Introduction
Cancer chemotherapy uses drugs to prevent the growth
of cancerous cells or kill cancerous cells in the human body
(1). In general, chemotherapy can
be applied for three primary purposes: As an adjuvant therapy, in
order to prevent the cancer cells from re-emerging following
initial surgery or radiation; as a neo-adjuvant therapy, shrinking
the tumour size for its easier removal with surgery; and as a
treatment for metastatic disease, to reduce the number of cancer
cells and kill cells that have spread to other parts of the body
from the primary cancer location, such as to the lymph nodes under
the arm in patients with breast cancer.
General chemotherapeutic drugs that are used to
treat breast cancer include the following: Anthracyclines, such as
doxorubicin (Adriamycin) and epirubicin (Ellence); taxanes, such as
paclitaxel (Taxol) and docetaxel (Taxotere); cyclophosphamide
(Cytoxan), capecitabine (Xeloda) and 5-fluorouracil; vinorelbine
(Navelbine), gemcitabine (Gemzar), trastuzumab (Herceptin) and
other anti-hormone drugs, as well as breast cancer drugs that
target human epidermal growth factor receptor-2 (2). Breast cancer chemotherapy is commonly
administered orally or by intravenous injection daily (3). In adjuvant and neo-adjuvant settings,
chemotherapeutic drugs are usually given as a combination of two or
more drugs, since single-drug chemotherapy is less effective
(3); however, the toxicity of
combined chemotherapy is also greater if the treatment programme is
not planned appropriately. An inappropriate combination of
chemotherapy may not be able to treat or reduce the spread of
breast cancer, and may continuously destroy other dividing cells
and affect surrounding healthy tissue. Indeed, the majority of the
current chemotherapies cause pain and adverse effects in patients,
including nausea and vomiting, loss of appetite, fatigue, mouth
soreness, hair loss, weight gain, premature menopause, reduced
resistance to infections and increased bleeding (4–6).
Therefore, it is important to seek effective treatment strategies
or combination therapies for breast cancer using novel
compounds/substances from natural products, which frequently have
reduced toxicity in comparison with traditional chemotherapies.
Naturally-occurring anti-cancer products may reduce pain, while
preventing the spread of cancer cells to other parts of the
body.
The conventional drug screening process can be
divided into two main stages: Discovery and development (7–11).
The discovery stage can be further divided into early discovery,
lead identification and lead optimization before the selected
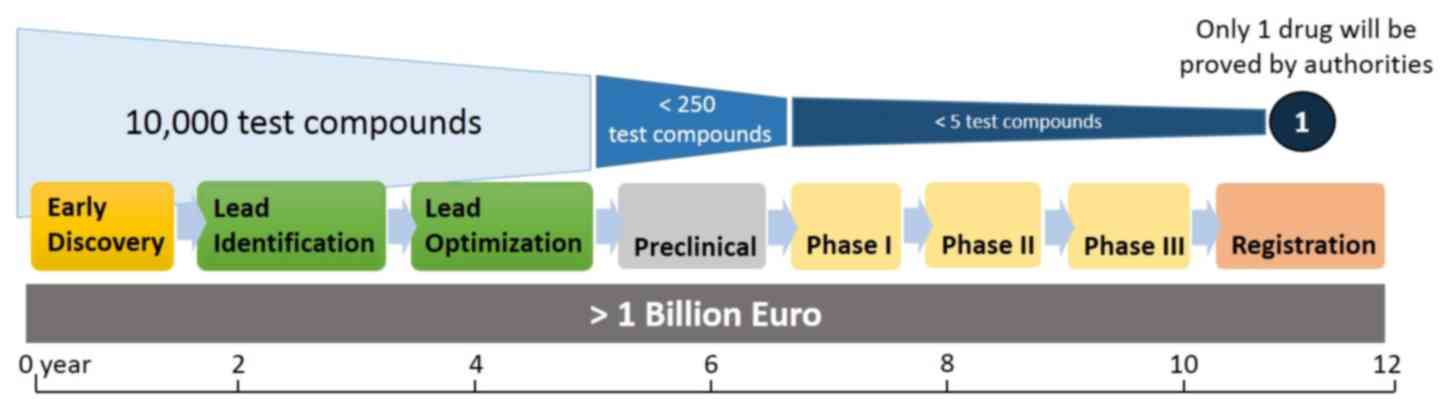
candidates proceed to the preclinical screening stages (Fig. 1). Early discovery is the research
phase of the screening process, during which thousands of potential
compounds are extracted and synthesised from various resources
annually, with the hope that certain of these compounds may possess
promising therapeutic effects. According to the Pharmaceutical
Research and Manufacturers of America (12), only one drug is selected from every
10,000 test compounds by the end of the screening process, with the
entire process costing >1 billion euros and requiring ~12 years
to complete. Therefore, the initial in vitro steps in the
screening process are particularly important, and increasing the
speed of the early high-throughput process to identify the specific
desired effects of compounds is crucial. As such, the current study
aimed to develop a novel screening assay to accelerate the
identification of specific compounds prior to animal studies and
preclinical stages.
Popular in vitro strategy to identify
preliminary growth inhibitory effects of potential agents
frequently involves the use of cell-based proliferation assays,
which include secondary metabolite detection in conditioned medium
using tetrazolium salts, such as the MTT assay; cell membrane
damage detection by assessing dehydrogenase release from damaged
cells; DNA fragmentation detection via an in situ
5′-bromo-2′-deoxyuridine assay; and other assays using cell
staining and flow cytometry. Conducting these assays requires
costly laboratory facilities and cell culture expertise, as the
culture is sensitive to impurities in the tested agents, which may
cause contamination unless the crude extracts used are dissolved in
an antimicrobial solvent, such as dimethyl sulfoxide. Furthermore,
these assays rely on slow-growing cell lines, in which multiple
passages may also change the genotype and phenotype of the cells.
Thus, a cost-effective approach is required to overcome these
limitations, as subjecting all unidentified compounds to cell-based
screening in the early screening process is costly.
The present study aimed to develop a yeast-based
screening assay using Pichia pastoris that was transformed
with a plasmid expressing DNA topoisomerase I (TOPOI), namely
SMD1168H-TOPOI. DNA topoisomerase is involved in cell
proliferation, and thus overexpression of this enzyme in yeast
enhances the proliferation, mimicking cancer cells. The use of a
yeast-based assay is more versatile, as it allows for the screening
of a larger number of compounds without the need for cell culture
facilities and expertise, while producing similar findings that are
comparable to the cell-based assays. Therefore, such an assay
results in a faster preliminary screening process. Only candidate
compounds that exhibit a positive effect at the early discovery
stages will then proceed to the next steps of the drug discovery
process.
Materials and methods
Preliminary design of the yeast-based
assay
A Pichia pastoris strain clone of SMD1168H
carrying TOPOI in a pPICZαA plasmid was generated in our previous
study (13), and was referred to
as SMD1168H-TOPOI. This clone was used for the development of the
yeast-based screening assay in the present study. The assay was
designed to have the following characteristics: i) Easy to operate
with no special skill required; ii) no specific equipment is
required, and can be performed with a simple laboratory set-up, for
instance using only a shaker flask system; iii) the yeast cell
density is the main component of the assay, therefore, no
additional detection kit, enzyme or reagent is needed other than
the chemical, medium and reagent to maintain the yeast cells; iv) a
short amount of time is required to complete the full assay (<1
week), and the assay can be performed at room temperature or at
least in a laboratory equipped with an air-conditioner; and v)
produces results that are comparable to cell-based screening
assays.
Yeast cultivation for the cell density
measurement
Yeast culture stocks (SMD1168H, SMD1168H-pPICZαA and
SMD1168H-TOPOI), which were constructed and stored in glycerol at
−80°C as previously described (13), were retrieved and enriched using 5
ml buffered glycerol-complex medium (BMGY) in a universal bottle.
The transformed yeast strain was incubated overnight at 15–20°C in
an incubator shaker at 250 rpm. Subsequently, 250 µl of the
overnight culture was transferred into a 250 ml conical flask that
contained 25 ml fresh BMGY. The culture was then incubated in the
shaker for another 16 h at 15–20°C with agitation at 250 rpm, until
the exponential growth phase was reached. The growth of the yeast
cells was induced using 1.0% (v/v) methanol. After 12 h of
incubation, culture medium with 100 µM glutamate or L-glutamic acid
(97% purity; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany; serving
as a growth agonist) was added, and cells were incubated for 96 h.
A small volume of the sample was withdrawn from the treated culture
every 12 h for cell density measurement, and the yeast cell density
was expected to be enhanced under these conditions. An optional
step involved replenishing the same volume of the medium containing
the drug solution in order to maintain the original culture volume.
Throughout the assay, methanol was added every 24 h. Alternatively,
camptothecin (97% purity; Sigma-Aldrich; Merck KGaA; serving as a
growth antagonist) was added to the culture instead of glutamate.
Similar to glutamate-treated cells, samples were withdrawn from the
yeast cell culture treated with 100 µM camptothecin every 12 h for
cell density measurement, and the cell density was expected to be
inhibited under these culture conditions.
Measurement of the glutamate- and
camptothecin-treated yeast cell density by spectrophotometry
The cell density (culture turbidity) of the
collected samples was assessed at an optical density of 600 nm
every 12 h for 72 h using a spectrophotometer, and the turbidity
level in each sample was recorded. The value of the samples was
then compared with the value of the background control. Next, the
growth profile of the yeast at each time-point was plotted. The
density (unit) of the yeast culture was expected to grow
continuously or to be reduced by ~40% following treatment with
glutamate or camptothecin, respectively, for 96 h compared with the
density of the background control and relative to the measurement
at 12 h of cultivation. The normal yeast (SMD1168H) and the yeast
transformed with only the pPICZαA plasmid (SMD1168H-pPICZαA;
without the inserted gene) were used as the controls. Subsequently,
the overall response of the yeast culture treated with camptothecin
was compared with the performance of the drug in a cell-based MTT
assay.
Analysis of the inhibitory effect on
camptothecin-treated cell lines by MTT assay
As SMD1168H-TOPOI was expected to mimic cancer
cells, an MTT assay was conducted to examine the effect of
camptothecin on various human cell lines and compare it with the
effect in yeast cells. The cell lines used in this assay included
highly metastatic MDA-MB-231 breast cancer cells, weakly metastatic
CAL-27 oral cancer and MCF-7 breast cancer cells, bone
marrow-derived mesenchymal stem cells and MCF-10a normal breast
cells. All cell lines used in the present study were previously
purchased from the American Type Culture Collection (Manassas, VA,
USA) and cultured in respective growth medium in the laboratory.
The cancerous cells were maintained in high glucose Dulbecco's
Modified Eagle's medium (DMEM) supplemented with 10% Fetal bovine
serum (FBS) and 100 µg/ml streptomycin/penicillin, whereas the
non-cancerous cells were maintained in DMEM/F12 medium supplemented
with 100 mg/ml epidermal growth factor, 1 mg/ml hydrocortisone, 10
mg/ml insulin, 10% FBS and 100 µg/ml penicillin/streptomycin. The
aforementioned culture reagents were all purchased from Gibco
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). Briefly,
2×104 cells/ml of each cell line were seeded in 96-well
plates and routinely cultured in a humidified incubator at 37°C
with 5% CO2 for 24 h or until the cells reached ~70%
confluence. Subsequently, the cells were treated with increasing
concentrations (0–100 µg/ml) of camptothecin solution for 24, 48
and 72 h. At the end of each incubation period, 24 µl MTT reagent
(5 mg/ml; Sigma-Aldrich; Merck KGaA) was added to each well, and
the reaction was incubated for 4 h. The solution was carefully
removed without disturbing the formazan crystals that had formed in
each well. Subsequently, 100 µl acidified isopropanol was added to
each well and agitated for homogeneous colour development.
Following colour development, the intensity of the colour in the
plate was measured at 570 nm using an ELISA plate reader. Dose
response curves were generated from cell viability (%) plotted
against the logarithmic scale (Log) concentration of drug used. The
responses of camptothecin in cell-based and yeast-based assays were
then compared.
Statistical analysis
All graphs were generated and statistical analysis
of data was performed using one-way analysis of variance via
Friedman test by GraphPad Prism software (version 7.04 for Windows;
GraphPad Software, Inc., La Jolla, CA, USA). Subsequent to the
initial measurement/analysis, the experiments were repeated twice
for cell density measurement (n=3 in total) and three times for
cell inhibitory effect analysis (n=4 in total) to confirm the
consistency, repeatability and reproducibility of the results. All
values are expressed as the mean ± standard deviation. P<0.05
was considered to indicate a statistically significant
difference.
Results
Cell density of untreated SMD1168H and
its recombinant forms
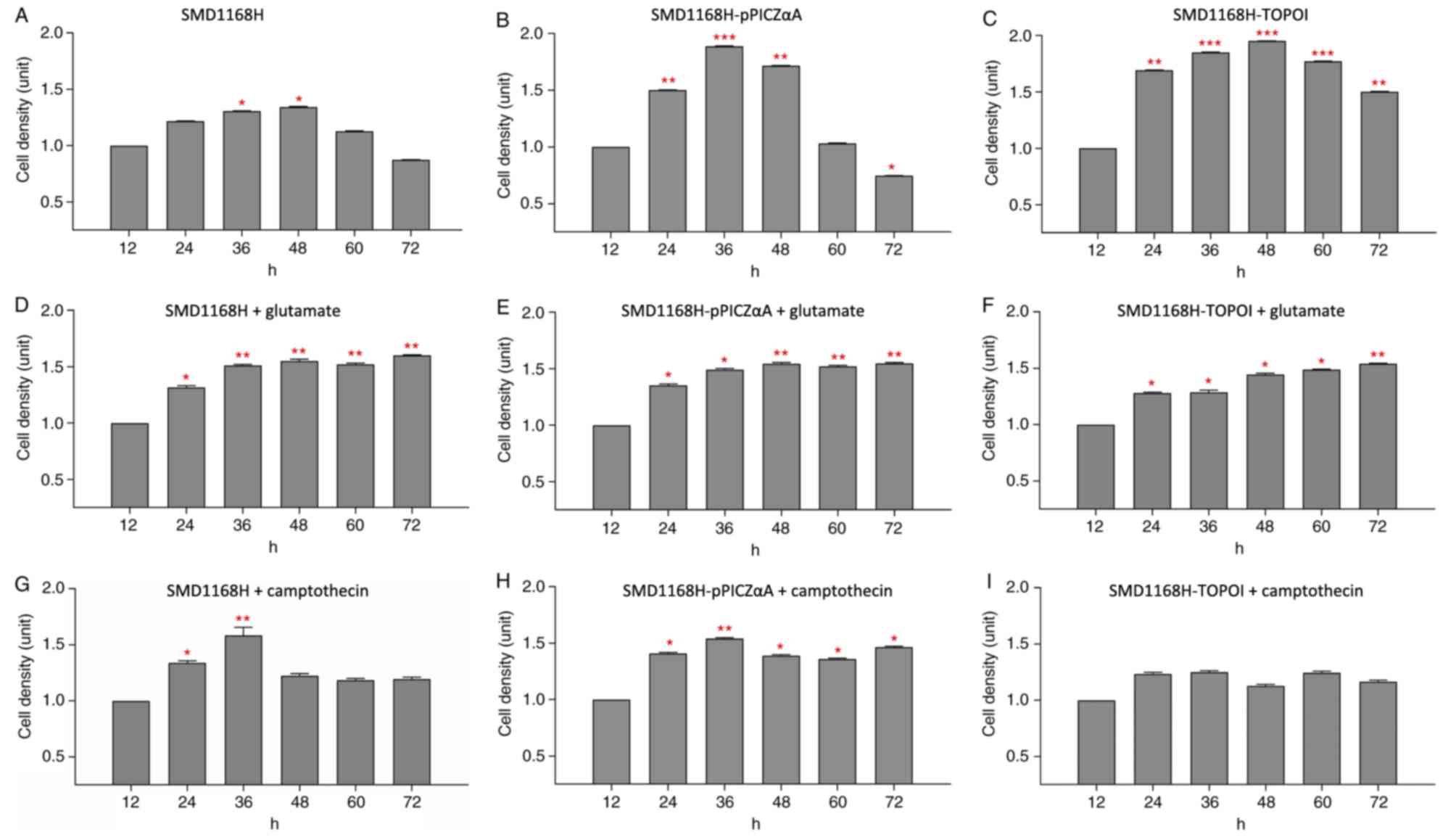
The cell density of the yeast strain SMD1168H was
gradually increased after 36 h of cultivation (1.306 units;
P<0.05) and reached a peak value of 1.343 units at ~48 h
(P<0.05; Fig. 2A). Following
this peak, the cell density was reduced from 60 h of cultivation
and onwards, with the yeast growth reaching the lowest level of
0.873 units at 72 h. A similar growth profile was observed when
SMD1168H was transformed with the empty pPICZαA plasmid
(SMD1168H-pPICZαA) and maintained in the culture without treatment
(Fig. 2B). The cell density was
significantly increased at 24 h (1.502 units; P<0.01), reaching
an optimum growth level of 1.888 units at 36 h (P<0.001) and
then reduced to 1.717 units at 48 h (P<0.01). The cell density
reached the lowest level of 0.747 units at 72 h (P<0.05), which
was lower than the minimal level observed for normal SMD1168H. This
phenomenon may be due to the transformation of the plasmid vector
into the yeast, causing the cells to be more fragile. By contrast,
a different growth curve was observed when the SMD1168H was
transformed with the pPICZαA plasmid carrying the TOPOI gene
(SMD1168H-TOPOI). A significantly higher cell density was observed
at 24 h (1.692 units; P<0.01), 36 h (1.851 units; P<0.001)
and 48 h (1.953 units; P<0.001) of cultivation, and the yeast
growth remained high at later time points (Fig. 2C). Although the growth of the yeast
did slightly reduce from 60 h onwards, the cells did not reach the
lowest level of growth observed in the other yeast strains. The
cell density at 60 h (1.774 units; P<0.001) and 72 h (1.504
units; P<0.01) remained significantly higher compared with the
12 h group, indicating that the transformed TOPOI gene enhanced the
growth of the yeast.
Cell density of glutamate-treated
SMD1168H and its recombinant forms
As shown in Fig.
2D, SMD1168H treated with 100 µM glutamate exhibited a
significantly higher cell density of 1.316 units at 24 h compared
with at 12 h. The cell density of the glutamate-treated SMD1168H
reached an optimum level of 1.511 units at ~36 h (P<0.01) and
was maintained at approximately this level until 72 h of
cultivation. The cell densities were 1.551 units (P<0.01), 1.520
units (P<0.01) and 1.600 units (P<0.01) at 48, 60 and 72 h of
cultivation, respectively. Similarly, in the glutamate-treated
SMD1168H-pPICZαA, the growth of the yeast reached 1.351 units at 24
h (P<0.05) and an optimum level at 1.492 units at 36 h of
cultivation (P<0.05), and the cells continuously grew at this
level until 72 h of cultivation (Fig.
2E). The cell densities were 1.545 units (P<0.01), 1.520
units (P<0.01) and 1.546 units (P<0.01) at 48, 60 and 72 h,
respectively. However, the growth profile of the glutamate-treated
SMD1168H-TOPOI was not observed to be as expected. The recombinant
yeast grew steadily during the early cultivation, and the cell
densities were 1.280 units (P<0.05), 1.288 units (P<0.05) and
1.446 units (P<0.05) at 24, 36 and 48 h of cultivation,
respectively (Fig. 2F). The cell
density reached 1.487 units at 60 h of cultivation, similar to the
SMD1168H and SMD1168H-pPICZαA, with the optimum level of 1.540
units observed at 72 h of cultivation (P<0.01). This phenomenon
indicates that using both glutamate and TOPOI in the culture may
create competition among the components. As such, the growth of
cells was slightly stunted, which resulted in a delay in reaching
the optimum growth level. The growth of these three yeast types
clearly responded to glutamate treatment in the culture, indicating
that the recombinant yeast may be useful in estimating the effect
of other potential agents for cancer treatment.
Cell density of camptothecin-treated
SMD1168H and its recombinant forms
SMD1168H treated with 100 µM camptothecin for 72 h
exhibited a similar optimum level of cell density as
glutamate-treated SMD1168H before 48 h of cultivation. The treated
yeast was still capable of maintaining a cell density of 1.340
units at 24 h of cultivation (P<0.05) and an optimum level of
1.583 units at ~36 h of cultivation (P<0.01; Fig. 2G). However, the cell density was
subsequently reduced to ~1.225 units, and the growth was gradually
maintained at this level until 72 h. In the camptothecin-treated
SMD1168H-pPICZαA, a similar growth profile was observed, with a
cell density of 1.408 units at 24 h (P<0.05) and a peak levels
of 1.540 units being reached at 36 h (P<0.01; Fig. 2H). The growth of the yeast was
reduced to ~1.389 units and growth continued at this level until 72
h of cultivation. Notably, camptothecin did not induce the same
inhibitory effects in SMD1168H-TOPOI as that demonstrated in the
other two yeast clones. Despite a markedly lower cell density in
the yeast expressing the camptothecin target (TOPOI) at 24 and 36 h
compared with SMD1168H and SMD1168H-pPICZαA, the recombinant yeast
grew steadily at a cell density of ~1.170 units throughout the
experiment, and the growth of the recombinant yeast was not
inhibited by camptothecin treatment, with no statistically
significant changes observed (Fig.
2I). Camptothecin treatment of SMD1168H-TOPOI was expected to
reduce the yeast cell density; however, this treatment did not
entirely inhibit the yeast growth, indicating the aggressiveness of
the SMD1168H-TOPOI, which may be comparable to that of highly
metastatic human cancer cells.
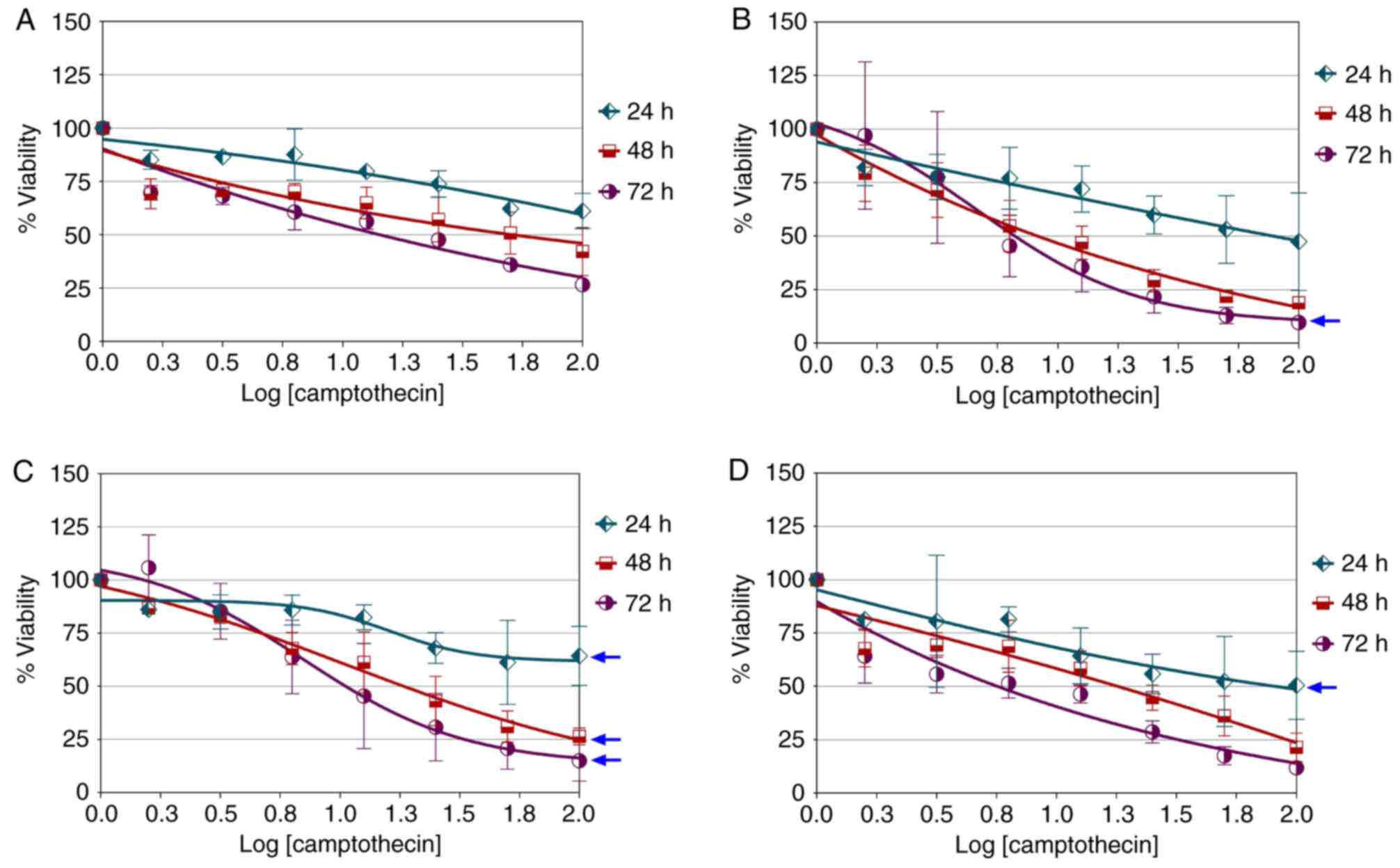
Camptothecin dose-response in various
cell lines
Treatment of MCF-10a cells with camptothecin for 72
h did not produce a typical dose-response curve (Fig. 3A). Similarly, no typical
dose-response was observed in CAL-27 cells following treatment with
camptothecin over 48 h. Camptothecin only produced a logistic
dose-response in CAL-27 cells when treatment was performed for 72 h
(hillslope=−1.29; maximal response, ≤25%; Fig. 3B). The maximal dose-response of
camptothecin in MCF-7 cells was observed to improve with treatment
for 24 h (hillslope=−2.42; maximal response, ≤50%), 48 h
(hillslope=−0.65; maximal response, ≤25%) and 72 h
(hillslope=−1.28; maximal response, ≤25%; Fig. 3C). In general, chemosensitivity to
camptothecin was better in MCF-7 cells in comparison with CAL-27
cells, although similar dose response was observed in MCF-7
(hillslope=−1.28; maximal response, ≤25%) and CAL-27
(hillslope=−1.29; maximal response, ≤25%) cells treated for 72 h.
As shown in Fig. 3D, camptothecin
only produced a typical dose-response in MDA-MB-231 cells treated
for 24 h (hillslope=−0.37; maximal response, ≤50%). However, the
effect was not sustained after 24 h of treatment. The
semi-parabolic growth profile, which is a general growth curve that
conveys resistance of an organism to a particular drug treatment,
was produced when treatment was performed for 72 h. This phenomenon
indicates that reduced growth at 48 h of treatment and onwards may
be due to by-product accumulation in the culture. These preliminary
findings indicate that there was a considerable similarity in the
initial response of MDB-MB-231 cells and the recombinant yeast
SMD1168H-TOPOI to camptothecin treatment.
Discussion
The present study demonstrated that common growth
profiles were observed in untreated SMD1168H and SMD1168H-pPICZαA,
whereas growth enhancement was observed in SMD1168H that had been
transformed with the TOPOI gene via a pPICZαA plasmid (namely the
SMD1168H-TOPOI). Glutamate enhanced the growth of the SMD1168H and
SMD1168H-pPICZαA; however, when combined with TOPOI expression in
the culture, this treatment did not multiply the growth effect
further. By contrast, camptothecin reduced the cell growth of the
SMD1168H after 36 h of cultivation, but showed no effect on the
SMD1168H-pPICZαA. The cell density was notably lower during the
early treatment of the yeast expressing TOPOI with camptothecin
compared with the other yeast strains; however, the cell growth
remained stable throughout the experiment. This phenomenon
indicates that it was difficult to reduce the growth of the
transformed SMD1168H-TOPOI, with a similar phenomenon observed in
the highly metastatic MDA-MB-231 breast cancer cells treated with
camptothecin.
SMD1168H-TOPOI was developed in our laboratory as a
newly simplified prototype for potential use in anti-growth
compound screening. Cell density or growth inhibition can be used
as the parameter for measuring the effect of a compound on the
yeast. This technique may be useful for screening potential growth
inhibitors in the future. A similar strategy has been used to
enable high-throughput screening for novel pharmacological
modulators of K+ channels. In a previous study, an assay
was developed based on the growth of yeast that functionally
expresses mammalian Kir2.1 channels (14). In another study, a less demanding
phenotypic yeast-based assay on 96-well microplates was established
using a methylamine-sensitive yeast strain, in which a
methylamine-permeable aquaporin was expressed to rescue
proliferation on selection plates, whereby specific inhibition of
the aquaporin directly correlated with reduced cell proliferation
(15). Engineered-fission yeast
strains have also been used in high-throughput screening for
phosphodiesterase (PDE) inhibitors that possess ‘drug-like’
characteristics over a 48-h period by assessing the growth
behaviour of the yeast to determine the activity of heterologous
cyclic nucleotide PDEs (16). This
system could be used to screen cDNA libraries for biological
regulators of target PDEs, and to identify whether different PDE
protein complexes exhibit distinct patterns of inhibitor
sensitivity. To be more specific, a high-throughput yeast-based
growth assay has also been used to screen for potential PDE11
inhibitors; however, identifying compounds that inhibit PDE11 is
required, as the biological roles of the enzyme are poorly
understood (17). In addition, a
robust yeast-based growth assay that is potentially applicable to
numerous equilibrative nucleoside transporters (ENTs) was developed
in order to identify inhibitors of ENT1 of the malarial parasite
Plasmodium falciparum (PfENT1) (18). These inhibitors were detected based
on their ability to rescue the growth of PfENT1-expressing fui1Δ
yeast in the presence of a cytotoxic PfENT1 substrate,
5-fluorouridine. The data supported the hypothesis that blocking
purine transport through PfENT1 may be a novel and compelling
approach for antimalarial drug development (18). By contrast, the expression of
Xenopus cyclin A1 was induced in a yeast-based growth
interference assay in order to elevate the activity of cell
division cycle 28 (Cdc28) kinase in the yeast (19). The hyperactivation of Cdc28 kinase
in yeast resulted in growth arrest, and thus compounds that can
rescue the cyclin A1-induced growth arrest may be potential novel
antitumour drug candidates that act on the cyclin-dependent
kinase-mediated cell cycle regulation pathways.
By assessing growth behaviour, the enzymes that are
targeted in cells do not need to be purified at specific time
points for the screening purposes. This strategy provides a simple
solution that meets general laboratory needs without requiring
specific facilities or equipment to carry out the preliminary
screening approach for potential growth inhibitors. Indeed, no
additional detection kits, enzymes or reagents are required other
than the chemical, medium and reagent required to maintain the
recombinant yeast. The assay can ideally be used for screening
synthetic compounds, plant extracts, nanoparticles, small molecules
and chemical compounds extracted from natural products, including
flavonoids. Only those agents that produce a positive effect in the
yeast-based approach will then proceed to the subsequent steps,
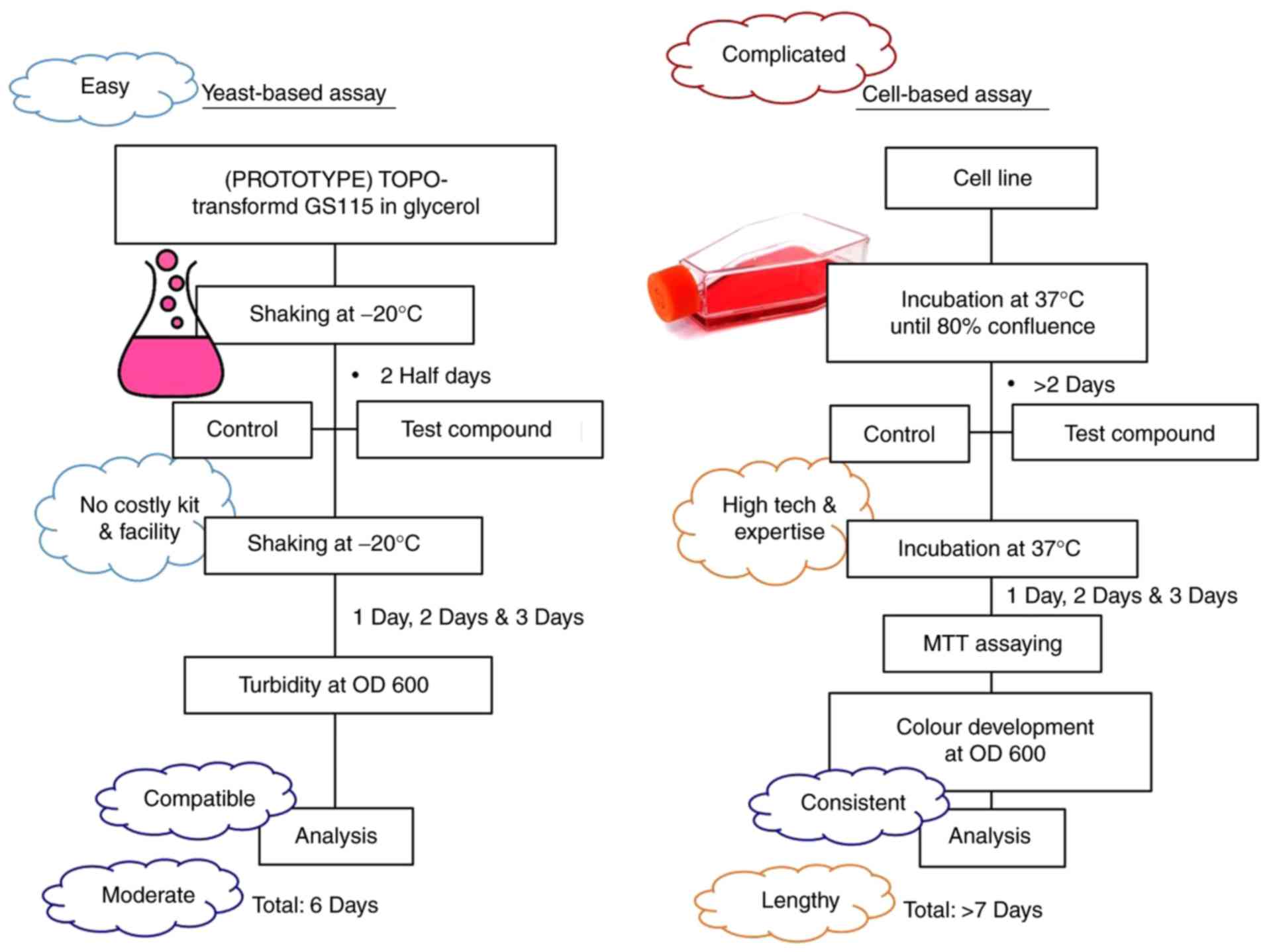
which require more costly gel-based or cell-based assays. To the
best of our knowledge, the majority of cell-based assays typically
require >7 days to be accomplished, whereas the yeast-based
assay requires only 6 days (Fig.
4), which reduces the duration of the screening process by
~15%. As such, this may save months of work in the 12 years
required to develop a drug from the original 10,000 test compounds.
Therefore, the yeast-based strategy saves time, money and
resources, and makes the screening procedures more practical,
versatile and competitive in the market, as well as more affordable
for the researchers in low-resource laboratories.
The yeast-based strategy described in the present
study was developed when constructing Pichia with multiple
copy numbers of TOPOI for the expression and purification of the
target enzyme (13,20). GS115 and SMD1168 yeasts were found
to be better than Pichia strains in accommodating the
exogenous recombinant TOPOI expression, and an enzyme activity of
~3.02×105 U/l of crude culture was obtained in the
recombinant yeast; however, only SMD1168 was able to stably express
the enzyme in the culture supernatant at room temperature (13). This prototype development is based
on our present original research, and provides innovation and
novelty to screening processes within the medical biotechnology
sector.
Yeast expressing higher levels of TOPOI exhibit
similarities to highly metastatic human cancer cells, such as
MDA-MB-231 (21,22); they exhibit similarly robust cell
growth, as determined by respective growth inhibition assays. As
such, this SMD1168H-TOPOI can be used as a cost-effective
yeast-based assay to conduct high-throughput screening for
target-specific growth inhibitors and accelerate the identification
of potential compounds. Novel antitumour drugs that have reduced
toxicity compared with current chemotherapies are required and may
improve the treatment of patients with breast cancer. A similar
strategy has previously been conducted using a simplified
yeast-based screening approach to search for activators of
caspase-3 and caspase-7 (23).
This was followed by evaluation of the activity of the selected
compounds in two human tumour cell lines, including the acute
promyelocytic leukaemia HL-60 and breast adenocarcinoma MCF-7
cells. This proof of principle strategy demonstrates the
effectiveness of the yeast assays in the discovery of caspase
activators, which may pave the way for a new class of caspase
activators with improved anticancer properties.
Similar yeast-based assays have been used as
platforms for screening and identifying various types of compounds.
For instance, a rapid two-step yeast-based assay was developed to
screen for anti-prion drugs (24).
This assay was used to identify compounds effective against budding
yeast prions that are responsible for the [PSI+] and
[URE3] phenotypes, and was an efficient high-throughput screening
approach for the identification of novel prion inhibitors. An
additional robust high-throughput yeast-based assay to determine
potential genotoxicity and mutagenicity of drug candidates early in
the discovery phase of drug development was created to replace the
most widely used Ames Salmonella test, which is not readily
adaptable for high-throughput screening, for the assessment of
mutagenicity and genotoxicity. The yeast-based system assaying DNA
repair was able to detect genotoxicity, which incorporated
metabolic activation (25). This
assay is efficient, requires little time and small amounts of the
compound, while it is adaptable to a high-throughput platform and
yields data that accurately and reproducibly detect DNA damage
based on metabolic activation. Furthermore, the use of genetically
tractable model yeast as a vehicle for target-based high-throughput
screening has overcome numerous limitations of in vitro
biochemical and phenotypic assay platforms for drug discovery by
allowing the identification of on-target compounds that function
within a eukaryotic cellular context (26).
A yeast-based assay for monitoring GAr-dependent
inhibition of translation was also established and identified
doxorubicin as a compound that specifically interferes with the GAr
effect on translation in yeast (27). This approach was, thus, validated
as an effective high-throughput screening assay for the
identification of drugs that interfere with Epstein-Barr virus
(EBV) immune evasion and may be candidates for treating
EBV-associated diseases, including cancer. Another yeast-based
system for identifying and screening inhibitors against coronavirus
N7-methyltransferase (MTase) was developed using 96-well and
384-well microtiter plates in order to examine MTase inhibitors
previously identified using in vitro biochemical assays,
such as sinefungin, which effectively suppressed N7-MTase (28). These results validated the
application of a yeast-based assay system for inhibitor screening,
while also demonstrating the difference between in vitro and
cell-based biochemical assays, whereby more potent inhibitors
reducing the activity of coronavirus based on the human N7-MTase
can be identified.
In addition to coronavirus, the development of a
fully automated anti-parasitic drug-screening yeast system has been
reported, which allows for multiple parasite targets to be assessed
simultaneously due to the expression of different fluorescent
proteins in the yeast strain. Compounds that specifically target
parasite enzymes can be selected through this assay, rather than
their host counterparts, thus enabling the early elimination of
compounds that carry potential side effects (29). In this system, compounds that
cannot discriminate between the host and parasite enzymes are
excluded by including a strain expressing the human target in the
multiplexed screen. The advantages of this type of assay include
the use of known targets and the lack of requirement for in
vitro culture of parasites.
Rapid yeast-based assays can also be established for
screening active drugs against human inherited mitochondrial
diseases affecting ATP synthase, in particular ataxia, neuropathy
and retinitis pigmentosa syndrome (30). The unique ability of yeast to
survive without the production of ATP by oxidative phosphorylation
was used to identify chlorhexidine by screening a chemical library
and oleate through a candidate approach (30). Furthermore, the high-throughput
yeast-based screening bioassays have also been used to detect
selective oestrogen receptor modulators and selective androgen
receptor (AR) modulators (31), to
screen commercial chemical libraries to identify PDE7 inhibitors
(32), to robustly identify new
lead compounds targeting specific enzymes and to detect the
toxicity of human CA isozyme II (hCAII) inhibitors (33), which can be achieved using a
multidrug-sensitive derivative of the Δnce103 strain expressing a
low level of hCAII. Finally, the use of yeast present the advantage
that it is a relevant surrogate model for eukaryotic cell processes
that can be miniaturised and automated.
To the best of our knowledge, the majority of
screening assays are developed using Saccharomyces (S.)
cerevisiae. For instance, a miniaturised short-term in
vivo genotoxicity screening assay based on genetically modified
S. cerevisiae was performed to explore the chronic
cytotoxicity and genotoxic effect of compounds in a eukaryotic
organism (34). In another
example, an efficient and reliable yeast-based detection system was
created to evaluate the androgenic activity of endocrine disruptors
from pulp and paper mill effluents (35). This system used S.
cerevisiae transformed with β-galactosidase genes, and the
reporter expression was driven by human AR and response elements,
in order to identify compounds that altered the reporter gene
induction by testosterone. The findings of the assay suggested that
the pulp and paper mill effluents are rich in androgenic chemicals,
and this detection system could be applicable as a primary
screening method for inhibitors/activators of AR functions
(35). S. cerevisiae has
also been used to screen for polymorphisms of human genes, such as
heat shock protein 90, which is essential for cell proliferation in
budding yeast (36). Speed and low
cost make yeast-based assays a useful tool for identifying human
polymorphisms and proteins. In addition, a sensitive, fast and
user-friendly progesterone receptor (PR) transactivation assay was
established using recombinant S. cerevisiae that was
modified to express green fluorescent protein driven by human PR
and progesterone response element (37). Stimulation of cells with increasing
concentrations of progesterone resulted in a significant elevation
in fluorescence activity, and this yeast-based bioassay provided a
robust and rapid method for high-throughput screening of
(anti)-progestative compounds from various sources. Another S.
cerevisiae model system has also been used to investigate the
regulation of human BRAFV600E (38). Under osmotic stress conditions,
hBRAFV600E can rescue the growth of strains carrying a double or
triple mitogen-activated protein kinase deletion in high osmolarity
glycerol. The results of this previous study demonstrated the
potential of using S. cerevisiae to investigate hBRAFV600E,
identify its functional interactors and, in doing so, uncover new
cancer-associated genes with therapeutic potential. In brief, live
yeast cell-based assays are rapid, inexpensive, sensitive and
amenable to high-throughput methods that can be used for a variety
of applications, including isolation of novel genes, directed
evolution and gene-specific drug screening, and will facilitate
various approaches in numerous research areas.
In the current study, Pichia was used as the
host to express TOPOI, rather than Saccharomyces, due to the
simplicity of the techniques required for molecular genetic
manipulation and the similarity to Saccharomyces (39). Furthermore, the ability of
Pichia to produce foreign proteins at a higher level,
intracellularly and extracellularly, and the ability to perform
eukaryotic post-translational modifications make this yeast strain
more suitable to produce proteins for human applications. The
SMD1168H-TOPOI also offers a competitive and low-cost strategy for
potential growth inhibitor screening that may be accessible in
low-resource laboratory settings, while still enabling a
high-throughput screening process. TOPOI is the focus of the
present study; this enzyme is a general target for screening
potential growth inhibitors that can be used as effective compounds
for combined use with breast cancer chemotherapy drugs, since
various anticancer drugs, including camptothecin, are TOPOI
inhibitors.
In conclusion, the current study developed a
preliminary form of a yeast-based system that is cost-effective,
fast, easy to operate and efficient for compound screening. This
system is expected to overcome certain limitations of the
cell-based proliferation assays, while maintaining similar
screening functions. Although yeast-based bioassays have been
established as powerful approaches to identify potential
therapeutic compounds, creating robust models that are amenable to
high-throughput screening remains a challenge. Therefore, further
studies should focus in this area to ensure that the function of
the assay can be implemented effectively in the future.
Acknowledgements
Not applicable.
Funding
The authors would like to thank the Ministry of
Higher Education of Malaysia for the scholarship awarded to the
students working on this project. The authors would also like to
thank Jian Xin for providing sponsorship during the preparation of
this manuscript.
Availability of data and materials
The datasets generated and/or analysed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
BYK and JX conceived and designed the study. BYK
drafted the manuscript and revised it critically for important
intellectual content. WNAWM, PCS and NM conducted the experiments
and data acquisition, interpreted the results and analysed all data
under the supervision of BYK. All authors have read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Brearley MJ: Chemotherapy. Feline Soft
Tissue and General Surgery. Langley-Hobbs SJ, Demetriou JL and
Ladlow JF: Elsevier Ltd.; Amsterdam: pp. 161–167. 2014, View Article : Google Scholar
|
|
2
|
Lukong KE: Understanding breast cancer-The
long and winding road. BBA Clin. 7:64–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cimino GD, Pan CX and Henderson PT:
Personalized medicine for targeted and platinum-based chemotherapy
of lung and bladder cancer. Bioanalysis. 5:369–391. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Suzuki H, Asakawa A, Amitani H, Nakamura N
and Inui A: Cancer cachexia-pathophysiology and management. J
Gastroenterol. 48:574–594. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferioli M, Zauli G, Martelli AM, Vitale M,
McCubrey JA, Ultimo S, Capitani S and Neri LM: Impact of physical
exercise in cancer survivors during and after antineoplastic
treatments. Oncotarget. 9:14005–14034. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilkes GM: Targeted therapy: Attacking
cancer with molecular and immunological targeted agents. Asia Pac J
Oncol Nurs. 5:137–155. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cragg GM, Boyd MR, Grever MR and Schepartz
SA: Pharmaceutical prospecting and the potential for pharmaceutical
crops. Natural product drug discovery and development at the United
States National Cancer Institute. Ann Missouri Bot Gard. 82:47–53.
1995. View
Article : Google Scholar
|
|
8
|
Hermiston TW and Kirn DH: Genetically
based therapeutics for cancer: Similarities and contrasts with
traditional drug discovery and development. Mol Ther. 11:496–507.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hughes JP, Rees S, Kalindjian SB and
Philpott KL: Principles of early drug discovery. Br J Pharmacol.
162:1239–1249. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Katiyar C, Gupta A, Kanjilal S and Katiyar
S: Drug discovery from plant sources: An integrated approach. Ayu.
33:10–19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan SY, Zhou SF, Gao SH, Yu ZL, Zhang SF,
Tang MK, Sun JN, Ma DL, Han YF, Fong WF and Ko KM: New perspectives
on how to discover drugs from herbal medicines: CAM's outstanding
contribution to modern therapeutics. Evid Based Complement Alternat
Med. 2013:6273752013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pharmaceutical Research and Manufacturers
of America: Pharmaceutical Industry Profile 2010. PhRMA;
Washington, DC: March. 2010
|
|
13
|
Chan MK, Lim SK, Miswan N, Chew AL,
Noordin R and Khoo BY: Expression of stable and active human DNA
topoisomerase I in Pichia pastoris. Protein Expr Purif.
141:52–62. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zaks-Makhina E, Kim Y, Aizenman E and
Levitan ES: Novel neuroprotective K+ channel inhibitor identified
by high-throughput screening in yeast. Mol Pharmacol. 65:214–219.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu B, Altmann K, Barzel I, Krehan S and
Beitz E: A yeast-based phenotypic screen for aquaporin inhibitors.
Pflugers Arch. 456:717–720. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Demirbas D, Ceyhan O, Wyman AR and Hoffman
CS: A fission yeast-based platform for phosphodiesterase inhibitor
HTSs and analyses of phosphodiesterase activity. Handb Exp
Pharmacol. 135–149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ceyhan O, Birsoy K and Hoffman CS:
Identification of biologically active PDE11-selective inhibitors
using a yeast-based high-throughput screen. Chem Biol. 19:155–163.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Frame IJ, Deniskin R, Rinderspacher A,
Katz F, Deng SX, Moir RD, Adjalley SH, Coburn-Flynn O, Fidock DA,
Willis IM, et al: Yeast-based high-throughput screen identifies
Plasmodium falciparum equilibrative nucleoside transporter 1
inhibitors that kill malaria parasites. ACS Chem Biol. 10:775–783.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Asai A, Tsujita T, Sharma SV, Yamashita Y,
Akinaga S, Funakoshi M, Kobayashi H and Mizukami T: A new
structural class of proteasome inhibitors identified by microbial
screening using yeast-based assay. Biochem Pharmacol. 67:227–234.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ang RP, Teoh LS, Chan MK, Miswan N and
Khoo BY: Comparing the expression of human DNA topoisomerase I in
KM71 and X33 strains of Pichia pastoris. Electron J
Biotechnol. 21:9–17. 2016. View Article : Google Scholar
|
|
21
|
Hong TB, Rahumatullah A, Yogarajah T,
Ahmad M and Yin KB: Potential effects of chrysin on MDA-MB-231
cells. Int J Mol Sci. 11:1057–1069. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xuan H, Li Z, Yan H, Sang Q, Wang K, He Q,
Wang Y and Hu F: Antitumor activity of Chinese propolis in human
breast cancer MCF-7 and MDA-MB-231 cells. Evid Based Complement
Alternat Med. 2014:2801202014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pereira C, Lopes-Rodrigues V, Coutinho I,
Neves MP, Lima RT, Pinto M, Cidade H, Vasconcelos MH and Saraiva L:
Potential small-molecule activators of caspase-7 identified using
yeast-based caspase-3 and −7 screening assays. Eur J Pharm Sci.
54:8–16. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bach S, Talarek N, Andrieu T, Vierfond JM,
Mettey Y, Galons H, Dormont D, Meijer L, Cullin C and Blondel M:
Isolation of drugs active against mammalian prions using a
yeast-based screening assay. Nat Biotechnol. 21:1075–1081. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu X, Kramer JA, Swaffield JC, Hu Y, Chai
G and Wilson AG: Development of a highthroughput yeast-based assay
for detection of metabolically activated genotoxins. Mutat Res.
653:63–69. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Denny PW: Yeast: Bridging the gap between
phenotypic and biochemical assays for high-throughput screening.
Expert Opin Drug Discov. 16:1–8. 2018.
|
|
27
|
Voisset C, Daskalogianni C, Contesse MA,
Mazars A, Arbach H, Le Cann M, Soubigou F, Apcher S, Fåhraeus R and
Blondel M: A yeast-based assay identifies drugs that interfere with
immune evasion of the Epstein-Barr virus. Dis Model Mech.
7:435–444. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun Y, Wang Z, Tao J, Wang Y, Wu A, Yang
Z, Wang K, Shi L, Chen Y and Guo D: Yeast-based assays for the
high-throughput screening of inhibitors of coronavirus RNA cap
guanine-N7-methyltransferase. Antiviral Res. 104:156–164. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bilsland E, Sparkes A, Williams K, Moss
HJ, de Clare M, Pir P, Rowland J, Aubrey W, Pateman R, Young M, et
al: Yeast-based automated high-throughput screens to identify
anti-parasitic lead compounds. Open Biol. 3:1201582013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Couplan E, Aiyar RS, Kucharczyk R, Kabala
A, Ezkurdia N, Gagneur J, St Onge RP, Salin B, Soubigou F, Le Cann
M, et al: A yeast-based assay identifies drugs active against human
mitochondrial disorders. Proc Natl Acad Sci USA. 108:11989–11994.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bovee TF, Thevis M, Hamers AR, Peijnenburg
AA, Nielen MW and Schoonen WG: SERMs and SARMs: Detection of their
activities with yeast-based bioassays. J Steroid Biochem Mol Biol.
118:85–92. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Alaamery MA, Wyman AR, Ivey FD, Allain C,
Demirbas D, Wang L, Ceyhan O and Hoffman CS: New classes of PDE7
inhibitors identified by a fission yeast-based HTS. J Biomol
Screen. 15:359–367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sangkaew A, Krungkrai J and Yompakdee C:
Development of a high throughput yeast-based screening assay for
human carbonic anhydrase isozyme II inhibitors. AMB Express.
8:1242018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lichtenberg-Fraté H, Schmitt M, Gellert G
and Ludwig J: A yeast-based method for the detection of cyto and
genotoxicity. Toxicol In Vitro. 17:709–716. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chatterjee S, Majumder CB and Roy P:
Development of a yeast-based assay to determine the
(anti)androgenic contaminants from pulp and paper mill effluents in
India. Environ Toxicol Pharmacol. 24:114–121. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
MacLean MJ, Llordella MM, Bot N and Picard
D: A yeast-based assay reveals a functional defect of the Q488H
polymorphism in human Hsp90alpha. Biochem Biophys Res Commun.
337:133–137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chatterjee S, Kumar V, Majumder CB and Roy
P: Screening of some anti-progestin endocrine disruptors using a
recombinant yeast-based in vitro bioassay. Toxicol In Vitro.
22:788–798. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lubrano S, Comelli L, Piccirilli C,
Marranci A, Dapporto F, Tantillo E, Gemignani F, Gutkind JS,
Salvetti A, Chiorino G, et al: Development of a yeast-based system
to identify new hBRAFV600E functional interactors. Oncogene.
38:1355–1366. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jacobs PP, Inan M, Festjens N, Haustraete
J, Van Hecke A, Contreras R, Meagher MM and Callewaert N: Fed-batch
fermentation of GM-CSF-producing glycoengineered Pichia
pastoris under controlled specific growth rate. Microb Cell
Fact. 9:932010. View Article : Google Scholar : PubMed/NCBI
|