Introduction
Warfarin is an anticoagulant widely used for
preventing thromboembolic events correlated with arterial and
venous thrombosis, particularly in the management of atrial
fibrillation (AF). The conventional clinical treatment strategy,
for example, experience-based dosing, may frequently cause higher
or lower dosages of warfarin than necessary as warfarin has a
limited therapeutic range and displays wide variable responses in
individuals. A high dose may elevate the risk of bleeding in the
initiation stage of the therapy while a low dose may not produce
adequate anticoagulation (1).
Hence, it is extremely important to administer the right dosage of
warfarin in the initial stage for patients with AF.
Clinically, patients who receive implantation with a
mechanical valve prosthesis have to be on anticoagulants such as
warfarin throughout their lifetime (2). Warfarin is a widely used
anticoagulant, but its therapeutic safety window is small and there
are variations in the dosage for individuals. One challenge for
physicians and surgeons is to determine the appropriate individual
dose of warfarin; for example, a dose adequate to produce the
favorable anti-clotting effect without causing adverse drug
complications (3). At present,
available data indicate that the initial dosage of warfarin dosing
in individuals are associated with a variety of factors, such as
heredity, weight, race, diet, gender, concomitant drug
interactions, the presence of comorbidities, body size and age
(4).
As a group of widely distributed non-coding small
RNAs, microRNAs (miRNAs) consist of approximately 19 to 22
nucleotides. miRNAs target the 3′untranslated regions (UTRs) of
mRNAs to inhibit the translation of genes post-transcriptionally
mainly through incomplete base-pairing in mammalian cells, leading
to reduced expression level of target genes (5). miRNAs are involved in the regulation
of cell differentiation, growth, stress and numerous other
biological processes (6–8).
DNA variations including single nucleotide
polymorphisms (SNPs) may be harbored in microRNA target sites
present in mRNAs, which are short substitutions, deletions or
nucleotide insertions, with an incidence of approximately 1% in a
population (9). miRNA binding to
target mRNAs and subsequent gene modulation can be impacted by SNPs
in miRNA target sites (10–12).
In a similar manner, SNPs that impact miRNA binding may also be
harbored by miRNA target sites on circRNA genes.
VKORC1 has been reported to be functionally involved
in the bioactivation of vitamin K, a key element in the coagulation
system (13). It was found that
VKORC1 is a target of miR-137 by using an online miRNA database.
One polymorphism in the promoter region of miR-137 is believed to
be able to compromise the transcription of the miRNA and reduce its
expression level (14). In the
present study, the miR-137/VKORC1 relationship in HepG2 cells was
verified and the association between miR-137 promoter rs2660304
polymorphism and the post-operative warfarin maintenance dose was
tested.
Materials and methods
Subjects
A total of 155 Han Chinese patients with AF were
enrolled in the present study between September 2016 and May 2017
at the China-Japan Union Hospital of Jilin University. The study
was conducted according to the Declaration of Helsinki. All
participants were diagnosed with AF, and also received long-term
therapy with warfarin for at least 3 months. Data related to the
demographic characteristics of the subjects including age, sex,
height, weight, warfarin dose and international normalized ratio
(INR) of each patient were collected prior to our research. Written
informed consent was obtained from all patients or their
first-degree relatives after explanation of all the potential
risks. The demographic characteristics are presented in Table I. The study was approved by The
Research Ethics Committees of China-Japan Union Hospital of Jilin
University (approval no. 201700347).
 | Table I.Demographic and clinicopathological
characteristics of the AF patients recruited in the present
study. |
Table I.
Demographic and clinicopathological
characteristics of the AF patients recruited in the present
study.
| Demographic data | Patient data |
|---|
| Total number | 155 |
| Mean age (years) | 52.12±10.23 |
| Male sex, n (%) | 86 (55.48) |
| Mean weight (kg) | 63.23±12.23 |
| Mean height (cm) | 165.2±18.23 |
| Stable warfarin dose
(mg/day) | 3.08±0.24 |
| INR | 2.28±0.55 |
Cell culture and transfection
Two different liver cancer cell lines, HepG2 or
HepG2.2.1.5 cells were purchased from the Cell Bank of the Chinese
Academy of Sciences (Shanghai, China) and incubated in Dulbecco's
modified Eagle's medium (DMEM) culture medium (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% fetal bovine serum (FBS) in an
incubator with 5% CO2 at 37°C. Lipofectamine 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was utilized
to perform the miR-137 mimic (5′-UUAUUGCUUAAGAAUACGCGUAG-3′)
transfection when the cells were grown to 70–90% confluence using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following standard protocol by the supplier
(Fig. 1). The transfection medium
was replaced 8 h after transfection. Three independent tests were
carried out.
RNA isolation and real-time PCR
Forty-eight hours post-transfection, the cells were
harvested, and TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was utilized to isolate the
total RNA from the liver cells following the manufacturer's
guidelines. RNase-free water (Promega, Southampton, UK) was
utilized to elute the RNA and stored at −80°C in a refrigerating
cabinet. BioTek PowerWave XS (SSi Robotics, Tustin, CA, USA)
spectrophotometer was utilized to detect the purity, concentration
and content of RNA. The miRNA cDNA synthesis kit (Invitrogen;
Thermo Fisher Scientific, Inc.) was utilized to synthesize the cDNA
(VKORC1 and miR-137). Fast SYBR Green Master Mix (Life
Technologies; Thermo Fisher Scientific, Inc.) was utilized to
perform real-time PCR in a 20 µl volume mix, and the protocol of
the reaction was carried out under the following conditions: 50°C
for 2 min (initial step), 95°C for 10 min, followed by 40 cycles of
95°C for 3 sec for denaturation and 60°C for 30 sec for
elongation/hybridization. RNU43 was served as the internal control
to normalize the expression of VKORC1 mRNA and miR-137. The
2−ΔΔCq method (15) was
utilized to analyze the data of real-time RT-PCR. The experiment
was repeated three times.
Genotype determination by TaqMan
genotyping kit
The Omega FFPE DNA kit (Omega, Shanghai, China) was
utilized to purify and isolate the genomic DNA based on the
manufacturer's guidelines. The TaqMan Genotyping kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was performed to carry
out the genotype determination.
Vector construction (test effect of
rs2660304 on transcription of the promoter)
The upstream region of miR-137 was amplified based
on the information collected from the UCSC genome browser
(http://genome.ucsc.edu/), and the PCR product was
inserted into the pGL3-Basic vector (Promega, Madison, WI, USA) to
generate the wild-type promoter of miR-137. The site-directed
mutagenesis was used to introduce the minor allele of the rs2660304
polymorphism.
Vector construction (test regulatory
relationship between miRNA and target)
The fragment of the 3′UTR (untranslated region) of
VKORC1 with the predicted miR-137 binding site was amplified
through PCR, the PCR products were inserted into the pGL3-Basic
vector (Promega) to yield the luciferase reporter constructs, and
then and the site-directed mutagenesis was used to replace the
potential binding site with the reverse and complementary sequences
to generate the mutant 3′UTR of VKORC1.
Luciferase assay
In brief, the HepG2 cells (2×106) were
seeded in 48-well plates. When the cells were grown to 80%
confluence, Lipofecamine2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was utilized to co-transfect the HepG2
cells with the wild-type/mutant type vector. Approximately 0.4 mg
of firefly luciferase reporter vector with the mutant target site
and wild-type and 0.02 mg of the control plasmid containing
Renilla luciferase pGL3 (Promega) were utilized in this
reaction. After incubation for 12 h, the transfection medium was
replaced, and passive lysis buffer (Promega) was utilized to lyse
the cells, and a 96-well plate luminometer (Berthold Detection
Systems, Pforzheim, Germany) was utilized to analyze the cell
lysates. The Dual-Luciferase Reporter Assay system (Promega) was
then utilized to measure the activities of firefly and
Renilla luciferase following the manufacturer's
recommendation. Three independent experiments were performed.
Western blot analysis
Western blotting was utilized to assess the
expression of VKORC1 protein in HepG2 cells. The HepG2 cells were
harvested, and RIPA lysis buffer (Invitrogen; Thermo Fisher
Scientific, Inc.) containing complete protease inhibitor mix, 0.1%
sodium dodecyl sulfate (SDS), 0.5% Na-DOC, 1% NP-40, 0.05%
NaN3 and 50 mM Tris (pH 7.4) was utilized to lyse the
HepG2 cells. A BCA assay kit (Thermo Fisher Scientific, Inc.) was
utilized to determine the protein concentration. Proteins (35
µg/lane) were separated by 12% SDS-PAGE and electroblotted to
polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA,
USA). Non-fat milk (5%) was utilized to block the membranes to
avoid unspecific binding. The primary antibodies anti-VKORC1 (cat.
no. ab206656; dilution 1:2,000; Abcam, Cambridge, MA, USA) and
anti-β-actin (cat. no. ab8226; dilution 1:10,000; Abcam, Cambridge,
MA, USA) were utilized to detect the target protein overnight at
4°C. Horseradish peroxidase linked anti-rabbit secondary antibody
(cat. no. sc-516087; dilution 1:15,000; Santa Cruz Biotechnology,
Santa Cruz, CA, USA) was utilized to treat the membranes at room
temperature for 60 min. ImageQuant LAS 4000 (GE Healthcare Life
Sciences, Piscataway, NJ, USA) and enhanced chemiluminescence (ECL)
(PerkinElmer, Norwalk, CT, USA) were utilized to detect the signal
following the manufacturer's instruction. All experiments were run
in triplicate.
Statistical analysis
All results are shown as mean ± standard deviation
(SD). P<0.05 was considered to indicate a statistically
significant difference. The differences in experiments were
analyzed with the use of a two-sided Student's t-test, and the
significant differences between two groups were identified using
the Mann-Whitney U test. Multiple variate linear analysis was
performed to analyze the clinicopathological characteristics of the
AF patients. SPSS 15.0 statistical package (SPSS, Inc., Chicago,
IL, USA) was utilized to perform all the statistical analyses.
Results
Characteristics of the AF
patients
One hundred and fifty-five participants were
recruited in the present study, and the clinical data were
collected at the same time. The participant baseline
characteristics are summarized in Table I. The frequency of males was
55.48%. The mean age was 50.12±10.23, and the average weight was
63.23±12.23 kg. The average height of the subjects was 165.2±18.23
cm, the average stable warfarin dose was 3.08±0.24 mg/day and the
average INR was 2.28±0.55.
VKORC1 is a direct target gene of
miR-137
Furthermore, based on the results of in
silicon analysis, VKORC1 was identified as a virtual target
gene of miR-137 with the binding site located at 358–365 bp of
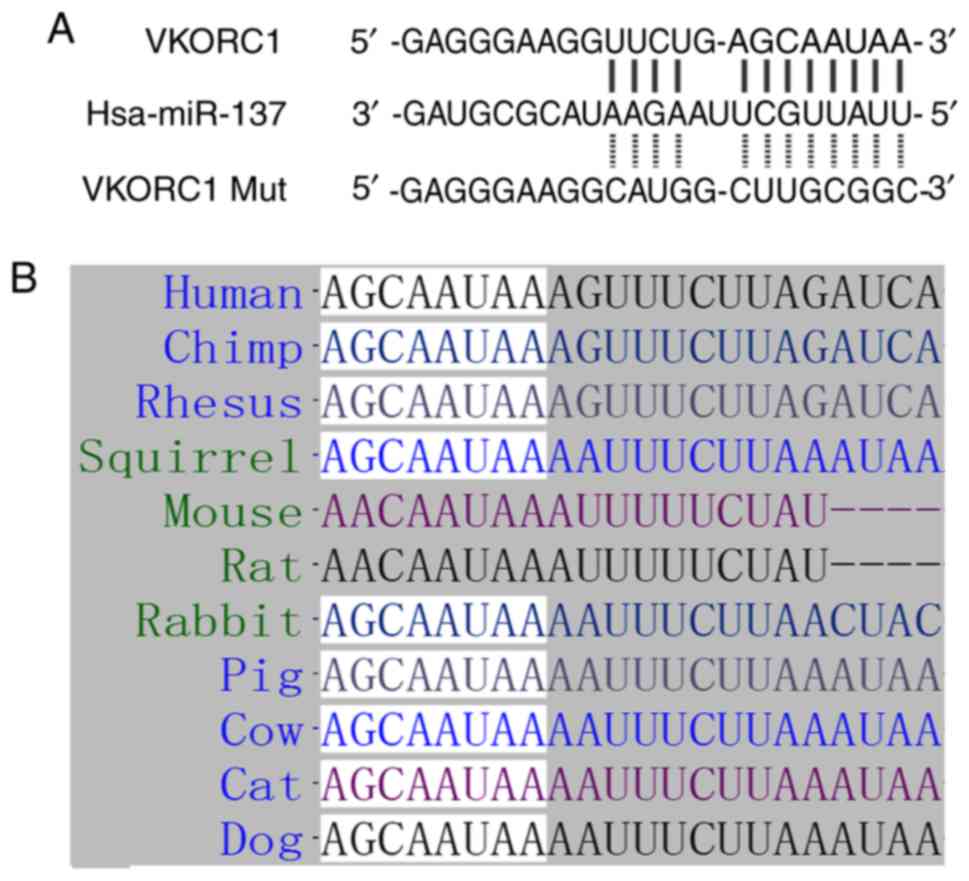
VKORC1 3′UTR (Fig. 2A). As shown
in Fig. 2B, the ‘seed sequence’ in
the VKORC1 3′UTR was highly conserved among the human, chimp,
rhesus, squirrel, mouse, rat, rabbit and cow, suggested that this
‘seed sequence’ may play a crucial role in the progression of AF in
humans.
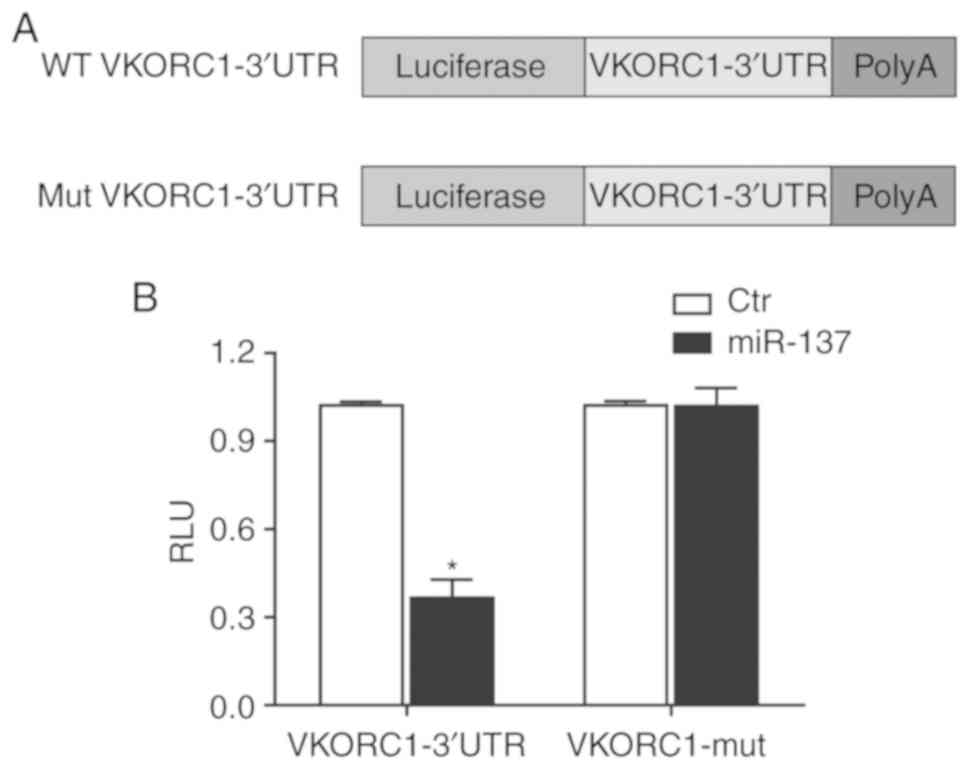
miR-137 directly regulates VKORC1, and the
interaction between VKORC1 3′UTR and miR-126 was next confirmed
using luciferase assay. The wild-type 3′UTR of VKORC1 was amplified
via PCR, and QuickChange XL Site-Directed Mutagenesis kit (Agilent
Technologies, Inc., Santa Clara, CA, USA) was utilized to mutate
the binding site of miR-137 on VKORC1 3′UTR to obtain mutant VKORC1
3′UTR. The wild-type and mutant 3′UTR of VKORC1 were inserted into
the downstream of the luciferase reporter. Subsequently, for the
HepG2 cells co-transfected with the luciferase conducts containing
wild-type and mutant 3′UTR of VKORC1 and miR-137 mimic, the
luciferase activity was detected 48 h post-transfection. As shown
in Fig. 3, miR-137 evidently
decreased the luciferase activity of the wild-type 3′UTR of VKORC1,
but the miR-137 binding site mutated completely abolished the
suppressive effect of miR-137 on luciferase activity. The data
collectively suggested that VKORC1 is the virtual target gene of
miR-137 and miR-137 suppresses the expression of VKORC1.
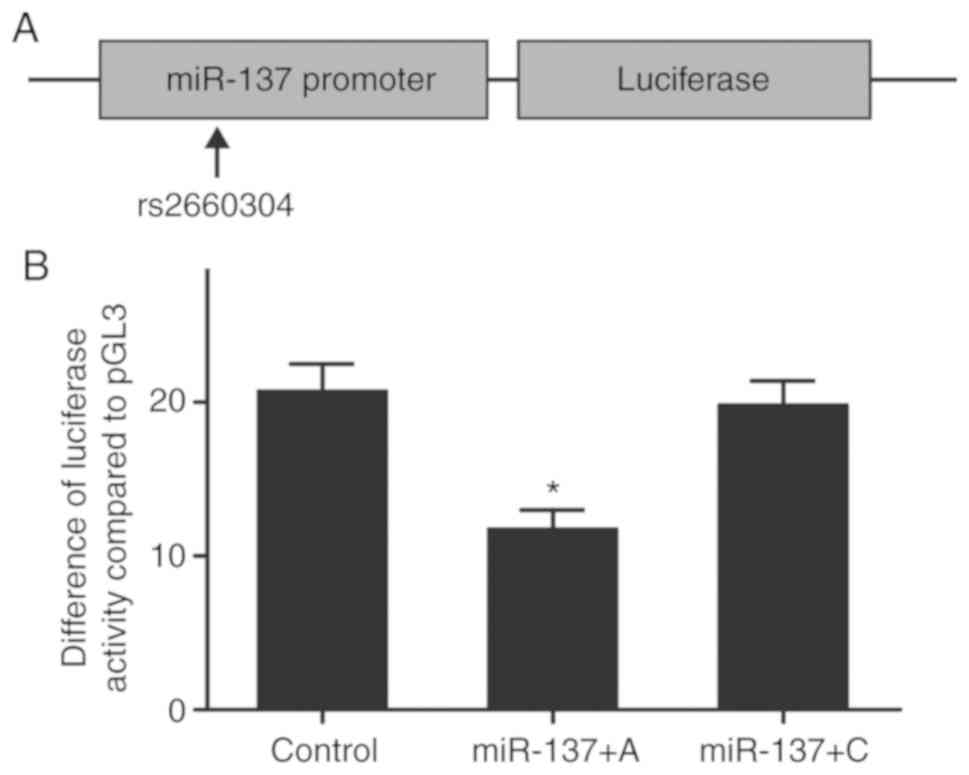
The rs2660304 minor allele reduces the
transcription activity of the miR-137 promoter
The effect of rs2660304 on the miR-137 transcription
activity of the promoter was explored using luciferase assay. As
shown in Fig. 4A, the rs2660304 A
allele and the rs2620304 C allele were respectively inserted into
the vector at the same site, and the above luciferase constructs
were transfected into HepG2 cells. The luciferase activity was
subsequently determined. As shown in Fig. 4B, it was found that the miR-137 SNP
with the presence of the A allele apparently reduced the activity
of luciferase when compared with miR-137, while C allele
replacement with A allele in the rs2660304 polymorphism abrogated
the suppressive effect of luciferase activity, indicating that the
presence of the minor allele of the polymorphism in the promotor
region of miR-137 substantially reduced the transcription activity
of the promoter.
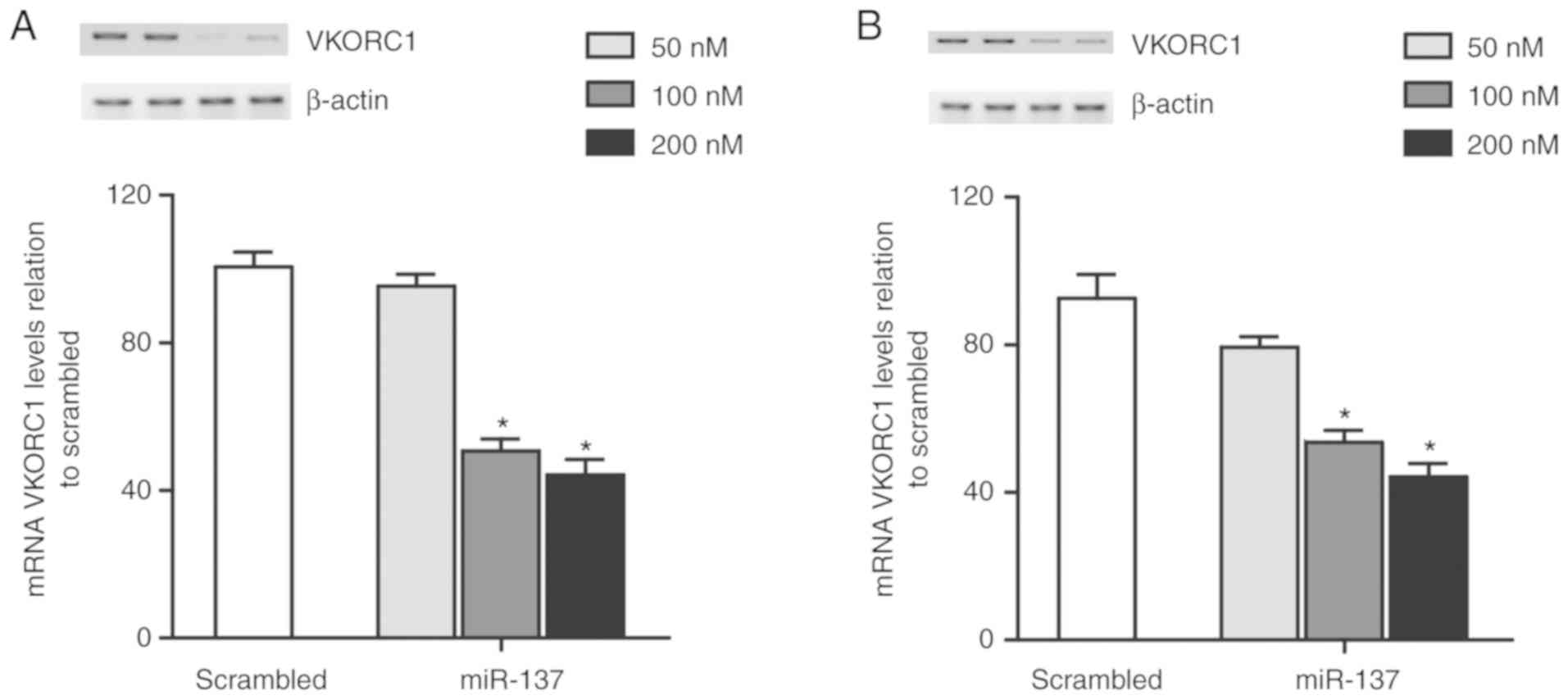
miR-137 suppresses VKORC1 expression
in vivo
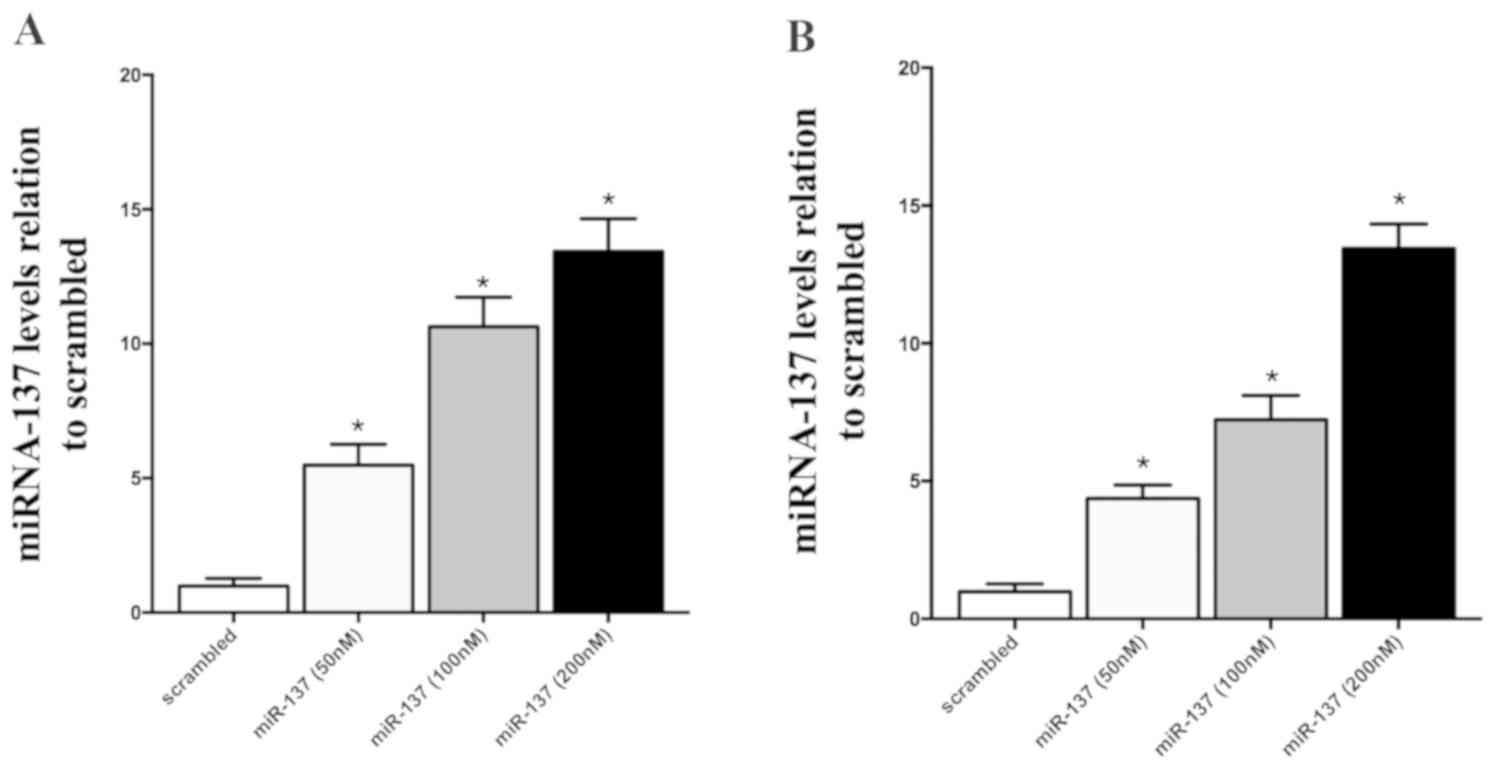
To specifically confirm that miR-137 suppressed the
expression of VKORC1 in two cell lines, HepG2 and HepG2.2.1.5 cells
respectively, using Lipofectamine 2000, the cultured cells were
transfected with different concentrations of miR-137 mimics (50,
100 and 200 nM; Fig. 5). A
concentration of 100 nM miR-137 mimics caused a significant
decrease in the VKORC1 expression level, and 200 nM miR-137 mimics
provoked ~50% decrease in VKORC1 expression related to that in the
scramble control. This confirmed that miR-137 is a direct regulator
of VKORC1, and miR-137 suppressed the VKORC1 expression in a
concentration-dependent manner in the two culture cell lines
(Fig. 5A, HepG2; Fig. 5B, HepG2.2.1.5).
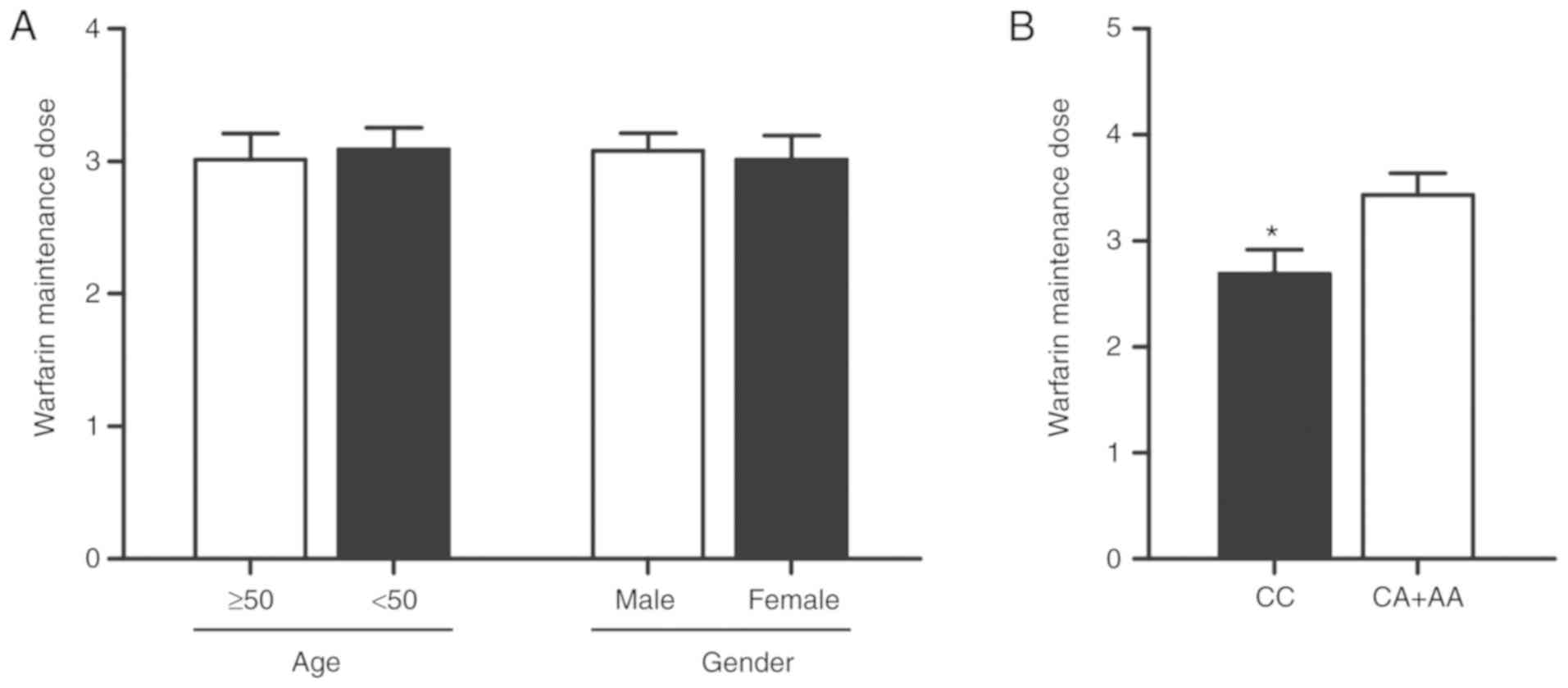
Warfarin maintenance dose in the
different groups
We used SPSS 15.0 statistical package (SPSS, Inc.,
Chicago, IL, USA) to analyze the maintenance dose in the different
groups, and as shown in Fig. 6, no
difference was noted regarding the warfarin maintenance dose
between the different age or sex groups (Fig. 6A). Furthermore, CC carriers showed
a significantly lower warfarin maintenance dose than the CA+AA
carriers (Fig. 6B).
Discussion
Studies have shown a correlation between
polymorphisms in the vitamin K epoxide reductase complex subunit 1
(VKORC1) gene and warfarin dose requirement (16,17).
Shomron (18), discovered 2 highly
evolutionarily conserved binding sites for miR-137 and miR-133 on
the VKORC1 gene by applying bioinformatic tools, indicating that
these miRNAs likely modulate VKORC1. A recent study by Perez-Andreu
et al (19), demonstrated
the possible binding sites for miR-137 and miR-133, but indicated
only co-expression of miR-133 with VKORC1 in human hepatocytes.
They also indicated that the VKORC1 mRNA expression was reduced by
overexpression of miR-133 in a dose-dependent manner. Furthermore,
in subjects who carry the G allele (rs9923231 in the promoter
region), this modulation of VKORC1 mRNA expression was also found,
which is correlated with an elevated transcription rate of VKORC1
mRNA. Hence, it is likely to predict that variants in miR-133
genes, resulting in an abnormal modulation of the VKORC1 gene,
could change the effectiveness of warfarin treatment. In this
study, we performed in silicon analysis, and identified that
VKORC1 is a virtual target gene of miR-137, and VKORC1 3′UTR is
highly conserved among species. A luciferase assay was then
conducted to verify that miR-137 directly mediates VKORC1 and the
interaction between miR-137 and VKORC1, and found that miR-137
evidently decreased the luciferase activity of wild-type 3′UTR of
VKORC1, while the luciferase activity of the mutant VKORC1 3′UTR
was comparable with the control in the miR-137-overexpressing
cells. Furthermore, it was specifically confirmed that miR-137
negatively regulated the expression of VKORC1 in the two liver
cancer cell lines, and it was found that in liver cancer cells,
miR-137 is a direct regulator of VKORC1, and miR-137
dose-dependently suppressed VKORC1 expression.
VKORC1 is known as an enzyme which is encoded by the
VKORC1 gene in humans. Vitamin K is critical for blood
clotting after being activated by enzymes (20). In some blood-clotting proteins,
this form of vitamin K is activated by enzymes and is a decreased
form necessary for the carboxylation of glutamic acid residues. The
product of this gene encodes the enzyme that contributes to
decreasing vitamin K 2,3-epoxide to the form being activated by
enzymes. The vitamin K antagonist known as warfarin and deficiency
of vitamin K can result in life-threatening bleeding, and the
product of this gene exhibits sensitivity to warfarin. There is a
correlation between the mutations in this gene in humans and
clotting factors dependent on vitamin K. In rats and humans, it has
also been correlated with resistance to warfarin; however, such
mutations are not common except in certain Jewish and Ethiopian
populations. Two pseudogenes have been found on the X chromosome
and chromosome 1. There is also a study which has described 2
alternatively spliced transcripts which encode distinct isoforms
(20).
VKOR complex, subunit 1 (VKORC1) is situated
on chromosome 16 at band p11.2. VKORC1 is primarily
expressed in the liver; but smaller VKORC1 amounts are found
in the pancreas and heart. A polymorphism within the promoter
region of VKORC1, a guanine to adenine conversion at
position-1639 (−1639 G>A, rs9923231), is related to a decreased
requirement of warfarin in individuals of Asian descent and
Caucasians. Reduced VKORC1 mRNA production by the −1639A allele and
decreased enzyme VKOR expression may have led to this (21). Patients with the variant of the
1639A promoter need a lower warfarin dose due to decreased VKOR
expression (22). The
VKORCI−1639A allele has been shown to be present in the
majority of people of Asian descent; however, is less common in
individuals of African descent and Caucasian individuals by
population studies. The VKORCI−1639G allele related to
increased expression and activity of VKORC1 is predominant in
individuals of African descent. Some research remains doubtful
whether the-1639G allele would lead to the dosing variability of
warfarin in individuals of African descent (23–25).
Warfarin dosing may be affected by other alleles in VKORC1;
for example, the Asp36Tyr amino acid substitution would result in a
mild to moderate warfarin dose increase (26). In this study, we found that no
difference was noted regarding the warfarin maintenance dose
between different age or sex groups (Fig. 6A), and furthermore, AC + AA
carriers showed a significantly higher warfarin maintenance dose
than CC carriers (Fig. 6B).
Furthermore, the effect of rs2660304 on the miR-137 transcription
activity of the promoter was investigated using luciferase assay.
The rs2660304 polymorphism A allele and rs2660304 polymorphism C
allele were inserted into the luciferase gene, and it was found
that miR-137 SNP with the presence of the A allele substantially
suppressed the activity of luciferase when compared with miR-137,
while C allele replacement with the A allele in the rs2660304
polymorphism abrogated the suppressive impact of luciferase
activity. Notably, miR-137 is not only involved in the control of
the coagulation system but also has been reported to be involved in
other systems such as the nervous system (27) and tumorigenesis (28), indicating a widespread and multiple
system involvement of miR-137 and this polymorphism may also be
associated with other function of different systems.
There are limitations in the present study. Firstly,
further research with a larger sample size is needed to confirm the
conclusions of this study. Secondly, further functional analysis is
warranted to confirm the interaction between miRNA-137 and
VKORC1.
In conclusion, these findings collectively provide
support that VKORC1 is a direct target of miR-137 and the
miR-137 rs2660304 polymorphism is associated with warfarin
maintenance dose in the management of AF. The rs2660304
polymorphism is a potential biomarker to predict the clinical
efficacy of warfarin in AF patients.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
ZT and YSY conceived and designed the study. ZT, ZHF
and DHW performed the experiments. ZHF, WY and DNL analyzed and
visualized the data. WY and DNL wrote the paper. ZT, YSY, ZHF and
DHW reviewed and edited the manuscript. All authors read and
approved the manuscript and agreed to be accountable for all
aspects of the research, ensuring that the accuracy or integrity of
any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Research Ethics Committees of China-Japan Union Hospital of Jilin
University (approval no. 201700347).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yoshizawa, Hayashi H, Tashiro Y, Sakawa S,
Moriwaki H, Akimoto T, Doi O, Kimura M, Kawarasaki Y, Inoue K and
Itoh K: Effect of VKORC1-1639 G>A polymorphism, body weight,
age, and serum albumin alterations on warfarin response in japanese
patients. Thromb Res. 124:161–166. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kidane AG, Burriesci G, Cornejo P, Dooley
A, Sarkar S, Bonhoeffer P, Edirisinghe M and Seifalian AM: Current
developments and future prospects for heart valve replacement
therapy. J Biomed Mater Res B Appl Biomater. 88:290–303. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Glurich I, Burmester JK and Caldwell MD:
Understanding the pharmacogenetic approach to warfarin dosing.
Heart Fail Rev. 15:239–248. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arellano-Rodrigo E: Home monitoring of
warfarin effects. N Engl J Med. 364:378–379. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Poy MN, Hausser J, Trajkovski M, Braun M,
Collins S, Rorsman P, Zavolan M and Stoffel M: miR-375 maintains
normal pancreatic alpha-and beta-cell mass. Proc Natl Acad Sci USA.
106:5813–5818. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hornstein E, Mansfield JH, Yekta S, Hu JK,
Harfe BD, McManus MT, Baskerville S, Bartel DP and Tabin CJ: The
microRNA miR-196 acts upstream of Hoxb8 and Shh in limb
development. Nature. 438:671–674. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Borel C and Antonarakis SE: Functional
genetic variation of human miRNAs and phenotypic consequences. Mamm
Genome. 19:503–509. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saetrom P, Biesinger J, Li SM, Smith D,
Thomas LF, Majzoub K, Rivas GE, Alluin J, Rossi JJ, Krontiris TG,
et al: A risk variant in an miR-125b binding site in BMPR1B is
associated with breast cancer pathogenesis. Cancer Res.
69:7459–7465. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: The implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gamazon ER, Ziliak D, Im HK, LaCroix B,
Park DS, Cox NJ and Huang RS: Genetic architecture of microRNA
expression: Implications for the transcriptome and complex traits.
Am J Hum Genet. 90:1046–1063. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sridharan K, Modi T, Bendkhale S, Kulkarni
D, Gogtay NJ and Thatte UM: Association of genetic polymorphisms of
CYP2C9 and VKORC1 with bleeding following warfarin: A case-control
study. Curr Clin Pharmacol. 11:62–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Warburton A, Breen G, Bubb VJ and Quinn
JP: A GWAS SNP for schizophrenia is linked to the internal MIR137
promoter and supports differential Allele-Specific expression.
Schizophr Bull. 42:1003–1008. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rieder MJ, Reiner AP, Gage BF, Nickerson
DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL and
Rettie AE: Effect of VKORC1 haplotypes on transcriptional
regulation and warfarin dose. N Engl J Med. 352:2285–2293. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
D'Andrea G, D'Ambrosio RL, Di Perna P,
Chetta M, Santacroce R, Brancaccio V, Grandone E and Margaglione M:
A polymorphism in the VKORC1 gene is associated with an
interindividual variability in the dose-anticoagulant effect of
warfarin. Blood. 105:645–649. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shomron N: MicroRNAs and pharmacogenomics.
Pharmacogenomics. 11:629–632. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peréz-Andreu V, Teruel R, Corral J, Roldán
V, García-Barberá N, Salloum-Asfar S, Gómez-Lechón MJ, Bourgeois S,
Deloukas P, Wadelius M, et al: miR-133a regulates vitamin K
2,3-epoxide reductase complex subunit 1 (VKORC1), a key protein in
the vitamin K cycle. Mol Med. 18:1466–1472. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ainle FN, Mumford A, Tallon E, McCarthy D
and Murphy K: A vitamin K epoxide reductase complex subunit 1
mutation in an Irish patient with warfarin resistance. Ir J Med
Sci. 177:159–161. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan HY, Chen JJ, Lee MT, Wung JC, Chen
YF, Charng MJ, Lu MJ, Hung CR, Wei CY, Chen CH, et al: A novel
functional VKORC1 promoter polymorphism is associated with
inter-individual and inter-ethnic differences in warfarin
sensitivity. Hum Mol Genet. 14:1745–1751. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang D, Chen H, Momary KM, Cavallari LH,
Johnson JA and Sadée W: Regulatory polymorphism in vitamin K
epoxide reductase complex subunit 1 (VKORC1) affects gene
expression and warfarin dose requirement. Blood. 112:1013–1021.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kealey C, Chen Z, Christie J, Thorn CF,
Whitehead AS, Price M, Samaha FF and Kimmel SE: Warfarin and
cytochrome P450 2C9 genotype: Possible ethnic variation in warfarin
sensitivity. Pharmacogenomics. 8:217–225. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schelleman H, Chen Z, Kealey C, Whitehead
AS, Christie J, Price M, Brensinger CM, Newcomb CW, Thorn CF,
Samaha FF and Kimmel SE: Warfarin response and vitamin K epoxide
reductase complex 1 in African Americans and Caucasians. Clin
Pharmacol Ther. 81:742–747. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kimmel SE, Christie J, Kealey C, Chen Z,
Price M, Thorn CF, Brensinger CM, Newcomb CW and Whitehead AS:
Apolipoprotein E genotype and warfarin dosing among Caucasians and
African Americans. Pharmacogenomics J. 8:53–60. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Loebstein R, Dvoskin I, Halkin H, Vecsler
M, Lubetsky A, Rechavi G, Amariglio N, Cohen Y, Ken-Dror G, Almog S
and Gak E: A coding VKORC1 Asp36Tyr polymorphism predisposes to
warfarin resistance. Blood. 109:2477–2480. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mahmoudi E and Cairns MJ: MiR-137: An
important player in neural development and neoplastic
transformation. Mol Psychiatry. 22:44–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Neault M, Mallette FA and Richard S:
miR-137 modulates a tumor suppressor network-inducing senescence in
pancreatic cancer cells. Cell Rep. 14:1966–1978. 2016. View Article : Google Scholar : PubMed/NCBI
|