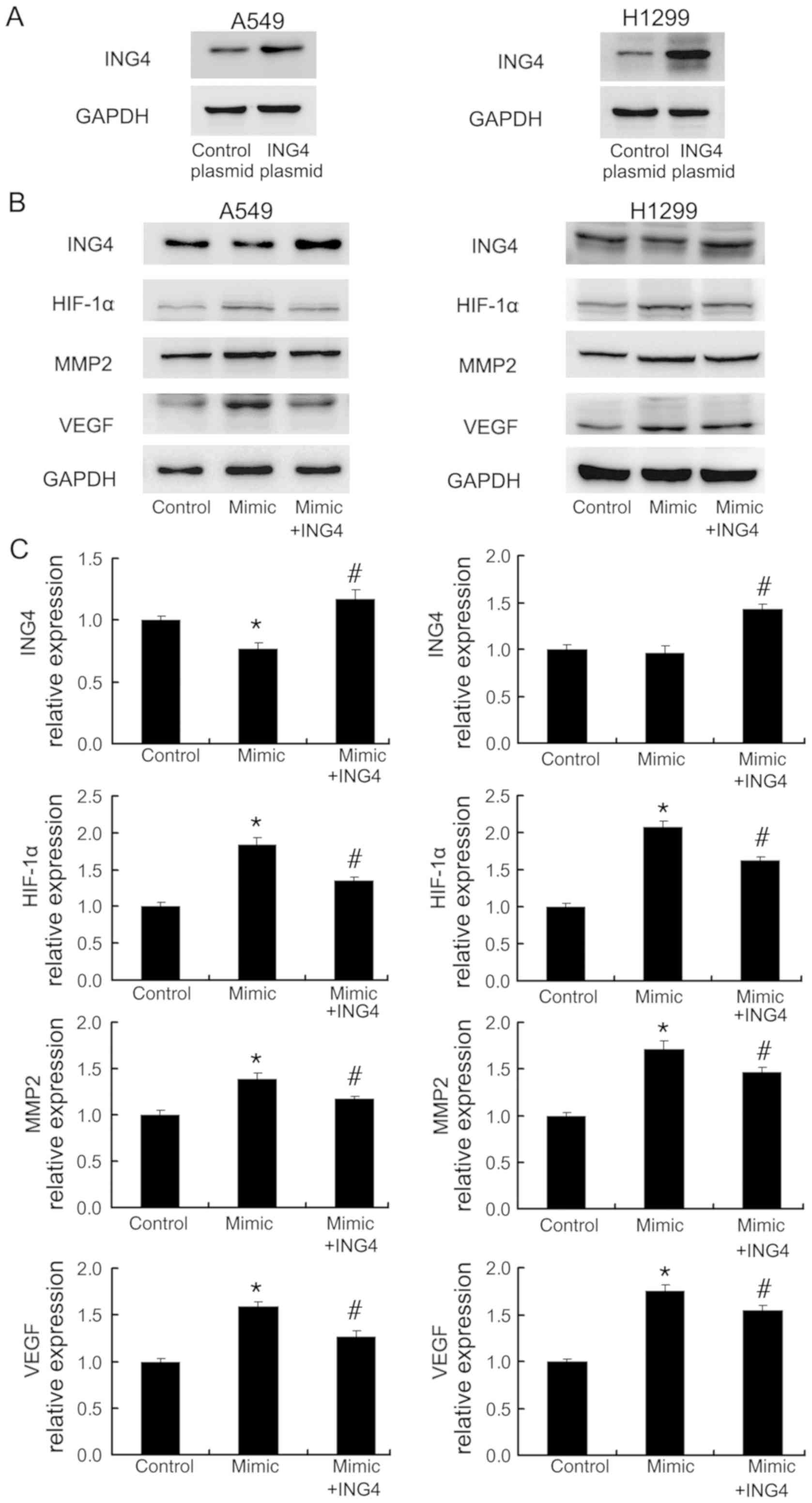
The effects of miR-214 on HIF-1α, MMP2
and VEGF are dependent on ING4
ING4 has been reported as a tumor suppressor which
inhibits MMP2 and HIF-1α expression (33–35).
To confirm the involvement of ING4 in miR-214-mediated regulation
of MMP2, VEGF and HIF-1α, ING4 plasmid was transfected into A549
and H1299 cells together with the miR-214 mimic. Western blotting
demonstrated the success of ING5 plasmid transfection in the two
cell lines (Fig. 5A). ING4
overexpression was demonstrated to abolish the miR-214-induced
upregulation of MMP2, VEGF and HIF-1α expression (Fig. 5B and C).
In addition, ING4 siRNA was introduced into A549 and
H1299 cells in combination with miR-214 inhibitor. ING4 expression
was markedly reduced by siRNA transfection, compared with the
control (Fig. 6A). As presented in
Fig. 6B, ING4 siRNA upregulated
MMP2, VEGF and HIF-1α expression. These data indicated that miR-214
induced the expression of MMP2, VEGF and HIF-1α by targeting tumor
suppressor ING4.
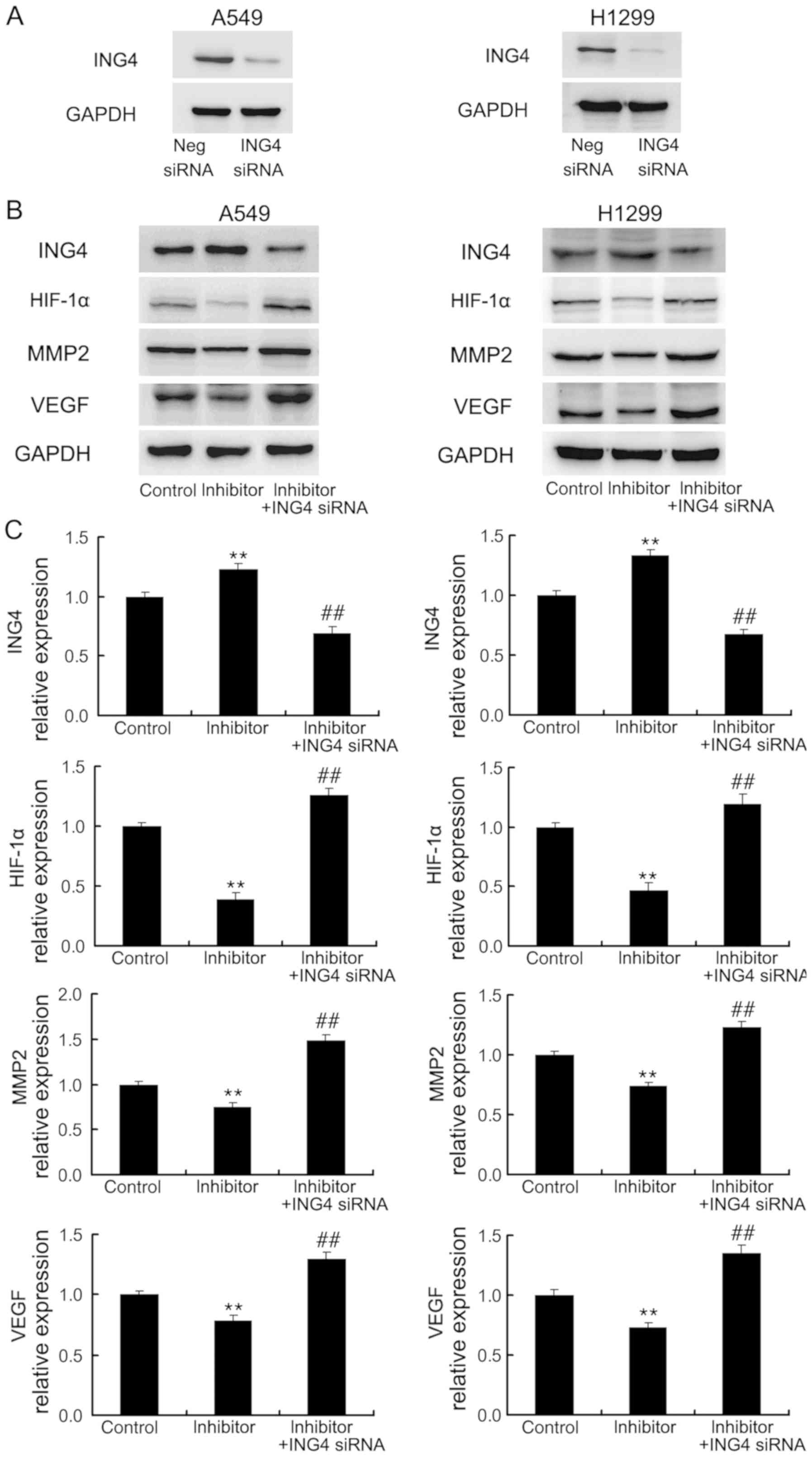 | Figure 6.ING4 siRNA transfection downregulates
MMP2, HIF-1α and VEGF expression. (A) Western blotting confirmed
that ING4 siRNA transfection downregulated ING4 protein expression
in A549 and H1299 cells. (B) A549 and H1299 cells were transfected
with the inhibitor control, miR-214 inhibitor or miR-214 inhibitor
together with ING4 siRNA. ING4 siRNA downregulated MMP2, HIF-1α and
VEGF, which were induced by miR-214. (C) Quantification of western
blotting results. These experiments were performed in triplicate.
**P<0.01 vs. control group; ##P<0.01 vs. mimic group. ING4,
inhibitor of growth family member 4; HIF-1α, hypoxia-inducible
factor 1α; AK3, adenylate kinase 3; VEGF, vascular endothelial
growth factor; MMP2, matrix metalloproteinase 2; miR-214,
microRNA-214. |
Discussion
Evidence indicates that miRNAs participate in cancer
development. miR-214 has been reported to be dysregulated in a
variety of human diseases, including cancer (36–38).
Downregulation of miR-214 has been implicated in several cancers
including gastric, cervical, esophageal and colorectal cancer
(22–24). However, studies have also
identified miR-214 as a promoter of growth and invasion in
non-small cell lung cancer (25,39).
Thus, the biological roles of miR-214 in human cancer are
contradicting. In the present study, it was confirmed that miR-214
expression was upregulated in lung cancer tissues and cell lines.
Using MTT, colony formation, Matrigel invasion and migration
assays, it was demonstrated that miR-214 served as a promoter of
cell growth invasion and migration in NSCLC cells. As miR-214 has
been reported to mediate hypoxia-induced cell proliferation in
pulmonary artery smooth muscle cells (27), the expression of HIF-1α and its
downstream factors were measured. It was revealed that miR-214
upregulated HIF-1α, while miR-214 inhibitor downregulated
HIF-1α.
HIF-1α is involved in adaptation to hypoxia and
angiogenesis during the development of various cancers (40). HIF-1α has been reported to be
overexpressed in human NSCLCs, and is associated with angiogenesis,
invasion, epithelial-mesenchymal transition and chemoresistance
(41–43). VEGF is a downstream protein of
HIF-1α, which has been implicated as an angiogenesis stimulating
protein during cancer progression (44–46).
AK3 is a target protein of HIF-1α (47), which is located exclusively in the
mitochondrial matrix (48). AK3
may participate in the high-energy phosphate transfer process
(49). The results of the present
study demonstrated that miR-214 induced HIF-1α and its target VEGF,
which may have promoted tumor angiogenesis and cell survival.
Next, the underlying mechanisms were explored.
Prediction software and reporter assays were used, which
demonstrated that miR-214 suppressed the mRNA and protein
expression of ING4, a well-defined tumor suppressor. ING4 has been
reported to inhibit tumor invasion by suppression of the MMP family
of proteins in osteosarcoma, gastric and lung cancer (50,51).
ING4 may also suppress HIF-1α and its downstream signaling proteins
(52,53). Thus, it was postulated that the
biological effects of miR-214 may be dependent on its regulation of
ING4 expression. To confirm this hypothesis, ING4 plasmid and siRNA
were co-transfected with miR-214 mimics or inhibitor. The results
demonstrated that ING4 siRNA prevented the effects of the miR-214
inhibitor by upregulating MMP2, VEGF and HIF-1α expression, whereas
ING4 plasmid suppressed the expression of these proteins.
Collectively, these data demonstrated that miR-214 induced MMP2 and
HIF-1α signaling in lung cancer cells by targeting tumor suppressor
ING4.
In conclusion, the present study demonstrated that
miR-214 functioned as an oncogene in lung cancer cells. miR-214
targeted tumor suppressor ING4, which in turn inhibited HIF-1α,
VEGF and MMP2 expression. Therefore, miR-214 may serve as a
potential therapeutic target in non-small cell lung cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
YL and LZ performed the experiments, evaluated the
data, wrote the manuscript and prepared the figures. YL designed
the experiments. YL and YQ evaluated the data and wrote the
manuscript. XY conceived and designed the study. All authors read
and approved the manuscript.
Ethics approval and consent to
participate
The present study was conducted with the approval of
the Ethics Committee of China Medical University (Shenyang, China).
Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gharibvand L, Beeson WL, Shavlik D,
Knutsen R, Ghamsary M, Soret S and Knutsen SF: The association
between ambient fine particulate matter and incident adenocarcinoma
subtype of lung cancer. Environ Health. 16:712017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sartorius B and Sartorius K: How much
incident lung cancer was missed globally in 2012? An ecological
country-level study. Geospat Health. 11:3962016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bailey C, Hewison A, Karasouli E,
Staniszewska S and Munday D: Hospital care following emergency
admission: A critical incident case study of the experiences of
patients with advanced lung cancer and chronic obstructive
pulmonary disease. J Clin Nurs. 25:2168–2179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tual S, Silverman DT, Koutros S, Blair A,
Sandler DP, Lebailly P, Andreotti G, Hoppin JA and Freeman LE: Use
of dieselized farm equipment and incident lung cancer: Findings
from the Agricultural Health Study Cohort. Environ Health Perspect.
124:611–618. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Akamatsu H, Mori K, Naito T, Imai H, Ono
A, Shukuya T, Taira T, Kenmotsu H, Murakami H, Endo M, et al:
Progression-free survival at 2 years is a reliable surrogate marker
for the 5-year survival rate in patients with locally advanced
non-small cell lung cancer treated with chemoradiotherapy. BMC
Cancer. 14:182014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tian T, Wang J and Zhou X: A review:
MicroRNA detection methods. Org Biomol Chem. 13:2226–2238. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Silva BOD, Lima KF, Gonçalves LR, Silveira
MBD and Moraes KCM: Correction: MicroRNA profiling of the effect of
the heptapeptide angiotensin-(1–7) in A549 lung tumor cells reveals
a role for miRNA149-3p in cellular migration processes. PLoS One.
12:e01902042017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pastorkova Z, Skarda J and Andel J: The
role of microRNA in metastatic processes of non-small cell lung
carcinoma. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub.
160:343–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang HM, Kuang S, Xiong X, Gao T, Liu C
and Guo AY: Transcription factor and microRNA co-regulatory loops:
Important regulatory motifs in biological processes and diseases.
Brief Bioinform. 16:45–58. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kraemer A, Anastasov N, Angermeier M,
Winkler K, Atkinson MJ and Moertl S: MicroRNA-mediated processes
are essential for the cellular radiation response. Radiat Res.
176:575–586. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang DQ, Zhou CK, Jiang XW, Chen J and
Shi BK: Increased expression of miR-222 is associated with poor
prognosis in bladder cancer. World J Surg Oncol. 12:2412014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Köhler CU, Bryk O, Meier S, Lang K,
Rozynek P, Brüning T and Käfferlein HU: Analyses in human
urothelial cells identify methylation of miR-152, miR-200b and
miR-10a genes as candidate bladder cancer biomarkers. Biochem
Biophys Res Commun. 438:48–53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Puerta-Gil P, García-Baquero R, Jia AY,
Ocaña S, Alvarez-Múgica M, Alvarez-Ossorio JL, Cordon-Cardo C, Cava
F and Sánchez-Carbayo M: miR-143, miR-222, and miR-452 are useful
as tumor stratification and noninvasive diagnostic biomarkers for
bladder cancer. Am J Pathol. 180:1808–1815. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deng H, Qianqian G, Ting J and Aimin Y:
miR-539 enhances chemosensitivity to cisplatin in non-small cell
lung cancer by targeting DCLK1. Biomed Pharmacother. 106:1072–1081.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luo D, Zhang Z, Zhang Z, Li JY, Cui J, Shi
WP, Dong XW, Yuan L, Lin P, Chen ZN, et al: Aberrant expression of
miR-362 promotes lung cancer metastasis through downregulation of
Sema3A. J Immunol Res. 2018:16870972018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Cui X, Li Y, Zhang T and Li S:
Upregulated expression of miR-421 is associated with poor prognosis
in non-small-cell lung cancer. Cancer Manag Res. 10:2627–2633.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin S, He J, Li J, Guo R, Shu Y and Liu P:
MiR-873 inhibition enhances gefitinib resistance in non-small cell
lung cancer cells by targeting glioma-associated oncogene homolog
1. Thorac Cancer. 9:1262–1270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng W, Zhao J, Tao Y, Guo M, Ya Z, Chen
C, Qin N, Zheng J, Luo J and Xu L: MicroRNA-21: A promising
biomarker for the prognosis and diagnosis of non-small cell lung
cancer. Oncol Lett. 16:2777–2782. 2018.PubMed/NCBI
|
|
21
|
Yu S, Geng S and Hu Y: miR-486-5p inhibits
cell proliferation and invasion through repressing GAB2 in
non-small cell lung cancer. Oncol Lett. 16:3525–3530.
2018.PubMed/NCBI
|
|
22
|
Yang TS, Yang XH, Wang XD, Wang YL, Zhou B
and Song ZS: MiR-214 regulate gastric cancer cell proliferation,
migration and invasion by targeting PTEN. Cancer Cell Int.
13:682013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wen Z, Lei Z, Jin-An M, Xue-Zhen L,
Xing-Nan Z and Xiu-Wen D: The inhibitory role of miR-214 in
cervical cancer cells through directly targeting mitochondrial
transcription factor A (TFAM). Eur J Gynaecol Oncol. 35:676–682.
2014.PubMed/NCBI
|
|
24
|
Cristóbal I, Caramés C, Madoz-Gúrpide J,
Rojo F, Aguilera O and García-Foncillas J: Downregulation of
miR-214 is specific of liver metastasis in colorectal cancer and
could play a role determining the metastatic niche. Int J
Colorectal Dis. 29:8852014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Long H, Wang Z, Chen J, Xiang T, Li Q,
Diao X and Zhu B: microRNA-214 promotes epithelial-mesenchymal
transition and metastasis in lung adenocarcinoma by targeting the
suppressor-of-fused protein (Sufu). Oncotarget. 6:38705–38718.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liao J, Lin J, Lin D, Zou C, Kurata J, Lin
R, He Z and Su Y: Down-regulation of miR-214 reverses erlotinib
resistance in non-small-cell lung cancer through up-regulating LHX6
expression. Sci Rep. 7:7812017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu H, Tao Y, Chen M, Yu J, Li WJ, Tao L,
Li Y and Li F: Upregulation of microRNA-214 contributes to the
development of vascular remodeling in hypoxia-induced pulmonary
hypertension via targeting CCNL2. Sci Rep. 6:246612016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shenoy SK, Han S, Zhao YL, Hara MR, Oliver
T, Cao Y and Dewhirst MW: β-arrestin1 mediates metastatic growth of
breast cancer cells by facilitating HIF-1-dependent VEGF
expression. Oncogene. 31:282–292. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Zhao Q, Ma S, Yang F, Gong Y and
Ke C: Sirolimus inhibits human pancreatic carcinoma cell
proliferation by a mechanism linked to the targeting of mTOR/HIF-1
alpha/VEGF signaling. IUBMB Life. 59:717–721. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
32
|
Grelet S, Link LA, Howley B, Obellianne C,
Palanisamy V, Gangaraju VK, Diehl JA and Howe PH: A regulated PNUTS
mRNA to lncRNA splice switch mediates EMT and tumour progression.
Nat Cell Biol. 19:1105–1115. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen Y, Huang Y, Hou P, Zhang Z, Zhang Y,
Wang W, Sun G, Xu L, Zhou J, Bai J and Zheng J: ING4 suppresses
tumor angiogenesis and functions as a prognostic marker in human
colorectal cancer. Oncotarget. 7:79017–79031. 2016.PubMed/NCBI
|
|
34
|
Wang CJ, Yang D and Luo YW: Recombinant
ING4 suppresses the migration of SW579 thyroid cancer cells via
epithelial to mesenchymal transition. Exp Ther Med. 10:603–607.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang QS, Li M, Zhang LY, Jin Y, Tong DD,
Yu Y, Bai J, Huang Q, Liu FL, Liu A, et al: Down-regulation of ING4
is associated with initiation and progression of lung cancer.
Histopathology. 57:271–281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peng R, Men J, Ma R, Wang Q, Wang Y, Sun Y
and Ren J: miR-214 down-regulates ARL2 and suppresses growth and
invasion of cervical cancer cells. Biochem Biophys Res Commun.
484:623–630. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu J, Li D, Dang L, Liang C, Guo B, Lu C,
He X, Cheung HY, He B, Liu B, et al: Osteoclastic miR-214 targets
TRAF3 to contribute to osteolytic bone metastasis of breast cancer.
Sci Rep. 7:404872017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu CX, Xu M, Tan L, Yang H, Permuth-Wey J,
Kruk PA, Wenham RM, Nicosia SV, Lancaster JM, Sellers TA and Cheng
JQ: MicroRNA miR-214 regulates ovarian cancer cell stemness by
targeting p53/Nanog. J Biol Chem. 291:228512016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang C, Ding M, Xia M, Chen S, Van Le A,
Soto-Gil R, Shen Y, Wang N, Wang J, Gu W, et al: A five-miRNA panel
identified from a multicentric case-control study serves as a novel
diagnostic tool for ethnically diverse non-small-cell lung cancer
patients. EBioMedicine. 2:1377–1385. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding G, Huang G, Liu HD, Liang HX, Ni YF,
Ding ZH, Ni GY and Hua HW: MiR-199a suppresses the hypoxia-induced
proliferation of non-small cell lung cancer cells through targeting
HIF1α. Mol Cell Biochem. 384:173–180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu Y, Bernauer AM, Yingling CM and
Belinsky SA: HIF1α regulated expression of XPA contributes to
cisplatin resistance in lung cancer. Carcinogenesis. 33:1187–1192.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Eleftheriadis SG, Sivridis E, Koutsopoulos
A, Galatoudis ZG, Chloropoulou PA, Koukourakis MI, Didilis VN,
Bougioukas GI and Giatromanolaki A: One-lung ventilation and
HIF1alpha expression in lung cancer and pneumothorax. Anticancer
Res. 30:1143–1148. 2010.PubMed/NCBI
|
|
44
|
Carbajo-Pescador S, Ordoñez R, Benet M,
Jover R, García-Palomo A, Mauriz JL and González-Gallego J:
Inhibition of VEGF expression through blockade of Hif1α and STAT3
signalling mediates the anti-angiogenic effect of melatonin in
HepG2 liver cancer cells. Br J Cancer. 109:83–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kafousi M, Vrekoussis T, Tsentelierou E,
Pavlakis K, Navrozoglou I, Dousias V, Sanidas E, Tsiftsis D,
Georgoulias V and Stathopoulos EN: Immunohistochemical study of the
angiogenetic network of VEGF, HIF1α, VEGFR-2 and endothelial nitric
oxide synthase (eNOS) in human breast cancer. Pathol Oncol Res.
18:33–41. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Giatromanolaki A, Koukourakis MI, Turley
H, Sivridis E, Harris AL and Gatter KC; Tumour; Angiogenesis
Research Group, : Phosphorylated KDR expression in endometrial
cancer cells relates to HIF1alpha/VEGF pathway and unfavourable
prognosis. Mod Pathol. 19:701–707. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
O'Rourke JF, Pugh CW, Bartlett SM and
Ratcliffe PJ: Identification of hypoxically inducible mRNAs in HeLa
cells using differential-display PCR. Role of hypoxia-inducible
factor-1. Eur J Biochem. 241:403–410. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Nobumoto M, Yamada M, Song S, Inouye S and
Nakazawa A: Mechanism of mitochondrial import of adenylate kinase
isozymes. J Biochem. 123:128–135. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chang X, Ravi R, Pham V, Bedi A,
Chatterjee A and Sidransky D: Adenylate kinase 3 sensitizes cells
to cigarette smoke condensate vapor induced cisplatin resistance.
PLoS One. 6:e208062011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li M, Zhu Y, Zhang H, Li L, He P, Xia H,
Zhang Y and Mao C: Delivery of inhibitor of growth 4 (ING4) gene
significantly inhibits proliferation and invasion and promotes
apoptosis of human osteosarcoma cells. Sci Rep. 4:73802014.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li S, Fan T, Liu H, Chen J, Qin C and Ren
X: Tumor suppressor ING4 overexpression contributes to
proliferation and invasion inhibition in gastric carcinoma by
suppressing the NF-κB signaling pathway. Mol Biol Rep.
40:5723–5732. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Colla S, Tagliaferri S, Morandi F, Lunghi
P, Donofrio G, Martorana D, Mancini C, Lazzaretti M, Mazzera L,
Ravanetti L, et al: The new tumor-suppressor gene inhibitor of
growth family member 4 (ING4) regulates the production of
proangiogenic molecules by myeloma cells and suppresses
hypoxia-inducible factor-1 alpha (HIF-1alpha) activity: Involvement
in myeloma-induced angiogenesis. Blood. 110:4464–4475. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ozer A, Wu LC and Bruick RK: The candidate
tumor suppressor ING4 represses activation of the hypoxia inducible
factor (HIF). Proc Natl Acad Sci USA. 102:7481–7486. 2005.
View Article : Google Scholar : PubMed/NCBI
|