Introduction
Lung cancer, the most common of malignant tumors, is
a leading cause of cancer-associated mortality all over the world
(1). There are two main types of
lung cancer: Non-small cell lung cancer (NSCLC) and SCLC (2). NSCLC is the most common type,
corresponding to 85% of all lung cancer cases (3). Approximately two-thirds of patients
diagnosed with NSCLC present the pathology of an already advanced
stage (4). Although new drugs have
been developed, including epidermal growth factor receptor and
anaplastic lymphoma kinase inhibitors, due to early tumor
recurrence and metastasis, the overall survival of patients with
NSCLC remains poor (5). Therefore,
it is essential to find novel biomarkers that can precisely predict
the prognosis of patients diagnosed with NSCLC, and to discover new
targets that may help treat this disease more effectively.
MicroRNAs (miRs/miRNAs) are small non-coding RNAs
(~18–25 nucleotides) that negatively regulate gene expression at
the post-transcriptional level by inhibiting mRNA translation or
causing its degradation, all via complementarities with the
3′-untranslated region (UTR) of their target genes (6,7). The
expression of miRs in tumor and normal tissues is known to be
different, a fact that has been described in a number of different
tumors (8). A previous study has
demonstrated that miRs are involved in the regulation of a number
of processes, including cell proliferation, metabolism,
differentiation and apoptosis (9).
Further research has confirmed that miRs also serve a pivotal role
in the onset and progression of cancer (9,10).
Other studies have demonstrated that multiple miRs are up-or
downregulated in NSCLC (11–13).
For example, Wang et al (14) reported that miR-124 suppresses cell
viability and enhances cell apoptosis by inhibiting signal
transducer and activator of transcription 3 in NSCLC. In addition,
Liu et al (15) reported
that the downregulation of miR-335 promotes cell viability or
inhibits cell apoptosis via the upregulation of transformer 2β
homolog, mediated by the activation of the protein kinase B (AKT)
signaling pathway in A459 lung cancer cells. Xue et al
(16) reported that miR-342-3p
inhibits cell proliferation and migration, and enhances cell
apoptosis in NSCLC cells by targeting anterior gradient 2.
Regarding miR-379, according to PubMed (www.ncbi.nlm.nih.gov/pubmed/) only six studies have
evaluated its association with lung cancer, and the mechanism of
miR-379 regulation in NSCLC remains unreported.
In the present study, miR-379 was observed to be
downregulated in NSCLC cell lines and tissues. Functional analyses
and experiments demonstrated that upregulation of miR-379
significantly suppressed proliferation, migration, invasiveness and
the epithelial-mesenchymal transition (EMT) process of NSCLC cells.
In addition, Transwell migration assays revealed that the
overexpression of miR-379 inhibited NSCLC cell migration and
invasion. Furthermore, conserved helix-loop-helix ubiquitous kinase
(CHUK), formally termed inhibitor of nuclear factor-κB (NF-κB)
kinase subunit α (IKKα) was confirmed to be a target of miR-379 and
that it has an oncogenic role in NSCLC progression via the
activation of the NF-κB signaling pathway. These results indicated
that miR-379 may inhibit NSCLC progression by directly targeting
CHUK and by activating the NF-κB signaling pathway.
Materials and methods
Patient tissue samples
A total of 30 pairs of human NSCLC and matched
normal tissues were obtained from 30 patients admitted to the
Department of Respiratory and Critical Care Medicine, Tianjin Chest
Hospital (Tianjin, China). All of the human NSCLC and matched
normal tissues were pathologically and histologically evaluated.
All samples were stored in liquid nitrogen before use. The present
study followed the guidelines of The Declaration of Helsinki. All
participants provided written informed consent, and the present
study was approved by the Ethical Oversight Committee of Department
of Respiratory and Critical Care Medicine, Tianjin Chest Hospital
(Tianjin, China).
Cell culture
The NSCLC cell lines, namely H1993 (NCI-H1993),
H1650 (NCI-H1650), H1299 (NCI-H1299), A549 (A-549) and SK-MES-1,
and a non-tumorigenic human bronchial epithelial cell line
(BEAS-2B) were purchased from American Type Culture Collection.
These were cultured in Dulbecco's modified Eagle's medium (DMEM;
HyClone; GE Healthcare Life Sciences) containing 10% fetal bovine
serum (FBS; HyClone; GE Healthcare Life Sciences), streptomycin
(100 µg/ml) and penicillin (100 µg/ml), and maintained in a
humidified atmosphere at 37°C with 5% CO2.
Cytosolic and nuclear protein
isolation
A549 and H1993 cells (1×107 cells/ml)
were harvested, washed with PBS, centrifuged (900 × g, for 6 min,
at room temperature), and re-suspended in ice-cold buffer A [10 mM
HEPES (pH 7.0), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT and
0.2 mM PMSF]. After 10 min of incubation on ice, the cells were
centrifuged again (900 × g, for 6 min, at room temperature), and
the supernatant (cytosolic fraction) was stored at −80°C. The
pellet containing the nuclear fraction was re-suspended in buffer C
[20 mM HEPES (pH 7.9), 20% glycerol, 420 mM NaCl, 1.5 mM
MgCl2, 0.2 mM EDTA, 0.5 mM DTT and 0.2 mM PMSF], and
incubated for 20 min at 0°C. Following vortex mixing, the resulting
suspension was centrifuged at 1,000 × g for 10 min at 4°C, and the
supernatant (nuclear extract) was stored at −80°C. The protein
concentration of both the cytosolic and nuclear extracts was
determined using the Bradford method via the Bio-Rad protein assay
kit (Bio-Rad Laboratories, Inc.).
Plasmid construction
The potential targets of miR-379 were predicted
using miRNA.org (http://www.microrna.org/microrna/home.do), TargetScan
7.1 (http://www.targetscan.org/vert_71/), miRbase
(http://www.mirbase.org/index.shtml)
and miRanda (http://www.mirdb.org/). The miRBase
Targets version 2.0 (http://www.mirbase.org/index.shtml) was used to search
for the potential miR-379 target sites in the CHUK 3′UTR.
The 3′UTR fragment of the CHUK gene containing the
predicted miR-3379 binding site was amplified by PCR from the A549
cell RNA. Total RNA was isolated from A549 cell using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. RNA (1 mg) was
reverse transcribed into cDNA using the Omniscript RT kit (Qiagen
GmbH), according to the manufacturer's protocol. The temperature
protocol for reverse transcription was as follows: 37°C for 60 min,
followed by a final step of 37°C for 5 sec. pCHUK, pSilencer, short
hairpin RNA CHUK (shR-CHUK), pri-miR-379, ASO-miR-379, pcDNA3 and
ASO-NC (Shanghai Jierui Biological Engineering Co., Ltd.) represent
overexpressed CHUK, empty vector, knockdown CHUK, overexpressed
miR-379, knockdown miR-379, empty vector and empty vector,
respectively. The sequences of the primers used in plasmid
construction were as follows: pCHUK forward,
5′-GTACCAGCATCGGGAACTTG-3′, and pCHUK reverse,
5′-ATGGCACCATCGTTCTCTGT-3′; shR-CHUK forward,
5′-GATCCGCAGTGCACTATGTGTCTGTTCAAGAGACAGACACATAGTGCACTGCTTTTTTGGAAA-3′,
and shR-CHUK reverse,
5′-AGCTTTTCCAAAAAAGCAGTGCACTATGTGTCTGTCTCTTGAACAGACACATAGTGCACTGCG-3′;
pri-miR-379 forward, 5′-CGGGGTACCGGTATAAGGCAGGGACTGGG-3′, and
pri-miR-379 reverse, 5′-CCGGAATTCGGATATGTGGGACCCGAAGG-3′;
ASO-miR-379 forward, 5′-CACUGGUACAAGGGUUGGGAGA-3′, and ASO-miR-379
reverse, 5′-CAGUACUUUUGUGUAGUACAA-3′. The thermocycling conditions
of the PCR amplification conditions were as follows: 95°C for 40
sec and 40 cycles of 95°C for 5 sec, 60°C for 40 sec, and finally
extend for 72°C 5 min.
For the luciferase assay, the sequence inserted into
the BamHI and EcoRI sites of the pcDNA3/enhanced
green fluorescent protein (EGFP) vector (Promega Corporation) were
immediately downstream from the stop codon of EGFP. The resulting
vector was named CHUK-3′UTR. A mutant version (CHUK-3′UTR mut) with
alterations in the seed sequence of the miR-379 binding site was
also constructed using the same PCR method (CHUK-3′UTR mut forward,
5′-CGCGGATCCCCTCAAAATAAAGAAGTATGGTAAT-3′, and CHUK-3′UTR mut
reverse, 5′-CCGGAATTCAGCTTTTTTATTTGTTAATGTCACA-3′). All the
insertions were confirmed upon sequencing.
Transfection assay
The pri-miR-379 or antisense oligonucleotide
(ASO)-miR-379 and respective controls [pcDNA3 and ASO-negative
control (NC)] were transfected into human NSCLC cell lines using
Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. A total of 100 nM miR-17-3P mimic,
inhibitor or negative control miRNA (Guangzhou RiboBio Co., Ltd.)
were transfected into cultured cells in accord with manufacturer's
instructions. After 48 h, the transfection efficacy was evaluated
by reverse transcription-quantitative PCR. The miR-379 expression
vector (pri-miR-379) was amplified from genomic DNA and cloned into
the pcDNA3 vector at KpnI and EcoRI sites. The
2′-O-methyl-modified miR-379 antisense oligo nucleotide
(ASO-miR-379) was commercially synthesized as an inhibitor of
miR-379.
RT-qPCR analysis
Total RNA and miR were isolated from cells or frozen
tissues with TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) and cDNA was synthesized with the miRVana miR
Isolation kit (Ambion; Thermo Fisher Scientific, Inc.) and
PrimeScript RT Master Mix (Takara Biotechnology Co., Ltd., Dalian,
China) according to the manufacturer's protocol. The temperature
protocol for the reverse transcription was as follows: 37°C for 60
min, followed by a final step of 37°C for 5 sec. The qPCR was used
to assay the expression levels of CHUK (forward,
5′-TGGAGCCCCTGAAGAAGAG-3′, and reverse, 5′-AAGTGCGTTGTGCGGTAGC-3′),
NFKB1A (forward, 5′-CTGCTCTCCCTTCCTCAGAC-3′, and reverse,
5′-TGAGGTAGGACCAGGAAACC-3′), β-actin (forward
5′-TAGTTGCGTTACACCCTTTCTTG-3′, and reverse,
5′-GCTGTCACCTTCACCGTTCC-3′), miRNA-379 (forward,
5′-GCGCTTATTGCTTAAGAATAC-3′, and reverse, 5′-CAGTGCAGGGTCCGAGGT-3′)
and U6 (forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′, and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3)′. These were performed on an ABI7300
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using the SYBR® Green PCR Master Mix (Thermo
Fisher Scientific, Inc.) and a Bio-Rad CFX-96 RT-PCR system
(Bio-Rad Laboratories, Inc.), according to the manufacturer's
instructions. The thermocycling conditions were as follows:
Preliminary denaturation at 96°C for 2 min, followed by 40 cycles
of denaturation at 96°C for 15 sec, annealing at 60°C for 1 min and
elongation at 60°C for 1 min. miR-379 levels were detected using a
miR-specific TaqMan MicroRNA Assays kit (Applied Biosystems, Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. The thermocycling conditions were: 95°C for 10 min;
40 cycles of 95°C for 1 min, 63°C for 2 min, 72°C for 1 min; final
72°C for 10 min. The relative expression levels of each gene were
calculated and normalized using the 2−∆∆Cq method
relative to U6 or β-actin (17).
All the reactions were run in triplicate.
Cellular proliferation and colony
formation
An MTT assay was used to determine the cell
viability. Cells were seeded 24 h after transfection was completed.
A total of 2×103 cells were seeded in a total volume of
100 µl in 96-well plates. The MTT assay was performed at 24, 48 and
72 h, and DMSO was used to dissolve the formazan and stop the
reaction. The optical density was measured using a Quant Universal
Microplate Spectrophotometer (BioTek Instruments, Inc.) at 490
nm.
For the colony formation assay, H1993 and A549 cells
(300 cells/well in 12-well plates) were seeded in complete medium
(containing DMEM, 10% fetal bovine serum, 100 µg/ml streptomycin
and 100 µg/ml penicillin) for 12 days, at 37°C, with 5%
CO2. Cells were fixed with 4% paraformaldehyde in PBS
for 10 min at room temperature, and stained with 0.1% crystal
violet for 30 min at room temperature. Only colonies containing
>50 cells were counted.
EGFP reporter assay
An EGFP reporter assay was performed to determine
the binding of miR-379 to the 3′UTR of CHUK mRNA. A549 and H1993
cells were co-transfected with pri-miR-379, ASO-miR-379 or their
respective control vectors and 3′UTR CHUK or 3′UTR CHUK mut,
performed as aforementioned. Following transfection in 48-well
plates, cells were cultured for 48 h at 37°C. Firefly and
Renilla luciferase activities were determined with the
Dual-Luciferase Reporter Assay System in a GloMax96 luminescence
reader (both from Promega Corporation), according to the
manufacturer's instructions. Relative luciferase activity was
expressed as the ratio of firefly luciferase activity to the
Renilla luciferase activity in each sample. High EGFP
intensity indicated enhanced promoter activity, reflecting the
binding of the UTR to the promoter.
Flow cytometry
A549 and H1993 cells were detached 48 h after
transfection. Cells were subsequently washed and fixed with PBS and
75% ethanol at 4°C overnight. A549 and H1993 cells were washed with
PBS after fixation, and treated with propidium iodide stain
(Beyotime Institute of Biotechnology) for 30 min at room
temperature or Annexin V-FITC (Sigma-Aldrich; Merck KGaA),
according to manufacturer's instructions. The cell cycle stage and
the levels of apoptosis of both cell types were analyzed with the
BD FACSCanto™ II flow cytometry system (BD Biosciences) and the
ModFit LT software package (version 3.1; Becton, Dickinson and
Company).
Transwell migration and invasion
assays
After 24 h from transfection with the associated
plasmids, A549 and H1993 were collected and re-suspended in
serum-free DMEM. A total of 6×105 cells/ml were added to
the upper Transwell chamber inserts, with or without matrix, and
the lower Transwell chamber was filled with DMEM supplemented with
20% FBS. Cells were incubated at 37°C for 48 h. The cells in the
lower chamber were fixed with 4% paraformaldehyde in PBS for 10 min
at room temperature, and stained with 0.1% crystal violet for 30
min at room temperature. The capacity of cell migration and
invasion were determined by measuring the number of cells in the
lower chamber under bright-field microscopy (magnification,
×200).
Immunofluorescence
Immunochemical staining was performed following the
manufacturer's instructions. Briefly, 5×103 cells (A549
and H1993 cells transfected with pcDNA3, pri-miR-379, ASO-NC and
ASO-miR-379) were fixed with 4% paraformaldehyde at 37°C for 30
min, and washed three times with PBS. Cells were washed with PBS
containing 0.2% Triton X-100 for 2 min, and incubated with PBS
containing 10% donkey serum (HyClone; GE Healthcare Life Sciences)
for 30 min, and incubated with a primary antibody against CHUK
(1:1,000; cat. no. 2078; Cell Signaling Technology) overnight at
4°C. Cells were washed three times with PBS, and then incubated for
60 min at 37°C in the dark with the corresponding FITC conjugated
secondary antibody (1:50; cat no. 9148; Cell Signaling Technology,
Inc.). Cells were rinsed three times with PBS, and incubated in the
dark with PBS containing DAPI (1:1,000) for 10 min at room
temperature to visualize nuclei. A total of five randomly selected
fields were then examined at an ×200 magnification using a phase
contrast fluorescence microscope (Olympus Corporation).
Western blotting
Cells were collected and the total protein content
of the transfected and control cells was extracted via lysis using
RIPA buffer including protease inhibitor cocktail (Roche
Diagnostics GmbH) for 30 min on ice. The total protein
concentration was measured with a bicinchoninic acid protein assay
before immunoblotting. Protein lysates (50 µg/lane) were separated
on a 10% SDS-PAGE and transferred onto polyvinylidene fluoride
membranes (EMD Millipore). The membranes were blocked with 5%
skimmed milk at room temperature for 1 h. The primary antibodies
were used according to the manufacturer's instructions, at 4°C
overnight. The primary antibodies used in the present study were
E-cadherin (1:1,000; cat. no. 3195; Cell Signaling Technology,
Inc.), cytokeratin (1:2,000; cat no. sc-15367; Santa Cruz
Biotechnology, Inc.), Vimentin (1:1,000; cat. no. 5741 Cell
Signaling Technology, Inc.), CHUK (IKKa; 1:1,000; cat. no. 2078;
Cell Signaling Technology, Inc.), NF-κB1 (P50; 1:1,000; cat no.
ab32360; Abcam), RELA (P65; 1:1,000; cat. no. ab16502; Abcam),
GAPDH (1:1,000; sc-47724; Santa Cruz Biotechnology, Inc.), Lamin A
(1:500; cat. no. sc-517580 Santa Cruz Biotechnology, Inc.), Bcl-2
(1:500; SAB4300340; Sigma-Aldrich; Merck KGaA), Bcl-XL (1:1,000;
cat. no. 2764; Cell Signaling Technology, Inc.), Survivin (1:2,000;
cat. no. 2808; Cell Signaling Technology, Inc.) and NFKBIA (Iκβα;
1:1,000; cat. no. 4814 Cell Signaling Technology, Inc.).
Subsequently, the membrane was incubated with a horseradish
peroxidase-conjugated secondary antibody (1:20,000, cat. no. 7074;
Cell Signaling Technology, Inc.) for 2 h at room temperature. The
blots were visualized using an enhanced chemiluminescent reagent
(Hanbio Biotechnology Co., Ltd.). Densitometric analyses of the
western blot bands were performed using Gel-Pro Analyzer software
version 6.0 (Media Cybernetics, Inc.). GAPDH and Lamin A were used
as internal controls. All of the experiments were performed in
triplicate.
Statistical analysis
All data reported are presented as the mean ±
standard deviation from at least three independent experiments,
unless otherwise noted. All statistical analyses were performed
using GraphPad PRISM version 5.0 (GraphPad Software, Inc.).
Differences between cancer tissues and the matched controls were
analyzed using a paired t-test. For comparisons between two
treatment groups, a Student's t-test was used. The correlation
between CHUK and miR-379 levels was analyzed using linear
regression analysis. Multiple groups were compared using one-way
analysis of variance, followed by Tukey's post-hoc test for
multiple comparisons. Differences between the expression levels of
miRNA-379 and different clinicopathological factors were calculated
using the χ2 test. P<0.05 was considered to indicate
a statistically significant difference.
Results
miR-379 expression is downregulated in
NSCLC tissues and cells
In order to confirm the role of miR-379 in human
NSCLC, the expression levels of miR-379 were analyzed in 30 paired
tumor tissues and adjacent non-cancerous lung tissues by RT-qPCR.
The results revealed that the expression of miR-379 was
significantly downregulated in human NSCLC tissues compared with
adjacent non-cancerous tissue samples (Fig. 1A). miR-379 expression in the human
NSCLC cell lines (H1993, H1650, H1299, SK-MES-1 and A549) and
BEAS-2B cell line was also evaluated. The miR-379 expression levels
were markedly downregulated in the human NSCLC cell lines compared
with BEAS-2B (Fig. 1B), which was
consistent with the data from the tissue samples (Fig. 1A). These results suggested that
miR-379 may be involved in the occurrence of NSCLC.
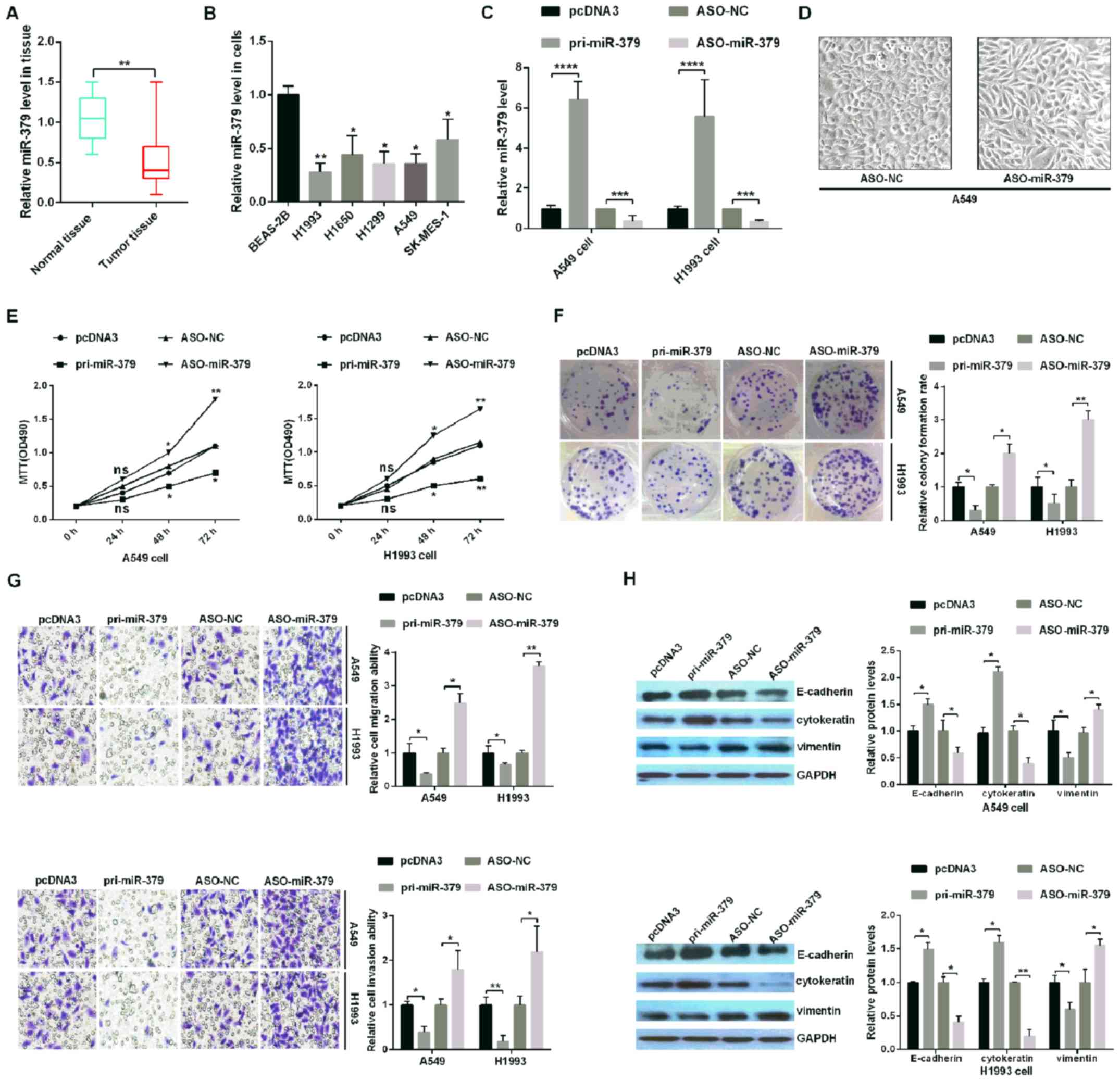 | Figure 1.miR-379 inhibits tumorigenesis in
human NSCLC tissues and cell lines. (A) The relative expression
levels of miR-379 in tumor and paired normal lung tissues. (B) The
expression of miR-379 across five NSCLC cell lines (H1993, H1650,
H1299, SK-MES-1 and A549) and a non-tumorigenic human bronchial
epithelial cell line (BEAS-2B) was analyzed by RT-qPCR. (C) The
relative expression of miR-379 was analyzed by RT-qPCR in A549 and
H1993 NSCLC cells transfected with pri-miR-379 (which upregulates
miR-379), ASO-miR-379 (which downregulates miR-379) and control
vectors (pcDNA3 and ASO-NC, respectively). (D) External cell
morphology was evaluated using microscopy (magnification, ×200).
(E) Analysis of A549 and H1993 cell viability following
transfection of the different plasmids was determined using an MTT
assay. (F) The colony formation ability of A549 and H1993 cells
following transfection with the different plasmids was determined
using a colony formation assay. (G) miR-379 suppressed cell
migration and cell invasion abilities, as demonstrated by the
Transwell assays. Magnification, ×10. (H) The protein levels of
E-cadherin, cytokeratin and Vimentin were verified by western
blotting assays in H1993 and A549 cells. All of the experiments
were repeated at least four times. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001, as indicated. miR, microRNA;
NSCLC, non-small cell lung cancer; RT-qPCR, reverse
transcription-quantitative PCR; ASO, antisense oligonucleotide; NC,
negative control. |
Upregulation of miR-379 inhibits the
proliferation, migration, invasion and EMT process of NSCLC
cells
To confirm whether miR-379 affects NSCLC
progression, H1993 and A549 cells were transfected with pcDNA3,
pri-miR-379, ASO-NC or ASO-miR-379. RT-qPCR assays demonstrated
that transfection with pri-miR-379 and ASO-miR-379 led to the up-
and downregulation, respectively, of miR-379 in H1993 and A549
cells (Fig. 1C). In addition, as
shown in Fig. 1D, A549-ASO-miR-379
cells appeared to be more mesothelial compared with the more
epithelial-like A549-ASO-NC cells, indicating that dysregulation of
miR-379 may be relevant to tumor metastasis. In addition, the
overexpression of miR-379 decreased cell viability, while the
knockdown of miR-379 appeared to promote the viability of H1993 and
A549 cells by MTT (Fig. 1E).
Colony formation assays further corroborated these results, as
indicated by the increase and decrease in the colony formation rate
following the down- and upregulation, respectively, of miR-379
(Fig. 1F).
To confirm whether miR-379 influences cell migration
and invasion, transfection with pri-miR-379 or ASO-miR-379 and its
corresponding control were performed. The results revealed that
miR-379 overexpression significantly decreased the invasion and
migration of H1993 and A549 cells compared with the pcDNA3-NC, and
miR-379 knockdown increased the invasion and migration of H1993 and
A549 cells compared with the ASO-NC (Fig. 1G). Finally, western blotting was
used to assess the levels of EMT markers, including E-cadherin,
cytokeratin and Vimentin, following manipulation of miR-379
expression. The expression levels of E-cadherin and cytokeratin
were increased following transfection with pri-miR-379 when
compared with the pcDNA3-NC, while the expression levels of
Vimentin were significantly decreased under the same conditions
(Fig. 1H). When transfected with
ASO-miR-379, the expression levels of E-cadherin and cytokeratin
were decreased compared with the ASO-NC, while the expression
levels of Vimentin were significantly increased under the same
conditions (Fig. 1H). This set of
results demonstrated that miR-379 may act as a tumor suppressor,
inhibiting the proliferation, migration and invasion of human NSCLC
cells.
CHUK is a direct target of miR-379 in
human NSCLC cells
To investigate the underlying mechanism of action of
miR-379 in human NSCLC cells, the potential targets of miR-379 were
predicted using miRNA.org, TargetScan 7.1, miRbase and
miRanda, and CHUK was identified as a potential target gene of
miR-379 in subsequent experiments (Fig. 2A). To prove that miR-379 may
directly target CHUK mRNA, EGFP reporter plasmids including the
3′UTR or the 3′UTR-mut of CHUK were constructed. The
co-transfection of pri-miR-379 and wild type 3′UTR of CHUK
significantly reduced the relative levels of EGFP activity, while
the co-transfection of ASO-miR-379 and wild-type 3′UTR of CHUK
significantly increased the relative EGFP activity (Fig. 2B). In addition, pri-miR-379 or
ASO-miR-379 did not affect the relative EGFP activity levels when
co-transfected with the mutant form of CHUK 3′UTR (Fig. 2C).
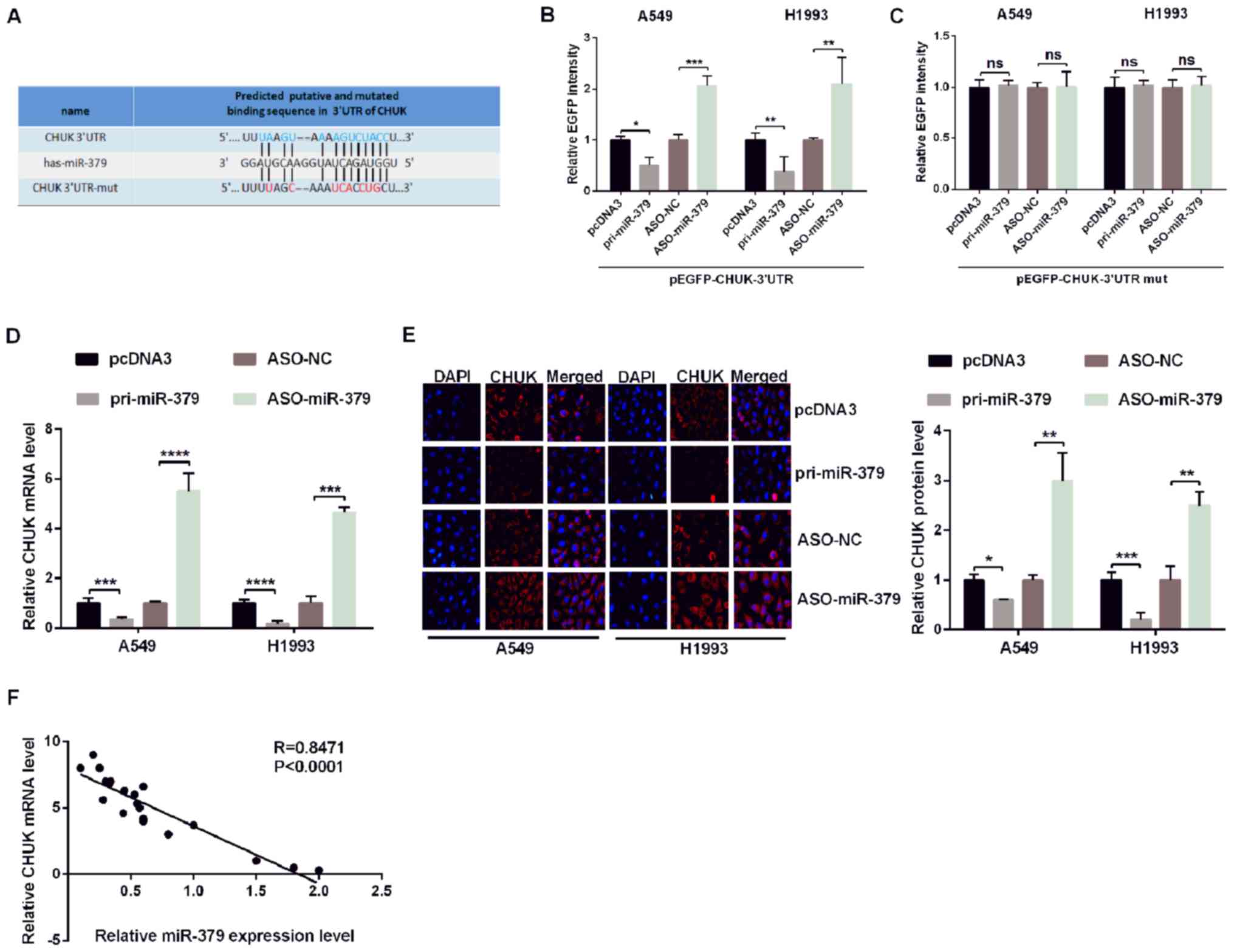 | Figure 2.miR-379 directly targets CHUK in
non-small cell lung cancer cells. (A) The predicted miR-379 binding
sites were identified using TargetScan 7.1. CHUK was identified as
a potential target mRNA, and its 3′UTR sequence, along with the
mutated sequence are presented. A549 and H1993 cells were
co-transfected with pri-miR-379 or ASO-miR-379 and with
pcDNA3/EGFP-CHUK (B) 3′UTR or (C) 3′UTR-mut. EGFP intensity was
measured by spectrophotometry, and the results revealed that CHUK
is a direct target of miR-379 as shown by the increase in
fluorescence in the presence of wild-type 3′UTR of CHUK. CHUK (D)
mRNA levels and (E) protein expression in A549 and H1993 cells
transfected with pri-miR-379 or ASO-miR-379 and the respective
controls were determined by reverse transcription-quantitative PCR
and immunofluorescence, respectively. (F) Correlation analysis of
the expression data revealed a negative correlation between the
expression of miR-379 and CHUK mRNA. All of the experiments were
repeated at least four times. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001, as indicated. miR, microRNA;
CHUK, inhibitor of nuclear factor-κB kinase subunit α; UTR,
untranslated region; mut, mutant; ASO, antisense oligonucleotide;
EGFP, enhanced green fluorescent protein; NS, not significant; NC,
negative control. |
To further confirm the association between miR-379
and CHUK, the endogenous CHUK mRNA and protein levels in A549 and
H1993 cells were evaluated following transfection with pri-miR-379
compared with ASO-miR-379 or the NCs pcDNA3 and ASO-NC. RT-qPCR and
immunofluorescence assays demonstrated that pri-miR-379
significantly reduced the expression of endogenous CHUK mRNA and
protein (Fig. 2D and E).
Additionally, as shown in Fig. 2F,
the levels of CHUK mRNA are negatively correlated with the levels
of miR-379. These results suggested that CHUK is a target gene of
miR-379, and that it may be negatively regulated by miR-379 in
human NSCLC cells.
CHUK functions as an oncogene in human
NSCLC cells
To confirm the results obtained, the expression
levels of CHUK mRNA were evaluated in 30 paired NSCLC tissues via
RT-qPCR. The results revealed that the expression of CHUK mRNA was
significantly upregulated in human NSCLC tissues compared with
adjacent non-cancerous tissue samples (Fig. 3A). CHUK mRNA expression in the
human NSCLC cell lines (H1993, H1650, H1299, SK-MES-1 and A549) and
the BEAS-2B control cell line was also evaluated. This revealed
that the expression levels of CHUK mRNA were markedly upregulated
in the NSCLC cell lines compared with BEAS-2B (Fig. 3B). To investigate whether CHUK may
affect NSCLC progression, H1993 and A549 cells were transfected
with pcDNA3, pCHUK, pSilencer or shR-CHUK. RT-qPCR and western
blotting assays demonstrated that the CHUCK mRNA and protein levels
were upregulated with pCHUK and downregulated with shR-CHUK in
H1993 and A549 cells when compared with the respective controls
(pcDNA3 and pSilencer, respectively; Fig. 3C and D). The overexpression of CHUK
promoted cell proliferation, while the knockdown of CHUK had the
opposite effect on both H1993 and A549 cells, as shown by the MTT
and colony formation assays (Fig. 3E
and F).
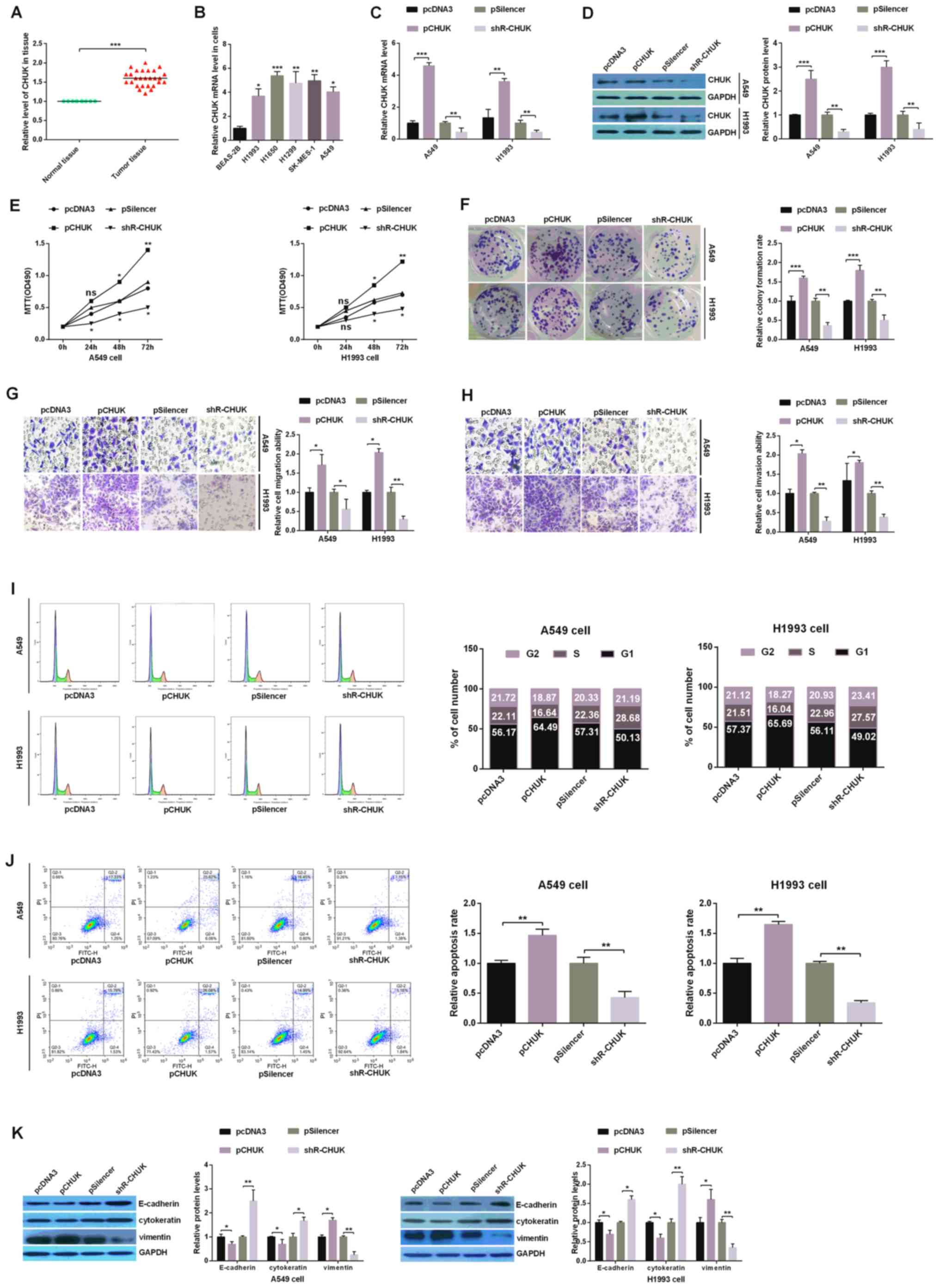 | Figure 3.CHUK leads to the development of
human NSCLC cells. (A) Relative expression levels of CHUK in tumor
and paired normal lung tissues. (B) The relative expression of CHUK
across five NSCLC cell lines (H1993, H1650, H1299, SK-MES-1 and
A549) and a non-tumorigenic human bronchial epithelial cell line
(BEAS-2B) was analyzed using RT-qPCR. A549 and H1993 cells were
transfected with a CHUK overexpressing plasmid (pCHUK), shR-CHUK or
an empty control plasmid (pcDNA3 or pSilencer, respectively). The
efficiency of overexpression and knockdown of CHUK was demonstrated
using (C) RT-qPCR and (D) western blotting. (E) Analysis of CHUK
expression manipulation on the viability of A549 and H1993 cells
was performed using an MTT assay. (F) The colony
formation/proliferation ability of A549 and H1993 cells with both
up- and downregulated levels of CHUK was determined using a colony
formation assay. The (G) migration and (H) invasion abilities of
transfected A549 and H1993 cells were determined using Transwell
assays (magnification, ×10). (I) Flow cytometry assay indicating
the percentage of A549 and H1993 cells in different phases of the
cell cycle following transfection with pCHUK or shR-CHUK and the
respective controls. (J) The rates of apoptosis were measured using
flow cytometry and by evaluating the levels of FITC and PI double
staining. (K) The protein levels of E-cadherin, cytokeratin and
Vimentin were analyzed using western blotting assays in H1993 and
A549 cells. All of the experiments were repeated at least four
times. *P<0.05, **P<0.01 and ***P<0.001, as indicated.
miR, microRNA; NSCLC, non-small cell lung cancer; RT-qPCR, reverse
transcription-quantitative PCR; CHUK, inhibitor of nuclear
factor-κB kinase subunit α; shR, short hairpin RNA; FITC,
fluorescein isothiocyanate; PI, propidium iodide. |
Regarding migration and invasion, H1993 and A549
cells transfected with pCHUK exhibited significantly greater
invasion and migration capabilities in Transwell assays compared
with pcDNA3-NC, while shR-CHUK significantly decreased invasion and
migration capabilities of cells compared with pSilencer (Fig. 3G and H). The flow cytometry assay
revealed that overexpression of CHUK may accelerate the cell cycle
process, induce G1-S arrest and decrease the apoptotic rate in
human NSCLC cells. On the other hand, the knockdown of CHUK
reversed the above results (Fig. 3I
and J). Lastly, the levels of E-cadherin and cytokeratin were
observed to be downregulated, while Vimentin was upregulated in
cells that overexpressed CHUK. Conversely, the knockdown of CHUK
promoted the expression of E-cadherin and cytokeratin, while
suppressing the expression of Vimentin (Fig. 3K). These results suggest that CHUK
may function as an oncogene in human NSCLC cells.
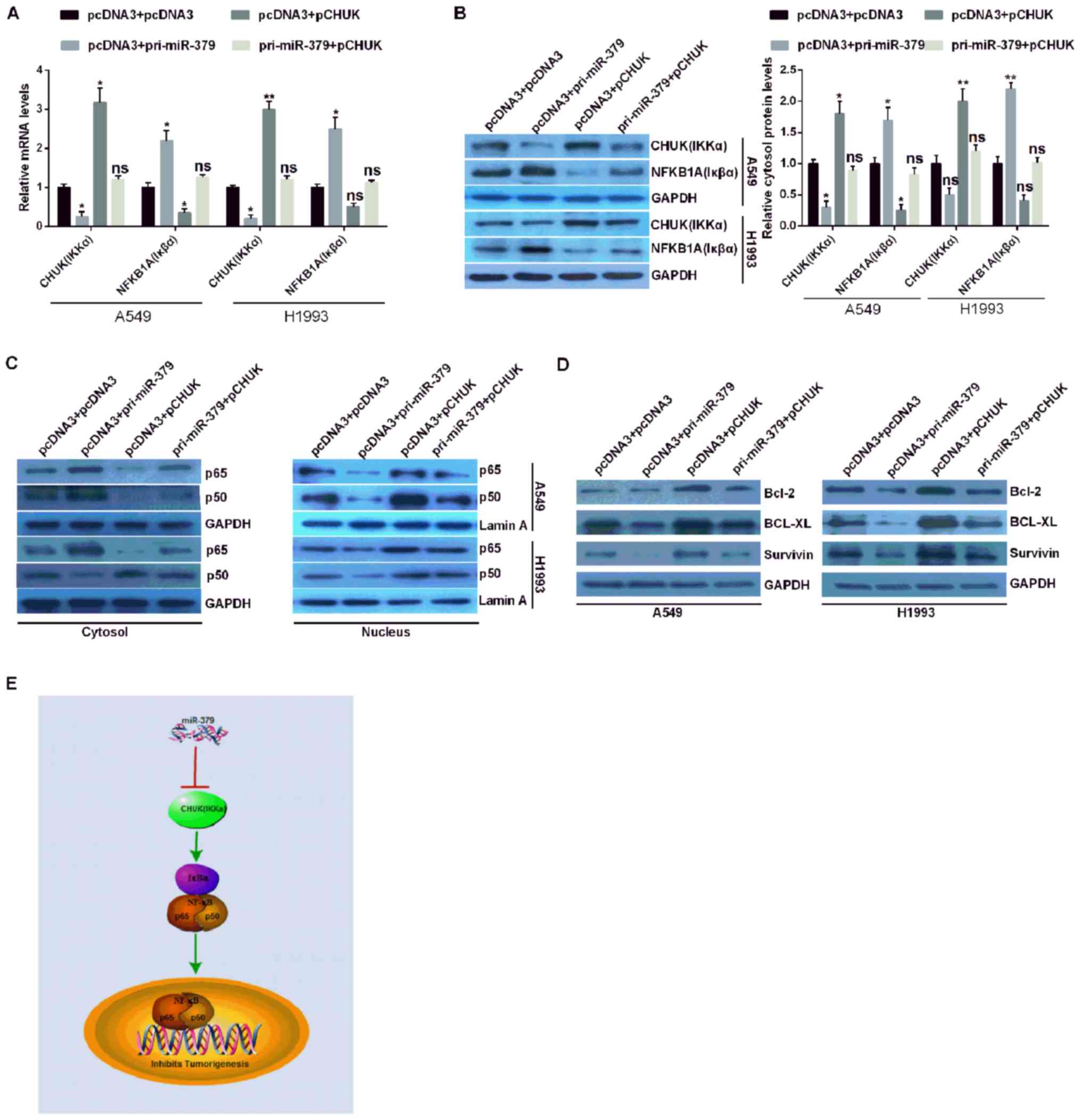
miR-379 inhibits the NF-κB pathway via
the downregulation of CHUK in NSCLC
In order to confirm whether miR-379 affected the
NF-κB signaling pathway in NSCLC cells, the expression levels of
the CHUK/IKKα genes and NFKBIA (Iκβα) were analyzed via RT-qPCR and
western blotting assays. The results demonstrated that NFKBIA
(Iκβα) was suppressed following CHUK (IKKα) overexpression, but
promoted following CHUK (IKKα) downregulation in the context of
miR-379 overexpression (Fig. 4A and
B). p65 (NF-κB3) and p50 (NF-κB1) are heterodimers identified
as constituting members of NF-κB (18). Therefore, the nuclear distribution
of p65 and p50 may have increased in pCHUK-transfected cells, and
decreased in pri-miR-379-transfected cells. The distribution levels
of p65 and p50 in the nucleus did not change in A549 and H1993
cells co-transfected with pri-miR-379 and pCHUK (Fig. 4C). However, under the same
conditions, the expression of p65 and p50 in the cytoplasm is
opposite to that of the nucleus (Fig.
4C). These results demonstrated that CHUK may increase the
expression of p65 and p50 in cell nuclei.
Therefore, it was hypothesized that CHUK may
activate NF-κB in NSCLC cells. To clarify this hypothesis, the
expression levels of Bcl-2, BCL-XL and survivin were detected. The
expression levels of these genes were higher in NSCLC cells
overexpressing CHUK and lower in NSCLC expressing miR-379. The
expression levels of Bcl-2, BCL-XL and survivin were similar in
miR-379 and CHUK co-transfected NSCLC cells compared with those in
pcDNA3 + pcDNA3 co-transfected NSCLC cells (Fig. 4D). These results suggested that
miR-379 may inhibit tumorigenesis by directly targeting CHUK 3′UTR,
inhibiting CHUK expression, and potentially suppressing the NF-κB
signaling pathway in NSCLC (Fig.
4E).
Discussion
NSCLC is one of the most common types of tumors and
has a high mortality rate (19).
The occurrence of NSCLC is a multi-stage process characterized by
significant changes in gene expression and in the physiological
structure of the lung tissue (20). Alterations in gene expression may
involve the inactivation of tumor suppressor genes or the
activation of oncogenes, which causes further abnormalities in gene
expression (21). Although
research on NSCLC has led to the development of several therapeutic
approaches, including radiation therapy, chemotherapy and surgical
resection, NSCLC remains difficult to treat and has a poor
prognosis (22). In order to fully
discern the progression of NSCLC, a deeper understanding of the
associated gene expression profiles and related biological
mechanisms is required to improve current treatments.
Previous studies have shown that miRs have crucial
roles during tumorigenesis (23–29),
and that miR-379 is downregulated in many types of cancer. For
example, Xie et al (30)
reported that miR-379 was downregulated in human osteosarcoma
specimens and cell lines, that miR-379 overexpression inhibited
cell proliferation and colony formation and that it promoted a
G0/G1 cell cycle arrest in human osteosarcoma cells. In addition,
Li et al (31) reported
that miR-379 inhibited vascular smooth muscle cell function and
survival by targeting insulin-like growth factor-1 through the
activation of extracellular signaling pathways in these cells.
Furthermore, Chen et al (32) reported that miR-379-5p may repress
cell invasion and metastasis, promote apoptosis and inhibit cell
cycle progression by targeting the protein tyrosine kinase 2/AKT
serine/threonine kinase signaling pathway in hepatocellular
carcinoma. These studies suggested that miR-379 may have important
functions in various types of cancer, and may constitute as a
therapeutic target in the treatment of these diseases. In the
present study, it was demonstrated that miR-379 is significantly
downregulated in NSCLC tissues and cell lines. The overexpression
of miR-379 may have significantly inhibited growth, migration and
invasion, promoted apoptosis, and arrested the cell cycle of NSCLC
cells in vitro by reducing the expression levels of Vimentin
and promoting the expression of cytokeratin and E-cadherin.
Overall, the results suggested that this miR may serve as a cancer
gene suppressor in human NSCLC cells. In addition, overexpression
of miR-379 may inactivate the NF-κB signaling pathway in NSCLC
cells.
miRs serve an important role across a variety of
biological processes, including cancer, via complementary binding
with the 3′UTR of their target genes (33). In the present study, bioinformatic
analysis was used to predict the target genes of miR-379, and CHUK
was chosen as a potential target. Furthermore, the expression level
of CHUK mRNA and protein was distinctly increased in the H1993,
H1650, H1299, SK-MES-1 and A549 cell lines, which was inversely
correlated with the levels of miR-379 in NSCLC cells. In addition,
a EGFP reporter assay revealed that miR-379 directly targeted the
3′UTR of the CHUK mRNA in A549 and H1993 cells. RT-qPCR and western
blot assays further demonstrated that the overexpression of miR-379
was associated with a significant decrease in the expression of
CHUK and that the knockdown of miR-379 increased the expression of
CHUK in both A549 and H1993 cells.
The NF-κB signaling pathway serves important roles
in a number of biological processes, including immunity,
inflammatory and apoptotic responses (34). The activation of the NF-κB pathway
is also involved in the pathogenesis of a variety of diseases,
including many types of human cancers. Notably, the activation of
NF-κB may also induce the expression of anti-apoptotic genes in
germ cells, which may in turn attenuate germ cell apoptosis
(35). The activation of NF-κB is
also involved in the cisplatin resistance of various types of
cancer (36,37). Naidu et al (38) reported that platelet-derived growth
factor receptor-modulated miR-23b and miR-125a-5p may inhibit lung
tumorigenesis by targeting multiple components of the KRAS and
NF-κB pathways. All of these studies confirmed that CHUK may be
involved in tumorigenesis; however, the function of CHUK in NSCLC
remained unclear. The results of the present study confirmed that
the expression levels of CHUK, at both the mRNA and protein levels,
were upregulated in NSCLC cell lines, and the overexpression of
CHUK may have enhanced cell viability, colony formation ability and
the EMT process in NSCLC cells. Furthermore, ectopic CHUK
expression may have activated the NF-κB signaling pathway in NSCLC
cells.
The present study demonstrated that the
downregulation of miR-379 in NSCLC tissues and cell lines is a
common phenomenon, and that miR-379 may serve an important role in
regulating the growth, migration and invasion of human NSCLC cells
by promoting EMT. In addition, miR-379 inhibited the expression of
CHUK by directly targeting the 3′-UTR of CHUK mRNA and miR-379 may
have inhibited the activation of the NF-κB signaling pathway by
regulating CHUK in NSCLC cells. In conclusion, the results of the
present study may have provided novel insights into NSCLC
tumorigenesis, and provided a potential new therapeutic target for
the treatment for NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BL, ZW and SC performed the experiments, analyzed
the data and wrote the manuscript. JZ designed and supervised the
study and wrote the manuscript. LD, YY and ZY assisted with data
collection and analysis, and wrote the manuscript.
Ethics approval and consent to
participate
All participants provided written informed consent.
The present study was approved by the Ethical Oversight Committee
of Department of Respiratory and Critical Care Medicine, Tianjin
Chest Hospital (Tianjin, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheng H, Shcherba M, Kandavelou K, Liang
Y, Liu H and Perez-Soler R: Emerging drugs for squamous cell lung
cancer. Expert Opin Emerg Drugs. 20:149–160. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karlsen TA, De Souza GA, Degaard B,
Engebretsen L and Brinchmann JE: MicroRNA-140 inhibits inflammation
and stimulates chondrogenesis in a model of interleukin 1β-induced
osteoarthritis. Mol Ther Nucleic Acids. 5:e3732016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yin J, Wang M, Jin C and Qi Q: MiR-101
sensitizes A549 NSCLC cell line to CDDP by activating caspase
3-dependent apoptosis. Oncol Lett. 7:461–465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zinner R, Visseren-Grul C, Spigel DR and
Obasaju C: Pemetrexed clinical studies in performance status 2
patients with non-small cell lung cancer (review). Int J Oncol.
48:13–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thayanithy V, Sarver AL, Kartha RV, Li L,
Angstadt AY, Breen M, Steer CJ, Modiano JF and Subramanian S:
Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma.
Bone. 50:171–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Makeyev EV and Maniatis T: Multilevel
regulation of gene expression by microRNAs. Science. 319:1789–1790.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu N, Zhang Q, Liu Q, Yang J and Zhang S:
A meta-analysis: microRNAs' prognostic function in patients with
nonsmall cell lung cancer. Cancer Med. 6:2098–2105. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang K, Han X, Zhang Z, Zheng L, Hu Z,
Yao Q, Cui H, Shu G, Si M, Li C, et al: The liver-enriched
lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch
pathways. Nat Commun. 8:1442017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Legras A, Pécuchet N, Imbeaud S, Pallier
K, Didelot A, Roussel H, Gibault L, Fabre E, Le Pimpec-Barthes F,
Laurent-Puig P and Blons H: Epithelial-to-mesenchymal transition
and microRNAs in lung cancer. Cancers (Basel). 9(pii): E1012017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang M, Meng B, Liu Y, Yu J and Chen Q:
MiR-124 inhibits growth and enhances radiation-induced apoptosis in
non-small cell lung cancer by inhibiting STAT3. Cell Physiol
Biochem. 44:2017–2028. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu J, Bian T, Feng J, Qian L, Zhang J,
Jiang D, Zhang Q, Li X, Liu Y and Shi J: miR-335 inhibited cell
proliferation of lung cancer cells by target Tra2β. Cancer Sci.
109:289–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xue X, Fei X, Hou W, Zhang Y, Liu L and Hu
R: miR-342-3p suppresses cell proliferation and migration by
targeting AGR2 in non-small cell lung cancer. Cancer Lett.
412:170–178. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Le F, Zhang JY, Liu W, Huang XM and Luo
WZ: The levels of NF-κB p50 and NF-κB p65 play a role in thyroid
carcinoma malignancy in vivo. J Int Med Res. 46:4092–4099. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Chen S, Hang W, Huang H and Ma H:
MiR-95 induces proliferation and chemo- or radioresistance through
directly targeting sorting nexin1 (SNX1) in non-small cell lung
cancer. Biomed Pharmacother. 68:589–595. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Zhao Y, Sun S, Liu Z, Zhang Y and
Jiao S: Overexpression of microRNA-221 is associated with poor
prognosis in non-small cell lung cancer patients. Tumour Biol.
37:10155–10160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun S, Schiller JH, Spinola M and Minna
JD: New molecularly targeted therapies for lung cancer. J Clin
Invest. 117:2740–2750. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun C, Liu Z, Li S, Yang C, Xue R, Xi Y,
Wang L, Wang S, He Q, Huang J, et al: Down-regulation of c-Met and
Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell
proliferation, migration and colony formation. Oncotarget.
6:25533–25574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bier A, Giladi N, Kronfeld N, Lee HK,
Cazacu S, Finniss S, Xiang C, Poisson L, Decarvalho AC, Slavin S,
et al: MicroRNA-137 is downregulated in glioblastoma and inhibits
the stemness of glioma stem cells by targeting RTVP-1. Oncotarget.
4:665–676. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun C, Huang C, Li S, Yang C, Xi Y, Wang
L, Zhang F, Fu Y and Li D: Hsa-miR-326 targets CCND1 and inhibits
non-small cell lung cancer development. Oncotarget. 7:8341–8359.
2016.PubMed/NCBI
|
|
26
|
Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y
and Li D: Long non-coding RNA NEAT1 promotes non-small cell lung
cancer progression through regulation of miR-377-3p-E2F3 pathway.
Oncotarget. 7:51784–51814. 2016.PubMed/NCBI
|
|
27
|
Sun CC, Li SJ, Zhang F, Zhang YD, Zuo ZY,
Xi YY, Wang L and Li DJ: The novel miR-9600 suppresses tumor
progression and promotes paclitaxel sensitivity in non-small-cell
lung cancer through altering STAT3 expression. Mol Ther Nucleic
Acids. 5:e3872016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang C, Liu J, Wang X, Wu R, Lin M,
Laddha SV, Yang Q, Chan CS and Feng Z: MicroRNA-339-5p inhibits
colorectal tumorigenesis through regulation of the MDM2/p53
signaling. Oncotarget. 5:9106–9117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xi Y, Wang L, Sun C, Yang C, Zhang F and
Li D: The novel miR-9501 inhibits cell proliferation, migration and
activates apoptosis in non-small cell lung cancer. Med Oncol.
33:1242016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xie X, Li YS, Xiao WF, Deng ZH, He HB, Liu
Q and Luo W: MicroRNA-379 inhibits the proliferation, migration and
invasion of human osteosarcoma cells by targetting EIF4G2. Biosci
Rep. 37(pii): BSR201605422017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li K, Wang Y, Zhang A, Liu B and Jia L:
miR-379 inhibits cell proliferation, invasion, and migration of
vascular smooth muscle cells by targeting insulin-like factor-1.
Yonsei Med J. 58:234–240. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen JS, Li HS, Huang JQ, Dong SH, Huang
ZJ, Yi W, Zhan GF, Feng JT, Sun JC and Huang XH: MicroRNA-379-5p
inhibits tumor invasion and metastasis by targeting FAK/AKT
signaling in hepatocellular carcinoma. Cancer Lett. 375:73–83.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Majidinia M, Aghazadeh J,
Jahanban-Esfahlani R and Yousefi B: The roles of Wnt/β-catenin
pathway in tissue development and regenerative medicine. J Cell
Physiol. 233:5598–5612. 2017. View Article : Google Scholar
|
|
34
|
Perkins ND: Integrating cell-signalling
pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol.
8:49–62. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wright A, Reiley WW, Chang M, Jin W, Lee
AJ, Zhang M and Sun SC: Regulation of early wave of germ cell
apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev
Cell. 13:705–716. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mabuchi S, Ohmichi M, Nishio Y, Hayasaka
T, Kimura A, Ohta T, Saito M, Kawagoe J, Takahashi K,
Yada-Hashimoto N, et al: Inhibition of NFkappaB increases the
efficacy of cisplatin in in vitro and in vivo ovarian cancer
models. J Biol Chem. 279:23477–23485. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Y, Ahmed F, Ali S, Philip PA, Kucuk O
and Sarkar FH: Inactivation of nuclear factor KB by soy isoflavone
genistein contributes to increased apoptosis induced by
chemotherapeutic agents in human cancer cells. Cancer Res.
65:6934–6942. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Naidu S, Shi L, Magee P, Middleton JD,
Laganá A, Sahoo S, Leong HS, Galvin M, Frese K, Dive C, et al:
PDGFR-modulated miR-23b cluster and miR-125a-5p suppress lung
tumorigenesis by targeting multiple components of KRAS and NF-κB
pathways. Sci Rep. 7:154412017. View Article : Google Scholar : PubMed/NCBI
|