Introduction
The prevalence of heart failure (HF) is >23
million worldwide at present and its number is continuing to
increase (1). Although the
management of HF has improved in the past 3 decades, the 5-year
mortality rate of patients with HF remains high at ~50% (2). A common cause of HF is acute
myocardial infarction (AMI), which often signals the onset of
cardiac dysfunction that may progress to HF (3). This progression is dependent on the
extent of myocardial damage, recurrent ischemia, the development of
myocardial stunning and hibernation, ventricle remodeling and
chronic neuroendocrine stimulation (4). The incidence of HF among patients
hospitalized for an AMI varies among studies, ranging between 14
and 36% (5–7). This highlights the requirement for
early and effective prediction tools and subsequent intervention
following AMI.
Biomarkers such as brain-type natriuretic peptide
and N-terminal pro-brain natriuretic peptide (NT-proBNP) are
associated with ventricle remodeling and the development of HF, and
are widely applied in the clinical diagnosis and prognosis of HF
(8,9); they have a fair prognostic value for
HF in patients with acute coronary syndromes (10). However, these biomarkers lack
specificity, as they are elevated in patients with renal failure,
primary aldosteronism, congestive HF and thyroid disease (11). Genome-wide gene expression
profiling has been extensively used for screening new potential
biomarkers for the diagnosis and/or prediction of disease severity,
such as breast cancer and gastric adenocarcinoma (12,13),
as well as the identification of novel drug targets (14). Maciejak et al (15) have previously utilized this
approach to identify new biomarkers that may have predictive value
for HF following AMI, including ribonuclease A family member 1
pancreatic (RNASE1), formin 1 (FMN1) and Jun dimerization protein 2
(JDP2). However, the dynamic expression changes of the key genes
and associated biological processes in the development of HF
following AMI have not been fully elucidated.
The present study used the microarray data of the
GSE59867 dataset deposited by Maciejak et al (15) to identify time-series
differentially expressed genes (DEGs) and pathways associated with
HF following AMI using comprehensive bioinformatics methods. The
time-series DEGs and their associated functions and pathways were
analyzed. The protein-protein interaction network (PPI) of these
DEGs was subsequently constructed and hub genes were identified.
Results from the current study may provide novel insights into
potential prognostic and therapeutic targets for HF following
AMI.
Materials and methods
Data sources
The gene expression profile data of GSE59867 were
downloaded from the Gene Expression Omnibus database (GEO;
www.ncbi.nlm.nih.gov/geo). The GeneChip
Human Gene 1.0 ST Array (Affymetrix; Thermo Fisher Scientific,
Inc.) platform was used. The dataset contained samples from 111
patients with ST-elevation myocardial infarction at 4 time points:
i) Admission, the day when AMI was confirmed; ii) discharge, 4–6
days post-AMI; iii) 1 month post-AMI; and iv) 6 months post-AMI.
The dataset included samples at the 4 aforementioned time points
from 46 control patients that had stable coronary artery disease on
the day of admission, and no history of myocardial infarction (MI).
Among the 111 patients with ST-elevation MI, 9 patients were
diagnosed with HF and 8 patients were considered no to have HF on
the basis of the first and fourth quartiles of plasma NT-proBNP
level and left ventricular ejection fractions (LVEF) at 6 months
post-AMI. The mean ± standard deviation of the age of the HF and
non-HF patients was 60.1±14.3 and 51.8±7.2, respectively. There
were no significant differences in mean age, sex, body mass index
or history of hypertension, diabetes, smoking,
hypercholesterolemia, MI and AMI. Additionally, there was no
significant difference in aspirin, clopidogrel, β-blockers,
angiotensin-converting-enzyme inhibitors and statins taken by the
two groups of patients. The percentage of HF and non-HF patients
taking diuretics was 77.8 and 12.5%, respectively. The level of
NT-proBNP was 918.3±848.5 and 62.0±14.1 pg/ml in HF and non-HF
patients respectively, while the level of LVEF was 39.3±8.4 and
66.8±1.9%, respectively. There were significant differences in
these three variables between the two groups (Table I). A total of 8 patients with HF
and 6 non-HF patients were followed across the aforementioned 4
time points and were used for subsequent analysis.
 | Table I.Demographic characteristics of HF and
non-HF patients. |
Table I.
Demographic characteristics of HF and
non-HF patients.
| Variables | HF patients
(n=9) | Non-HF patients
(n=8) | P-value |
|---|
| Age, years (mean ±
standard deviation) | 60.1±14.3 | 51.8±7.2 | 0.147 |
| Male sex, n
(%) | 6 (66.7) | 7 (87.5) | 0.576 |
| Body mass index,
kg/m2 (mean ± standard deviation) | 26.8±3.1 | 25.6±1.6 | 0.323 |
| Hypertension, n
(%) | 3 (33.3) | 1 (12.5) | 0.576 |
| Diabetes, n
(%) | 2 (22.2) | 1 (12.5) | >0.999 |
| Smoking, n (%) | 3 (33.3) | 5 (62.5) | 0.347 |
|
Hypercholesterolemia, n (%) | 5 (55.6) | 4 (50.0) | >0.999 |
| Previous MI, n
(%) | 0 (0.0) | 0 (0.0) | NA |
| AMI, n (%) | 8 (88.9) | 3 (37.5) | 0.106 |
| Aspirin, n (%) | 9 (100.0) | 8 (100.0) | NA |
| Clopidogrel, n
(%) | 8 (88.9) | 8 (100.0) | >0.999 |
| β-blockers, n
(%) | 9 (100.0) | 8 (100.0) | NA |
| ACE inhibitors, n
(%) | 9 (100.0) | 8 (100.0) | NA |
| Statins, n (%) | 9 (100.0) | 8 (100.0) | NA |
| Diuretics, n
(%) | 7 (77.8) | 1 (12.5) | 0.015 |
| NT-proBNP, pg/ml
(mean ± standard deviation) | 918.3±848.5 | 62.0±14.1 | <0.001 |
| LVEF, % (mean ±
standard deviation) | 39.3±8.4 | 66.8±1.9 | 0.001 |
Data preprocessing
The raw data were quantile normalized using the
robust multiarray average in the Affy package (www.bioconductor.org/packages/release/bioc/html/affy.html)
(16) in R Bioconductor. The
probes were converted into gene symbols according to probe
annotation information. If multiple probes corresponded to the same
gene symbol, the median was calculated as the gene expression value
of the gene. The probes that matched with multiple genes were
deleted. Additionally, the expression values of the genes with
unknown specific functions were removed.
DEGs analysis
The DEGs in peripheral blood samples from HF
patients at different time points compared with non-HF patients
were identified using the Limma package (www.bioconductor.org/packages/release/bioc/html/limma.html)
(17) in R Bioconductor. A
fold-change of gene expression ratio of >1.5 and P<0.05 were
used as the cut-off criteria. The hierarchical clustering analysis
of these DEGs was performed using Genesis (genome.tugraz.at)
(18). Additionally, a Venn
diagram was created using Venny software (version 2.1; bioinfogp.cnb.csic.es/tools/venny/index.html).
Functional enrichment analysis
To explore the potential biological processes and
pathways that may be involved in patients with post-AMI HF, the
Database for Annotation, Visualization and Integrated Discovery
(DAVID; david.ncifcrf.gov.uk) (19) was used to perform Gene Ontology
(GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analysis for the DEGs. P<0.05 was used as the threshold
value, and the number of genes enriched in each pathway was ≥2. The
differences in biological processes between the non-HF and HF
patients at different time points were investigated.
PPI construction
PPI networks are used to organize all protein-coding
genes into a large network that provides a better understanding of
the functional organization of the proteome (20). The Search Tool for the Retrieval of
Interacting Genes (STRING; string-db.org/cgi/input.pl) (21) database provides information
regarding predicted and experimental interactions of proteins in a
given cell. In the present study, the DEGs were mapped into the
STRING database to identify significant protein pairs with a
combined score of >0.4. The PPI network was subsequently
constructed using Cytoscape software, version 3.6.1 (www.cytoscape.org) (22). The nodes with a higher degree of
interaction were considered as hub genes. Additionally, cluster
analysis for identifying significant function modules with a degree
cutoff >3 in the PPI network was performed using the Molecular
Complex Detection plug-in (23) in
Cytoscape. GO biological process enrichment analysis for the DEGs
in modules of the PPI network was performed using the Biological
Networks GO plug-in (24) in
Cytoscape. P<0.05 was considered to indicate a statistically
significant difference.
Statistical analysis
All continuous data are expressed as the mean ±
standard deviation while categorical data are presented as
frequencies and percentages, and both were analyzed using SPSS
software, version 19.0 (IBM Corp.). Each experiment was repeated
three times. A Student's t-test was used to compare the continuous
data between the two patient groups. Categorical data between the
two patient groups were compared using Fisher's exact test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of candidate DEGs in HF
following AMI
A total of 108 DEGs (46 upregulated and 62
downregulated genes) on admission, 32 DEGs (16 upregulated and 16
downregulated genes) on discharge, 41 DEGs (22 upregulated and 19
downregulated genes) at 1 month post-AMI and 19 DEGs (2 upregulated
and 17 downregulated genes) at 6 months post-AMI were identified
between peripheral blood samples obtained from patients with HF and
non-HF patients (Fig. 1). Among
these DEGs, 4 genes were identified at each of the 4 time points
(Fig. 2A), namely, fatty acid
desaturase 2 (FADS2), leucine-rich repeat neuronal protein 3
(LRRN3), G-protein coupled receptor 15 (GPR15) and adenylate kinase
5 (AK5). Compared with those of samples obtained from non-HF
patients, the gene expression values of the aforementioned 4 genes
were significantly downregulated at the different time points
following AMI in patients with HF (P<0.05, Fig. 2B).
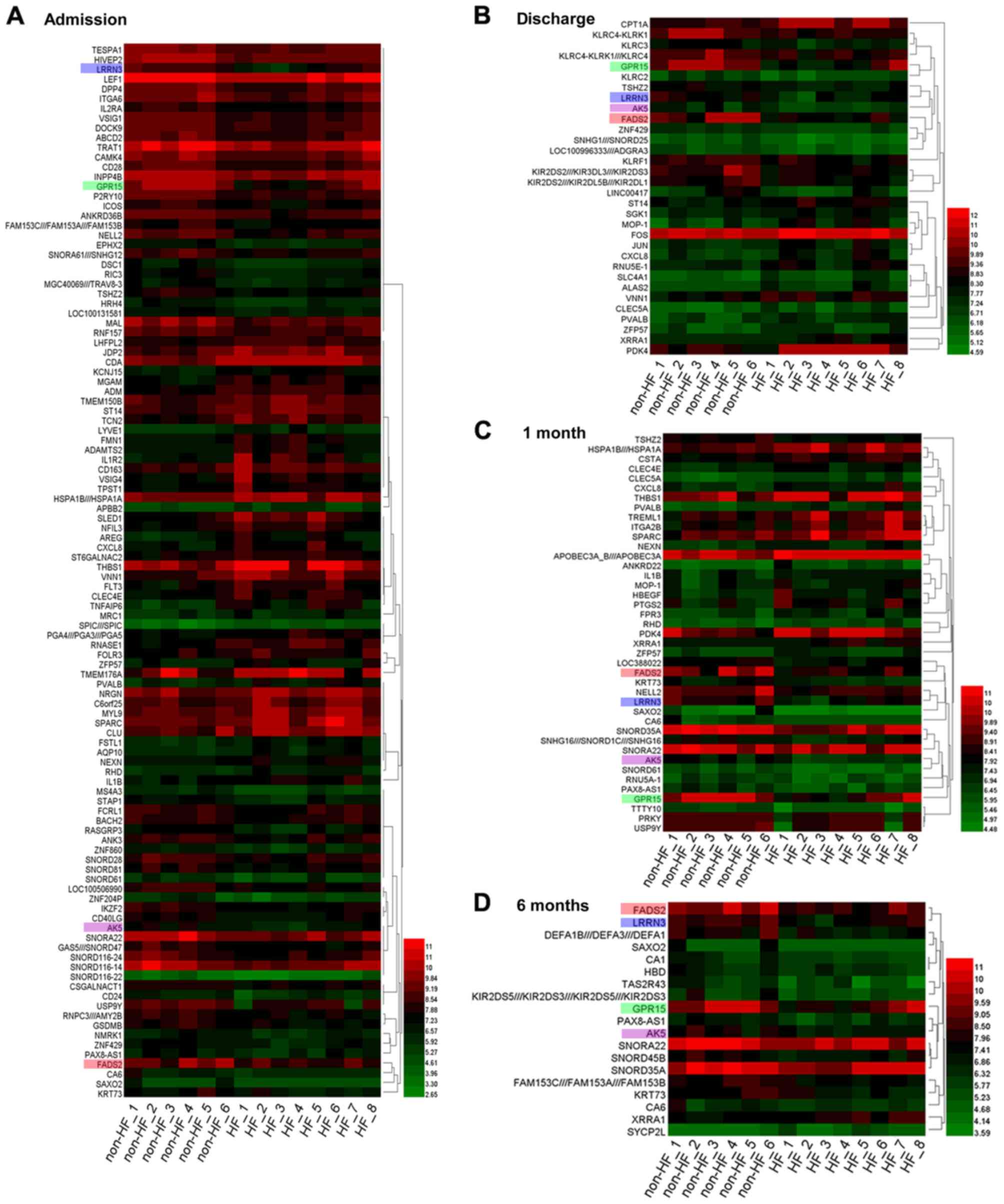 | Figure 1.Heat map of the DEGs identified in HF
and non-HF patients at different time points. DEGs identified (A)
on admission, (B) on discharge, (C) at 1 month post-AMI and (D) at
6 months post-AMI. Green represents lower expression, and red
represents higher expression. A set of 4 genes was identified at
each time point. Each gene is presented by a different color: i)
FADS2 (pink); ii) LRRN3 (blue); iii) GPR15 (green); and iv) AK5
(purple). DEGs, differentially expressed genes; HF, heart failure;
AMI, acute myocardial infarction; FADS2, fatty acid desaturase 2;
LRRN3, leucine-rich repeat neuronal protein 3; GPR15, G-protein
coupled receptor 15; AK5, adenylate kinase 5. |
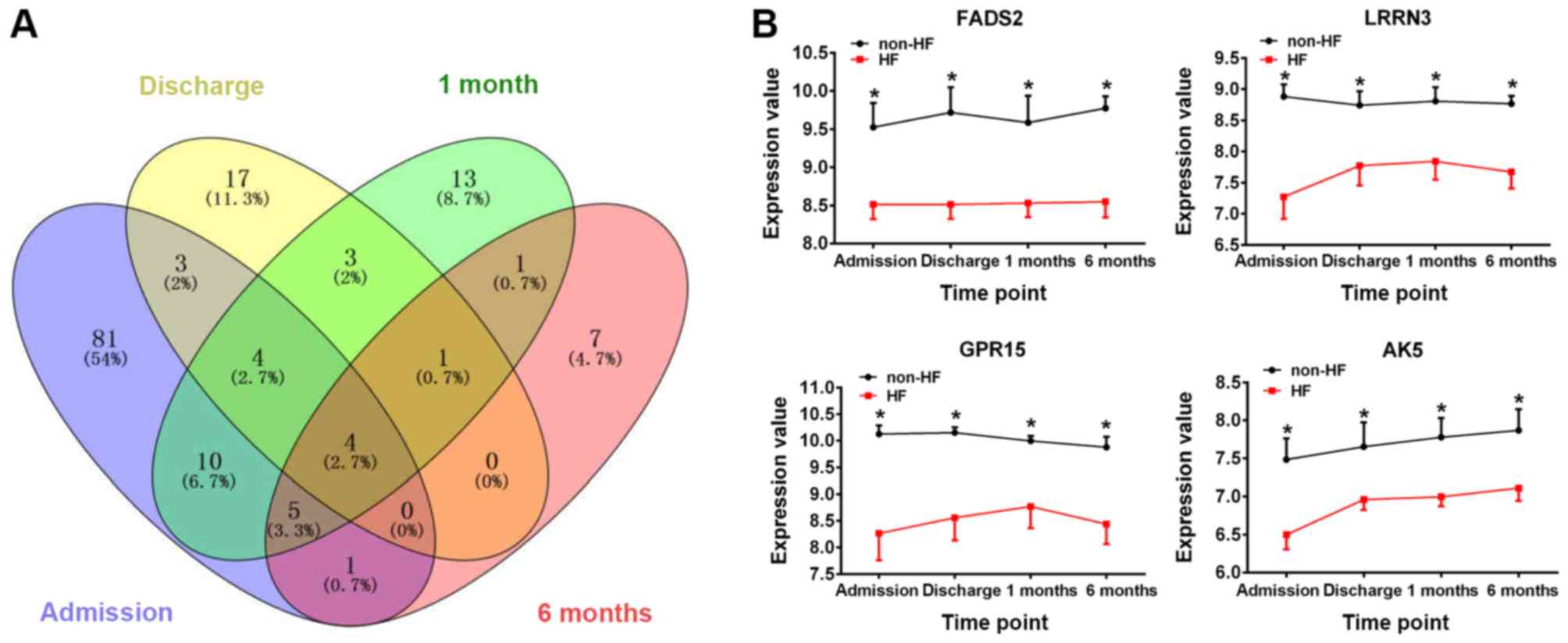 | Figure 2.Identification of DEGs in HF
following AMI. (A) Venn diagram of the DEGs. Different colours
represent different time points: i) Blue, admission; ii) yellow,
discharge; iii) green, 1 month post-AMI; and iv) pink, 6 months
post-AMI. (B) The changes in expression levels of the genes FADS2,
LRRN3, GPR15 and AK5 in HF and non-HF patients at the different
time points. *P<0.05 vs. HF group. DEGs, differentially
expressed genes; HF, heart failure; AMI, acute myocardial
infarction; FADS2, fatty acid desaturase 2; LRRN3, leucine-rich
repeat neuronal protein 3; GPR15, G-protein coupled receptor 15;
AK5, adenylate kinase 5. |
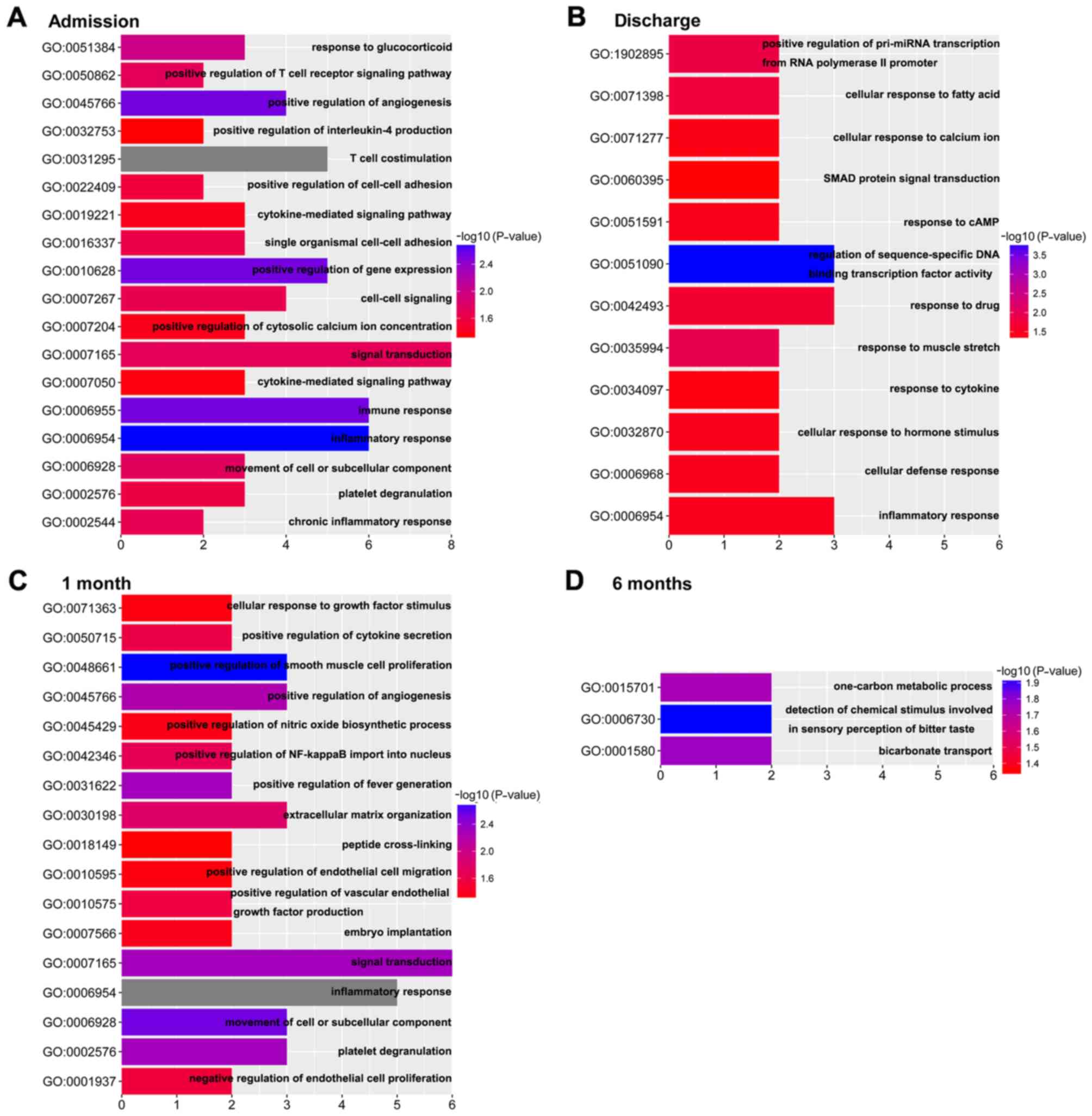
Functional enrichment analysis of
biological processes
The potential functions and pathways of the
time-series DEGs were analyzed using DAVID. The dynamic change of
biological processes in the development of HF following AMI is
presented in Fig. 3. The results
revealed 18 biological processes on admission, 12 biological
processes on discharge, 17 biological processes at 1 month post-AMI
and 3 biological processes at 6 months post- AMI. Compared with the
biological processes at 6 months post-AMI, which is considered the
stable phase (15), the
‘inflammatory response’ and ‘immune response’ were enriched on
admission. On discharge, the biological processes were mainly
enriched in response to stress, such as ‘response to drug’,
‘response to muscle stretch’, ‘cellular response to fatty acid’ and
so on. The biological processes at 1 month post-AMI were mainly
enriched in the regulation of cells and cytokines, including
‘positive regulation of smooth muscle cell proliferation’ and,
‘positive regulation of endothelial cell migration’, ‘cellular
response to growth factor stimulus’ and ‘positive regulation of
vascular endothelial growth factor production’. Notably, biological
processes involved in the ‘inflammatory response’ were enriched in
the first three time points.
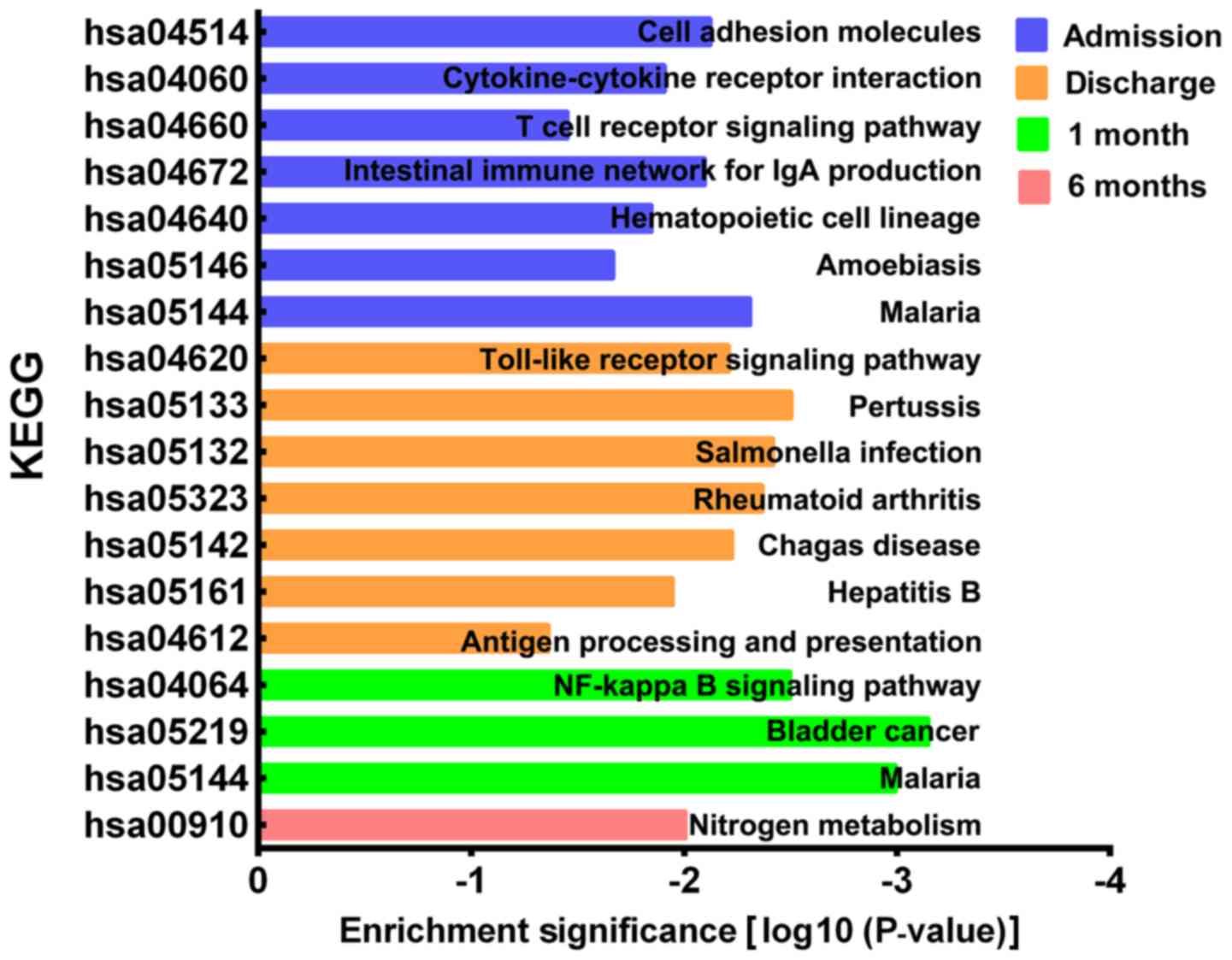
Functional enrichment analysis of KEGG
pathways
The KEGG pathways of the DEGs were analyzed using
DAVID. A total of 7 KEGG pathways were significantly enriched on
admission, including ‘cell adhesion molecules’, ‘cytokine-cytokine
receptor interaction’, ‘T cell receptor signaling pathway’,
‘intestinal immune network for IgA production’, ‘hematopoietic cell
lineage’, ‘amoebiasis’ and ‘malaria’ (Fig. 4). A total of 7 KEGG pathways were
significantly enriched on discharge, including ‘toll-like receptor
signaling pathway’, ‘pertussis’, ‘salmonella infection’,
‘rheumatoid arthritis’, ‘Chagas disease’, ‘hepatitis B’ and
‘antigen processing and presentation’. A total of 3 KEGG pathways
were significantly enriched at 1 month post-AMI and included the
‘NF-κβ signaling pathway’, ‘bladder cancer’ and ‘malaria’. Finally,
1 KEGG pathway, ‘nitrogen metabolism’, was significantly enriched
at 6 months post-AMI.
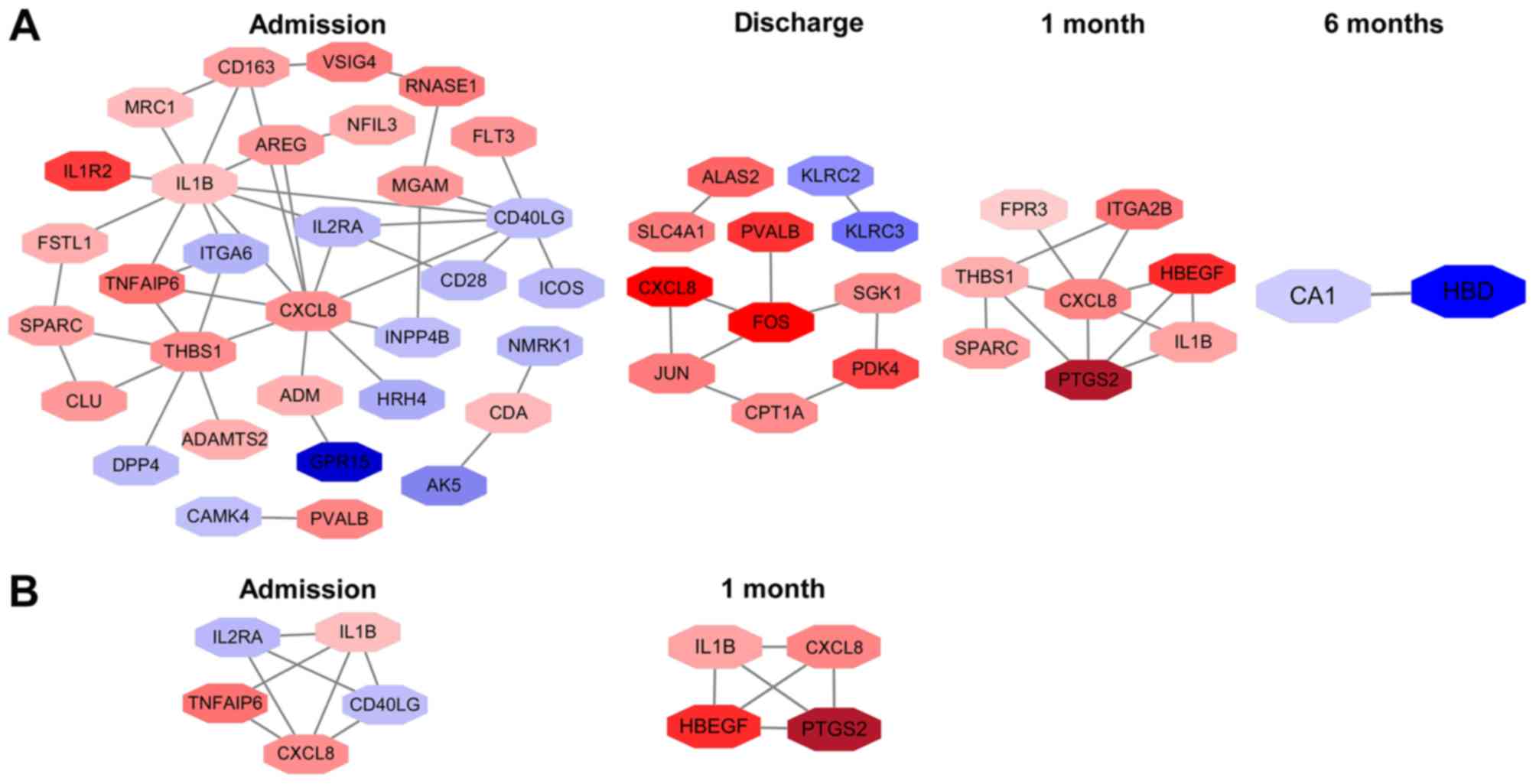
PPI network analysis
A total of 32 nodes and 44 protein pairs on
admission, 11 nodes and 10 protein pairs on discharge, 8 nodes and
12 protein pairs at 1 month post-AMI, and 2 nodes and 1 protein
pair at 6 months post-AMI, with a PPI score of >0.4 based on the
STRING database, were obtained (Fig.
5A). The results revealed that interleukin 1β (IL1β; degree,
10), C-X-C motif chemokine ligand 8 (CXCL8; degree, 10) and
thrombospondin 1 (THBS1; degree, 7) on admission, Fos
proto-oncogene AP-1 transcription factor subunit (degree, 4), Jun
proto-oncogene AP-1 transcription factor subunit (degree, 3) and
CXCL8 (degree, 2) on discharge, and CXCL8 (degree, 6), THBS1
(degree, 4) and prostaglandin-endoperoxide synthase 2 (PTGS2;
degree, 4) at 1 month post-AMI were the top three hub genes in the
first three time points. Notably, CXCL8 was the hub gene identified
in all of the first three time points. Additionally, carbonic
anhydrase (CA1; degree, 1) and hemoglobin subunit δ (HBD; degree,
1) were identified at 6 months post-AMI.
Additionally, 1 module network on admission and 1
module network at 1 month post-AMI were identified. Nodes,
including CXCL8, IL1β, CD40 ligand, tumor necrosis factor α
(TNFα)-induced protein 6 and interleukin 2 receptor subunit α, were
highlighted on admission (Fig.
5B). The highlighted nodes at 1 month post-AMI were CXCL8,
IL1β, PTGS2 and heparin binding EGF-like growth factor. IL1β and
CXCL8 were enriched in the 2 modules. However, no significant
biological processes and pathways were enriched in these module
networks.
Discussion
Maciejak et al (15) identified 3 upregulated genes on
admission following AMI, including RNASE1, FMN1 and JDP2, which
were associated with the increase of NT-proBNP and the decrease of
LVEF at 6 months post AMI. The study demonstrated that these 3
genes may have prognostic value for patients with HF. These 3
upregulated genes were identified in the present study when
comparing patients with HF patients with non-HF patients on
admission. Additionally, the present study identified 4 genes,
namely FADS2, LRRN3, GPR15 and AK5, which were downregulated on
admission, on discharge and at 1 and 6 months post-AMI. This
suggested that these four genes may serve roles in the progression
of HF following AMI and may be potential therapeutic targets for
preventing the development of HF following AMI.
Additionally, by analyzing the gene expression
profiles in peripheral blood samples obtained from patients with HF
patients and non-HF patients at different time points, the present
study identified a series of DEGs enriched in pathways of the
inflammatory response, immune response, the toll-like receptor
signaling pathway and the NF-κβ signaling pathway. CXCL8 and IL1β
were identified as hub genes in the PPI network.
FADS2 is a member of the fatty acid desaturase
family, which are important enzymes in fatty acid metabolism
(25). FADS2, also known as
∆-6-desaturase, is one of the 3 main desaturases in humans, and is
required for the synthesis of highly unsaturated fatty acids (HUFA)
(25). Previous studies indicated
that reduced serum concentrations of omega-3 and omega-6 HUFA were
associated with the progression of HF (26–28).
Alter et al (29) suggested
that increased ventricular wall stress and a reduced LVEF were
associated with decreased docosahexaenoic acid, a HUFA.
Furthermore, a clinical study demonstrated that dietary
supplementation with the HUFA eicosapentaenoic and docosahexaenoic
acids may prevent sudden cardiac death, acute coronary syndrome and
HF (30). The current study
revealed that the expression level of FADS2 was downregulated in
patients with HF when compared with that in patients with non-HF
during the entire follow-up period, suggesting that FADS2 may serve
an important role in the development of HF following AMI.
As a member of the neuronal leucine-rich repeat
family, LRRN3 is a type I transmembrane protein (31). Fukamachi et al (32) reported that LRRN3 may enhance the
phosphorylation of mitogen-activated protein kinase (MAPK) and
therefore activate its signaling pathway in COS-7 cell lines,
originally isolated from the kidney of the African green monkey.
Previous studies demonstrated that the MAPK signaling pathway is
implicated in the development of HF (33–35).
Zhang et al (34) revealed
that the MAPK signaling pathway regulated cardiomyocyte apoptosis
during post-infarction HF in mice. Fei et al (35) revealed that the MAPK signaling
pathway contributed to angiotensin II-induced cardiac fibrosis in
rats. Therefore, LRRN3 may mediate the development of HF
following AMI via the MAPK signaling pathway and its downstream
effectors.
GRP15 is an orphan receptor member of the G-protein
coupled receptor family that was initially identified as a
co-receptor for the human immunodeficiency virus (36). GPR15 was required for the
recruitment of T helper 17, T helper 1 effector and regulatory T
cells in a colitis model in mice (37). Koks et al (38) reported that the expression level of
GPR15 was upregulated in the blood of smokers and served a
potential role in chronic inflammatory pathologies. Pan et
al (39) demonstrated that
GPR15 binds the fifth epidermal growth factor-like region of
thrombomodulin and mediates angiogenesis and protection of vascular
endothelial cells by activating extracellular signal-regulated
kinase and increasing production of anti-apoptotic protein and NO.
Thrombomodulin did not protect vascular endothelial cells isolated
from GPR15 knockout mice from tacrolimus-induced vascular injury
(39). Based on the results
obtained in the aforementioned studies, GRP15 may serve a role in
the progression of HF following AMI.
AK5 belongs to adenylate kinase family (40). Adenylate kinases catalyze the
reversible nucleotide phosphoryl exchange reaction AMP+ATP→2ADP,
and thus control the cellular energy supply (40,41).
Unlike the other members of the adenylate kinase family, AK5 is a
cytosolic isoform highly expressed in the brain, and it serves an
important role in neurogenesis and neuronal-specific metabolism
(40). Lai et al (42) reported that the complex of AK5 and
copine VI was involved in epileptogenesis, which was caused by
neuronal hyperexcitability. However, to the best of our knowledge,
the association between AK5 and HF has not been investigated. The
decreased expression of AK5 in the blood of patients with HF may be
an adaptive response of the brain to ischemic stress present in the
heart. Future studies are required to elucidate the role of AK5 in
the development of HF following AMI.
Prolonged coronary occlusion leads to the death of
cardiomyocytes and damage of the extracellular matrix in the
tissues, triggering activation of an intense inflammatory response
(43). The inflammatory response
following AMI may aid in the repair of the infarcted myocardium
through scar formation to maintain cardiac integrity; however,
excessive inflammatory response may contribute to adverse left
ventricle remodeling and HF (44).
Both human and animal studies demonstrated that the inflammatory
response is involved in the pathogenesis of HF (45,46).
Compared with the healthy controls, patients with HF exhibited
increased levels of circulating inflammatory cytokines, including
tumor necrosis factor (TNF)-α, C-reactive protein, IL1β and IL6
(47–49). Previous studies suggested that
inhibition of IL1 with anakinra or anti-IL1β antibodies may reduce
chamber dilation and improve cardiac dysfunction (50,51).
Furthermore, a clinical trial reported that inhibition of TNF-α
with a recombinant chimeric soluble TNF receptor type 2 had
beneficial effects on cardiac function in patients with chronic HF
(52). The aforementioned studies
suggested that the inflammatory response may be implicated in the
development of HF following AMI.
The immune response was revealed to participate in
positive and negative ventricle remodeling following MI (53,54).
The immune response may synergistically function with the
inflammatory response to repair the infarcted myocardium by
removing dead cardiomyocytes and matrix debris, and producing
mediators that activate fibroblast growth and angiogenesis
(55). However, an overactive
immune response may increase dilative remodeling and result in
chamber dilation, systolic dysfunction and HF in patients surviving
AMI (56). A double blind,
placebo-controlled study of intravenous immunoglobulin induction
and maintenance therapy for 26 weeks resulted in a 5% improvement
of LVEF independent of the etiology of HF in 40 patients with
ischemic and non-ischemic cardiomyopathy (57). Animal experiments revealed that
regulation of the immune response by targeting interferon
regulatory factors may regulate cardiac hypertrophy in vitro
and in vivo (58,59). These studies demonstrate the
potential role of the immune response in the development of HF
following AMI.
Toll-like receptors are transmembrane proteins that
are part of the innate immune system used to identify pathogens and
sense endogenous danger-associated molecular patterns released from
necrotic or dying cells (60).
Toll-like receptor 4, one of the 13 known mammalian toll-like
receptors, may mediate the inflammatory response in the infarcted
heart (44). In a rat model of HF
following AMI, the expression level of toll-like receptor 4 was
found to be upregulated in chronic HF, and blockade of toll-like
receptor 4 improved left ventricle function (61). In addition, the activation of
toll-like receptors triggered a downstream signaling cascade
involving the activation of NF-κβ, interferon regulatory factors,
transcription factors and activator protein 1, leading to the
enhanced expression of a number of inflammatory genes (62). Therefore, the pathways of toll-like
receptor signaling and NF-κβ signaling, which were enriched in the
current study, may be implicated in the development of HF following
AMI.
CXCL8 and IL1β were hub genes analyzed by the PPI
network. CXCL8 belongs to the CXC chemokine family and serves an
important role in mediating neutrophil invasion in a number of
inflammatory processes (63). The
Controlled Rosuvastatin Multinational Trial in Heart Failure study
revealed that increased expression levels of CXCL8 were associated
with adverse outcome in patients with chronic HF (64). Husebye et al (65) demonstrated that high levels of
CXCL8 in patients with ST-elevation myocardial infarction
complicated with HF were associated with poor recovery of left
ventricular function. IL1β is a member of the IL1 cytokine family
and serves important roles in the regulation of cardiac
inflammation and repair (43).
IL1β promotes cardiac fibrosis by secreting cytokines, chemokines
and matrix metalloproteinases (66). Furthermore, increased plasma IL1β
levels have been associated with impaired myocardial function and
left ventricle hypertrophy following reperfusion after MI (67). Based on the results obtained in the
aforementioned studies, the upregulated expression levels of CXCL8
and IL1β may contribute to the development of HF following AMI.
In the present study, time-series DEGs between
peripheral blood samples obtained from HF and non-HF patients were
identified. A number of DEGs, including FADS2, LRRN3, GPR15, AK5,
CXCL8 and IL1β, were identified as possible target genes for the
development of HF following AMI. Furthermore, the present study
revealed that biological processes involved in the inflammatory and
immune responses and the toll-like receptor and NF-κβ signaling
pathways may serve important roles in the development of HF
following AMI. However, future experimental studies are required to
substantiate the results obtained in the present study.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available in the National Center of Biotechnology
Information's GEO database (www.ncbi.nlm.nih.gov/geo) with the accession number
GSE59867.
Authors' contributions
XL and HJ conceived and designed the study. BL and
XL performed the data analysis. XL, BL and HJ wrote the paper. All
authors approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AMI
|
acute myocardial infarction
|
|
AK5
|
adenylate kinase 5
|
|
CXCL8
|
C-X-C motif chemokine ligand 8
|
|
DEGs
|
differentially expressed genes
|
|
DAVID
|
Database for Annotation, Visualization
and Integrated Discovery
|
|
FADS2
|
fatty acid desaturase 2
|
|
FMN1
|
formin 1
|
|
GPR15
|
G-protein coupled receptor 15
|
|
GO
|
gene ontology
|
|
HF
|
heart failure
|
|
HUFA
|
highly unsaturated fatty acids
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
IL
|
interleukin
|
|
JDP2
|
Jun dimerization protein 2
|
|
LRRN3
|
leucine-rich repeat neuronal protein
3
|
|
LVEF
|
left ventricular ejection
fractions
|
|
MAPK
|
mitogen-activated protein kinase
|
|
NT-proBNP
|
N-terminal pro-brain natriuretic
peptide
|
|
PPI
|
protein-protein interaction
|
|
PTGS2
|
prostaglandin-endoperoxide synthase
2
|
|
RNASE1
|
ribonuclease A family member 1
|
|
STRING
|
search tool for the retrieval of
interacting genes
|
|
THBS1
|
thrombospondin 1
|
|
TNFα
|
tumor necrosis factor α
|
References
|
1
|
Roger VL: Epidemiology of heart failure.
Circ Res. 113:646–659. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roger VL, Weston SA, Redfield MM,
Hellermann-Homan JP, Killian J, Yawn BP and Jacobsen SJ: Trends in
heart failure incidence and survival in a community-based
population. JAMA. 292:344–350. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hoydal MA, Kirkeby-Garstad I, Karevold A,
Wiseth R, Haaverstad R, Wahba A, Stølen TL, Contu R, Condorelli G,
Ellingsen Ø, et al: Human cardiomyocyte calcium handling and
transverse tubules in mid-stage of post-myocardial-infarction heart
failure. ESC Heart Fail. 5:332–342. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McMurray JJ, Adamopoulos S, Anker SD,
Auricchio A, Böhm M, Dickstein K, Falk V, Filippatos G, Fonseca C,
Gomez-Sanchez MA, et al: ESC Guidelines for the diagnosis and
treatment of acute and chronic heart failure 2012: The Task Force
for the Diagnosis and Treatment of Acute and Chronic Heart Failure
2012 of the European Society of Cardiology. Developed in
collaboration with the Heart Failure Association (HFA) of the ESC.
Eur Heart J. 33:1787–1847. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hellermann JP, Jacobsen SJ, Gersh BJ,
Rodeheffer RJ, Reeder GS and Roger VL: Heart failure after
myocardial infarction: A review. Am J Med. 113:324–330. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Borghi C, Bacchelli S, Degli Esposti D and
Ambrosioni E; Survival of Myocardial Infarction Long-Term
Evaluation Study, : Effects of early angiotensin-converting enzyme
inhibition in patients with non-ST-elevation acute anterior
myocardial infarction. Am Heart J. 152:470–477. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
O'Connor CM, Hathaway WR, Bates ER,
Leimberger JD, Sigmon KN, Kereiakes DJ, George BS, Samaha JK,
Abbottsmith CW, Candela RJ, et al: Clinical characteristics and
long-term outcome of patients in whom congestive heart failure
develops after thrombolytic therapy for acute myocardial
infarction: Development of a predictive model. Am Heart J.
133:663–673. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
de Lemos JA, Morrow DA, Bentley JH, Omland
T, Sabatine MS, McCabe CH, Hall C, Cannon CP and Braunwald E: The
prognostic value of B-type natriuretic peptide in patients with
acute coronary syndromes. N Engl J Med. 345:1014–1021. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Haeck JD, Verouden NJ, Kuijt WJ, Koch KT,
Van Straalen JP, Fischer J, Groenink M, Bilodeau L, Tijssen JG,
Krucoff MW and De Winter RJ: Comparison of usefulness of N-terminal
pro-brain natriuretic peptide as an independent predictor of
cardiac function among admission cardiac serum biomarkers in
patients with anterior wall versus nonanterior wall ST-segment
elevation myocardial infarction undergoing primary percutaneous
coronary intervention. Am J Cardiol. 105:1065–1069. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tousoulis D, Kampoli AM, Stefanadi E,
Antoniades C, Siasos G, Papavassiliou AG and Stefanadis C: New
biochemical markers in acute coronary syndromes. Curr Med Chem.
15:1288–1296. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moe KT and Wong P: Current trends in
diagnostic biomarkers of acute coronary syndrome. Ann Acad Med
Singapore. 39:210–215. 2010.PubMed/NCBI
|
|
12
|
Hall PA, Reis-Filho JS, Tomlinson IP and
Poulsom R: An introduction to genes, genomes and disease. J Pathol.
220:109–113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heidecker B and Hare JM: The use of
transcriptomic biomarkers for personalized medicine. Heart Fail
Rev. 12:1–11. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gurwitz D: Expression profiling: A
cost-effective biomarker discovery tool for the personal genome
era. Genome Med. 5:412013. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maciejak A, Kiliszek M, Michalak M, Tulacz
D, Opolski G, Matlak K, Dobrzycki S, Segiet A, Gora M and Burzynska
B: Gene expression profiling reveals potential prognostic
biomarkers associated with the progression of heart failure. Genome
Med. 7:262015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sturn A, Quackenbush J and Trajanoski Z:
Genesis: Cluster analysis of microarray data. Bioinformatics.
18:207–208. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for Annotation,
Visualization, and Integrated Discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stelzl U, Worm U, Lalowski M, Haenig C,
Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A,
Koeppen S, et al: A human protein-protein interaction network: A
resource for annotating the proteome. Cell. 122:957–968. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li SW, Lin K, Ma P, Zhang ZL, Zhou YD, Lu
SY, Zhou X and Liu SM: FADS gene polymorphisms confer the risk of
coronary artery disease in a Chinese Han population through the
altered desaturase activities: Based on high-resolution melting
analysis. PLoS One. 8:e558692013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rupp H, Rupp TP, Alter P and Maisch B: N-3
polyunsaturated fatty acids and statins in heart failure. Lancet.
373:378–380. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rupp H, Rupp TP, Alter P and Maisch B:
Inverse shift in serum polyunsaturated and monounsaturated fatty
acids is associated with adverse dilatation of the heart. Heart.
96:595–598. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rupp H, Rupp TP, Alter P and Maisch B:
Mechanisms involved in the differential reduction of omega-3 and
omega-6 highly unsaturated fatty acids by structural heart disease
resulting in ‘HUFA deficiency’. Can J Physiol Pharmacol. 90:55–73.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alter P, Gluck T, Figiel JH, Koczulla AR,
Vogelmeier CF and Rupp H: From heart failure to highly unsaturated
fatty acid deficiency and vice versa: Bidirectional heart and liver
interactions. Can J Cardiol. 32:217–225. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Anderson JS, Nettleton JA, Hundley WG,
Tsai MY, Steffen LM, Lemaitre RN, Siscovick D, Lima J, Prince MR
and Herrington D: Associations of plasma phospholipid omega-6 and
omega-3 polyunsaturated Fatty Acid levels and MRI measures of
cardiovascular structure and function: The multiethnic study of
atherosclerosis. J Nutr Metab. 2011:3151342011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hamano S, Ohira M, Isogai E, Nakada K and
Nakagawara A: Identification of novel human neuronal leucine-rich
repeat (hNLRR) family genes and inverse association of expression
of Nbla10449/hNLRR-1 and Nbla10677/hNLRR-3 with the prognosis of
primary neuroblastomas. Int J Oncol. 24:1457–1466. 2004.PubMed/NCBI
|
|
32
|
Fukamachi K, Matsuoka Y, Ohno H, Hamaguchi
T and Tsuda H: Neuronal leucine-rich repeat protein-3 amplifies
MAPK activation by epidermal growth factor through a
carboxyl-terminal region containing endocytosis motifs. J Biol
Chem. 277:43549–43552. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yokota T and Wang Y: p38 MAP kinases in
the heart. Gene. 575:369–376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Q, Lu L, Liang T, Liu M, Wang ZL and
Zhang PY: MAPK pathway regulated the cardiomyocyte apoptosis in
mice with post-infarction heart failure. Bratisl Lek Listy.
118:339–346. 2017.PubMed/NCBI
|
|
35
|
Fei AH, Wang FC, Wu ZB and Pan SM:
Phosphocreatine attenuates angiotensin II-induced cardiac fibrosis
in rat cardiomyocytes through modulation of MAPK and NF-κB pathway.
Eur Rev Med Pharmacol Sci. 20:2726–2733. 2016.PubMed/NCBI
|
|
36
|
Deng HK, Unutmaz D, KewalRamani VN and
Littman DR: Expression cloning of new receptors used by simian and
human immunodeficiency viruses. Nature. 388:296–300. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nguyen LP, Pan J, Dinh TT, Hadeiba H,
O'Hara E III, Ebtikar A, Hertweck A, Gökmen MR, Lord GM, Jenner RG,
et al: Role and species-specific expression of colon T cell homing
receptor GPR15 in colitis. Nat Immunol. 16:207–213. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Koks G, Uudelepp ML, Limbach M, Peterson
P, Reimann E and Koks S: Smoking-induced expression of the GPR15
gene indicates its potential role in chronic inflammatory
pathologies. Am J Pathol. 185:2898–2906. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pan B, Wang X, Nishioka C, Honda G,
Yokoyama A, Zeng L, Xu K and Ikezoe T: G-protein coupled receptor
15 mediates angiogenesis and cytoprotective function of
thrombomodulin. Sci Rep. 7:6922017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dzeja P and Terzic A: Adenylate kinase and
AMP signaling networks: Metabolic monitoring, signal communication
and body energy sensing. Int J Mol Sci. 10:1729–1772. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hardie DG and Hawley SA: AMP-activated
protein kinase: The energy charge hypothesis revisited. Bioessays.
23:1112–1119. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lai Y, Hu X, Chen G, Wang X and Zhu B:
Down-regulation of adenylate kinase 5 in temporal lobe epilepsy
patients and rat model. J Neurol Sci. 366:20–26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang S and Frangogiannis NG:
Anti-inflammatory therapies in myocardial infarction: Failures,
hopes and challenges. Br J Pharmacol. 175:1377–1400. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fang L, Moore XL, Dart AM and Wang LM:
Systemic inflammatory response following acute myocardial
infarction. J Geriatr Cardiol. 12:305–312. 2015.PubMed/NCBI
|
|
45
|
Nakayama H and Otsu K: Translation of
hemodynamic stress to sterile inflammation in the heart. Trends
Endocrinol Metab. 24:546–553. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yndestad A, Damas JK, Oie E, Ueland T,
Gullestad L and Aukrust P: Systemic inflammation in heart
failure-the whys and wherefores. Heart Fail Rev. 11:83–92. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Levine B, Kalman J, Mayer L, Fillit HM and
Packer M: Elevated circulating levels of tumor necrosis factor in
severe chronic heart failure. N Engl J Med. 323:236–241. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pye M, Rae AP and Cobbe SM: Study of serum
C-reactive protein concentration in cardiac failure. Br Heart J.
63:228–230. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Testa M, Yeh M, Lee P, Fanelli R,
Loperfido F, Berman JW and LeJemtel TH: Circulating levels of
cytokines and their endogenous modulators in patients with mild to
severe congestive heart failure due to coronary artery disease or
hypertension. J Am Coll Cardiol. 28:964–971. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Abbate A, Salloum FN, Vecile E, Das A,
Hoke NN, Straino S, Biondi-Zoccai GG, Houser JE, Qureshi IZ, Ownby
ED, et al: Anakinra, a recombinant human interleukin-1 receptor
antagonist, inhibits apoptosis in experimental acute myocardial
infarction. Circulation. 117:2670–2683. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Toldo S, Mezzaroma E, Van Tassell BW,
Farkas D, Marchetti C, Voelkel NF and Abbate A: Interleukin-1β
blockade improves cardiac remodelling after myocardial infarction
without interrupting the inflammasome in the mouse. Exp Physiol.
98:734–745. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Deswal A, Bozkurt B, Seta Y,
Parilti-Eiswirth S, Hayes FA, Blosch C and Mann DL: Safety and
efficacy of a soluble P75 tumor necrosis factor receptor (Enbrel,
etanercept) in patients with advanced heart failure. Circulation.
99:3224–3226. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Frieler RA and Mortensen RM: Immune cell
and other noncardiomyocyte regulation of cardiac hypertrophy and
remodeling. Circulation. 131:1019–1030. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nahrendorf M and Swirski FK: Innate immune
cells in ischaemic heart disease: Does myocardial infarction beget
myocardial infarction? Eur Heart J. 37:868–872. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Frangogiannis NG: The immune system and
cardiac repair. Pharmacol Res. 58:88–111. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Frangogiannis NG: The immune system and
the remodeling infarcted heart: Cell biological insights and
therapeutic opportunities. J Cardiovasc Pharmacol. 63:185–195.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gullestad L, Aass H, Fjeld JG, Wikeby L,
Andreassen AK, Ihlen H, Simonsen S, Kjekshus J, Nitter-Hauge S,
Ueland T, et al: Immunomodulating therapy with intravenous
immunoglobulin in patients with chronic heart failure. Circulation.
103:220–225. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang DS, Li L, Huang L, Gong J, Xia H,
Liu X, Wan N, Wei X, Zhu X, Chen Y, et al: Interferon regulatory
factor 1 is required for cardiac remodeling in response to pressure
overload. Hypertension. 64:77–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Jiang DS, Wei X, Zhang XF, Liu Y, Zhang Y,
Chen K, Gao L, Zhou H, Zhu XH, Liu PP, et al: IRF8 suppresses
pathological cardiac remodelling by inhibiting calcineurin
signalling. Nat Commun. 5:33032014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yang Y, Lv J, Jiang S, Ma Z, Wang D, Hu W,
Deng C, Fan C, Di S, Sun Y and Yi W: The emerging role of Toll-like
receptor 4 in myocardial inflammation. Cell Death Dis. 7:e22342016.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu L, Wang Y, Cao ZY, Wang MM, Liu XM,
Gao T, Hu QK, Yuan WJ and Lin L: Up-regulated TLR4 in
cardiomyocytes exacerbates heart failure after long-term myocardial
infarction. J Cell Mol Med. 19:2728–2740. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mann DL, Topkara VK, Evans S and Barger
PM: Innate immunity in the adult mammalian heart: For whom the cell
tolls. Trans Am Clin Climatol Assoc. 121:34–51. 2010.PubMed/NCBI
|
|
63
|
de Winter RJ, Manten A, de Jong YP, Adams
R, van Deventer SJ and Lie KI: Interleukin 8 released after acute
myocardial infarction is mainly bound to erythrocytes. Heart.
78:598–602. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Nymo SH, Hulthe J, Ueland T, McMurray J,
Wikstrand J, Askevold ET, Yndestad A, Gullestad L and Aukrust P:
Inflammatory cytokines in chronic heart failure: Interleukin-8 is
associated with adverse outcome. Results from CORONA. Eur J Heart
Fail. 16:68–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Husebye T, Eritsland J, Arnesen H,
Bjørnerheim R, Mangschau A, Seljeflot I and Andersen GØ:
Association of interleukin 8 and myocardial recovery in patients
with ST-elevation myocardial infarction complicated by acute heart
failure. PLoS One. 9:e1123592014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Saxena A, Chen W, Su Y, Rai V, Uche OU, Li
N and Frangogiannis NG: IL-1 induces proinflammatory leukocyte
infiltration and regulates fibroblast phenotype in the infarcted
myocardium. J Immunol. 191:4838–4848. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Orn S, Ueland T, Manhenke C, Sandanger Ø,
Godang K, Yndestad A, Mollnes TE, Dickstein K and Aukrust P:
Increased interleukin-1β levels are associated with left
ventricular hypertrophy and remodelling following acute ST segment
elevation myocardial infarction treated by primary percutaneous
coronary intervention. J Intern Med. 272:267–276. 2012. View Article : Google Scholar : PubMed/NCBI
|