Introduction
Breast cancer (BC) is one of the most common
malignancies worldwide, accounting for 29% of all female cancer
cases, and has a high mortality rate (1–3).
Although advanced treatments, such as early detection, mastectomy,
radiotherapy, chemotherapy, endocrine treatment, targeted therapy
and even systemic treatment, have greatly improved, the 5-year
overall survival rate for BC is low, especially when the cancer
becomes metastatic (4). A high
proportion, ~90%, of BC-related mortality is attributed to the
formation of metastatic lesions (5). Thus, this malignancy is a threat to
the health of women worldwide. Emerging molecular biomarkers that
can predict patient outcomes and therapy responses have been
identified, and researchers have shown that numerous molecular
triggers are important in the development of BC (6–10).
Long non-coding RNAs (lncRNAs), with a length of
>200 nucleotides, are a class of RNA molecule that lack the
ability to produce functional proteins (11). They have been demonstrated to be
involved in complex biological processes, including cell cycle
regulation, cell differentiation, transcriptional modification,
chromosome remodelling, epigenetic regulation and tumour
progression (12–15). lncRNAs, which play an important
role in human cancer, are an area of emerging focus for clinical
applications (8,16,17).
lncRNAs can function as competing endogenous RNAs
(ceRNAs) by sequestering microRNAs (miRNAs/miRs) and regulating
downstream targets (18). In BC,
previous studies have revealed that the expression of lncRNAs is
related to the clinical features and overall survival rate of BC.
Dong et al (19) found that
the lncRNA AGAP2 antisense RNA 1 promoted BC growth and
chemoresistance, by regulating the expression of myeloid
differentiation primary response protein MyD88 in vitro and
in vivo. ATXN8OS is a lncRNA with a size of 1,236 bp located
on chromosome 13q21 (20). Koob
et al (21) reported that
abnormal expression of ATXN8OS occurs in various brain tissues. A
previous study revealed that ATXN8OS was up-regulated in BC and was
involved in the lncRNA-miRNA-mRNA ceRNA network. However, the
specific function of this lncRNA, or its role in the development
and progression of BC, was not investigated (22). Therefore, the aim of the present
study was to explore the underlying molecular mechanism of ATXN8OS
in BC, and to identify its potential role as a putative diagnostic
biomarker and therapeutic target.
Materials and methods
Patient samples
Human BC samples (n=120) and matched non-tumour
tissues were collected from patients who underwent a radical
mastectomy at 900th Hospital of the Joint Logistics Support Force
between August 2010 and October 2017. A 60-month follow-up survey
was performed. The BC tissues were reviewed blind by two
pathologists based on the American Society of Clinical Oncology
guidelines (23). Fresh surgical
samples were frozen in liquid nitrogen and stored at −80°C. No
interventional or other treatments were performed on the patients
prior to surgery. The diagnoses of these samples were verified by
pathologists in the hospital. Written informed consent was obtained
from all of the patients, and the study protocol (no. 20171026) was
approved by the Ethics Committee of 900th Hospital of the Joint
Logistics Support Force.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
In accordance with the manufacturer's instructions,
total RNA was isolated from tissues and cells using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). RT
was performed using a Thermo Scientific RT kit (Thermo Fisher
Scientific, Inc.). RT was performed by sequential incubations at 50
min at 42°C, 15 min at 70°C and 20 min at 37°C. RT-qPCR was
performed using an ABI7500 qPCR instrument (Applied Biosystems;
Thermo Fisher Scientific, Inc.). In total, 5 µl SYBR Premix Ex Taq
II (Takara Biotechnology Co., Ltd.), 0.4 µl forward primer (10 µM),
0.4 µl reverse primer (10 µM), 0.2 µl ROX Reference Dye (Takara
Biotechnology Co., Ltd.), 1.0 µl cDNA template and 3.0 µl ddH2O
were mixed in the reaction solution. qPCR was performed as follows:
Initial denaturation for 10 sec at 95°C, followed by 45 cycles of 5
sec at 95°C and 34 sec at 60°C. Relative ATXN8OS expression was
calculated using the comparative quantification cycle (Cq) method.
Fold changes were calculated using the 2−ΔΔCq method
(24). GAPDH and U6 served as
endogenous controls for mRNA or lncRNA/miRNA expression,
respectively. The primer sequences used in the present study were
as follows: ATXN8OS primer forward, 5′-GCGCGAGAGCCCCGTGTTA-3′ and
reverse, 5′-TCTCTTGCCCTTCTGCCTTCTACT-3′; tyrosine protein kinase
JAK2 primer forward, 5′-GGGAGGTGGTCGCTGTAAAA-3′ and reverse,
5′-ACCAGCACTGTAGCACACTC-3′; forkhead box A1 (FOXA1) primer forward,
5′-AATCATTGCCATCGTGTG-3′ and reverse, 5′-CGCGGCTTAAAATCTGGTAT-3′;
miR-204 primer forward, 5′-CTGTCACTCGAGCTGCTGGAATG-3′ and reverse,
5′-ACCGTGTCGTGGAGTCGGCAATT-3′; GAPDH primer forward,
5-GTCTCCTCTGACTTCAACAGCG-3 and reverse,
5′-ACCACCCTGTTGCTGTAGCCAA-3; U6 primer forward,
5′-CTCGCTTCGGCAGCACATA-3′ and reverse,
5′-AACGATTCACGAATTTGCGT-3′.
Cell culture
MCF-10A, a normal breast epithelial cell line, and
two human BC cell lines, MCF7 and MDA-MB-231, were obtained from
the Type Culture Collection of the Chinese Academy of Sciences. All
cells were cultured in DMEM (Biological Industries) supplemented
with 50 U/ml penicillin, 0.1 mg/ml streptomycin and 10% FBS
(Biological Industries) at 37°C in a 5% CO2 atmosphere
in a humidified incubator.
RNA oligonucleotides and
transfection
The ATXN8OS small interfering (si)RNA (si-ATXN8OS;
5′-TAAATGTTTTTCTTCCCTCTC-3′) and scrambled siRNA (si-ctrl;
5′-CGAGAATTTGGTCTAAAGAGAA-3′) were purchased from Shanghai
GenePharma Co., Ltd. miR-204 mimic (5′-AAGGGAAACAGTAGGAAGCGGA-3′),
miR-204 inhibitor (5′-TCCGCTTCCTACTGTTTCCCTT-3′), control miRNA
mimic (miR-con; 5′-AAGTGACGGTGAGATCCAGGCT-3′) and miR-con inhibitor
(5′-AGCCTGGATCTCACCGTCACTT-3′) were purchased from Shanghai
GenePharma Co., Ltd. Briefly, 1×106 cells were seeded
into a 6-well plate and transfected until they were 90% confluent.
Transfection of the cells with si-ATXN8OS or si-ctrl (10 nM) and
the miR-204 or miR-con mimic/inhibitor (10 nM) was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) with Opti-MEM serum-free medium (Gibco; Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol. The
efficiency of transfection was determined in each experiment using
RT-qPCR 24 h post-transfection. All functional experiments were
carried out 48 h post-transfection.
Cell viability assay
The Cell Counting Kit-8 (CCK-8, Dojindo Molecular
Technologies, Inc.) was used to evaluate MCF7 and MDA-MB-231
proliferation. Cells were collected 48 h after transfection and
seeded at a density of 1,000 cells/well in 96-well plates. At 0, 2
or 4 days, the CCK-8 reagent was added to the 96-well plate and the
cells were cultured for a further 2 h at 37°C, according to the
manufacturer's protocol. The absorbance was measured at a
wavelength of 450 nm using a Benchmark Microplate Reader (Bio-Rad
Laboratories, Inc.). Each assay was performed in triplicate and
repeated three times.
Flow cytometry
Flow cytometry was used to analyse the cell cycle.
MCF7 and MDA-MB-231 cells transfected with si-ATXN8OS and si-ctrl
were removed from the culture plates using trypsin and washed three
times with cold PBS until they were 90% confluent. The cells were
fixed with cold 70% ethanol at 4°C overnight. The cells were then
incubated with 20 µg/ml propidium iodide (Sigma-Aldrich; Merck
KGaA) for 30 min at room temperature using the BD Cycletest Plus
DNA Reagent kit (BD Biosciences) to stain the cells. Cell cycle
distribution profiles were generated using a FACSCalibur Flow
Cytometry system (BD Biosciences) with FlowJo software version
7.6.1 (Tree Star, Inc.).
Transwell invasion assay
A Transwell invasion assay was carried out to test
the invasion ability of cells. The upper Transwell chamber
containing DMEM without FBS was precoated with 100 µl of Matrigel
and 4×103 cells in 0.1 ml cell suspension were then
added to the coated membrane in the chamber. The lower chamber was
filled with 600 µl of 20% FBS DMEM. After 24 h, the cells that did
not pass through the membrane were cleared using cotton swabs and
4% paraformaldehyde was added to fix the cells at room temperature
for 20 min, followed by staining the cells with 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA) for 15 min at room temperature. Images
of five random fields were captured using an optical microscope
(magnification, ×400).
Western blot analysis
Western blot analysis was performed as described
previously (25). The primary
antibodies were incubated at 4°C overnight and then incubated with
the secondary goat antibodies horseradish peroxidase-conjugated
(1:2,000; cat. no. ab6112; Abcam) for 2 h at room temperature. The
following primary antibodies were used: JAK2 (1:1,000; cat. no.
3230; Cell Signaling Technology, Inc.), FOXA1 (1:1,000; cat. no.
53528; Cell Signaling Technology, Inc.), angiopoietin 1 (ANGPT1;
1:500; cat. no. ab8451; Abcam), TGF-β receptor type 2 (TGFβR2;
1:500; cat. no. ab186838; Abcam) and GAPDH (1:1,000; cat. no.
ab181602; Abcam).
Reporter vector construction and
luciferase reporter assay
StarBase V3.0 (http://starbase.sysu.edu.cn/index.php) was used to
search for potential miRNAs that can bind to ATXN8OS. Through this
analysis, ATXN8OS fragments containing the proposed binding site of
miR-204 were identified. The ATXN8OS fragments were then amplified
and integrated into the pGL3-promoter vector (Promega Corporation),
between the NheI and BglII sites, to construct the
reporter vector ATNX8OS-wild-type (Wt ATXN8OS). In parallel with Wt
ATXN8OS, the related mutant fragments were also cloned to construct
the reporter vector ATXN8OS-mutant (Mut ATXN8OS). MCF7 and
MDA-MB-231 cells were co-transfected with 100 ng Wt ATXN8OS or Mut
ATXN8OS and 10 nM miR-204 or miR-con mimic/inhibitor using
Lipofectamine® 2000. Luciferase activity was measured 48
h after transfection using a dual-luciferase reporter assay system
(Promega Corporation) according to the manufacturer's protocol.
Firefly luciferase activity was normalized to Renilla
(Promega Corporation) luciferase gene activity.
Statistical analysis
All statistical data were analysed using SPSS 19.0
software (IBM Corp.) and GraphPad Prism 5.0 software (GraphPad
Software, Inc.). Clinicopathological characteristics were evaluated
using the χ2 test. The overall survival rate was
assessed using the Kaplan-Meier method. The log-rank test was used
to compare the survival data. Student's t-test or the Wilcoxon
signed-rank test was employed to compare parameters between two
groups, and one-way ANOVA and the Dunnett's post hoc test was used
to evaluate the differences among three or more groups. Pearson
analysis was used to assess the correlation between the expression
of the related miR-204 and ATXN8OS sequences. Data are presented as
the mean ± SD. P<0.05 was considered to indicate a statistically
significant difference. The experiments were performed >5
times.
Results
Clinical significance of ATXN8OS
expression in BC
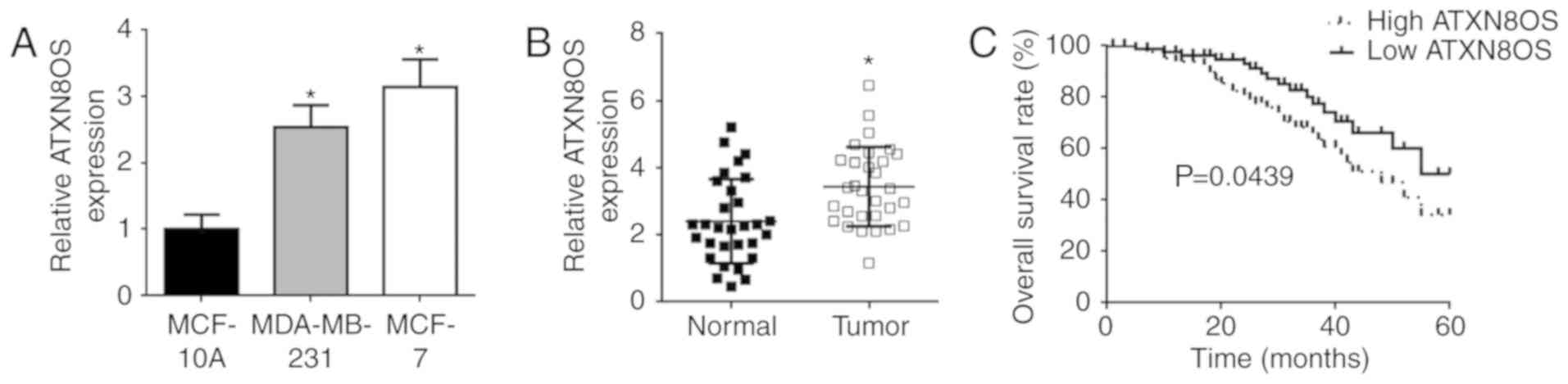
The expression of ATXN8OS in BC tissues and matched
non-tumour tissues was analysed using RT-qPCR. The expression of
ATXN8OS was found to be significantly higher in the human
MDA-MB-231 and MCF-7 BC cell lines than in the MCF-10A normal
breast cell line (Fig. 1A). The
expression of ATXN8OS was also significantly higher in BC tissues
than in the matched normal breast tissues (Fig. 1B). Moreover, Kaplan-Meier analysis
showed that the overall survival rate of patients with BC was lower
in the group with high ATXN8OS expression and higher in the group
with low ATXN8OS expression (P=0.0439; Fig. 1C). The correlations between the
expression of ATXN8OS and the clinicopathological characteristics
were assessed with the χ2 test. High ATXN8OS was
correlated with the size of the carcinoma (P=0.0017),
tumour-node-metastasis (TNM) stage (P=0.0038) and lymphatic
metastasis (P=0.0004). However, no significant differences were
found between the expression of ATXN8OS and sex, age, oestrogen
receptor status, progesterone receptor status or human EGF receptor
2(Her2)/neu status (Table I).
 | Table I.Correlation between ATXN8OS
expression in breast cancer and clinical characteristics. |
Table I.
Correlation between ATXN8OS
expression in breast cancer and clinical characteristics.
|
| Relative ATXN8OS
expression |
|---|
|
|
|
|---|
| Characteristic | No. | Low | High | P-value |
|---|
| Sex |
|
Male | 10 | 6 | 4 | 0.7445 |
|
Female | 110 | 56 | 54 |
|
| Age, years |
|
>50 | 74 | 41 | 33 | 0.2602 |
|
≤50 | 46 | 20 | 26 |
|
| Size, cm |
|
>2 | 65 | 20 | 45 | 0.0017 |
| ≤2 | 55 | 33 | 22 |
|
|
Tumour-node-metastasis stage |
|
I–II | 58 | 38 | 20 | 0.0038 |
|
III–IV | 62 | 24 | 38 |
|
| Lymphatic
metastasis |
|
Negative | 72 | 47 | 25 | 0.0004 |
|
Positive | 48 | 15 | 33 |
|
| Oestrogen receptor
status |
|
Negative | 36 | 12 | 24 | 0.3151 |
|
Positive | 84 | 37 | 47 |
|
| Progesterone
receptor status |
|
Negative | 42 | 26 | 16 | 0.3367 |
|
Positive | 78 | 40 | 38 |
|
| Human EGF receptor
2/neu status |
|
Negative | 84 | 44 | 40 | 0.6915 |
|
Positive | 36 | 17 | 19 |
|
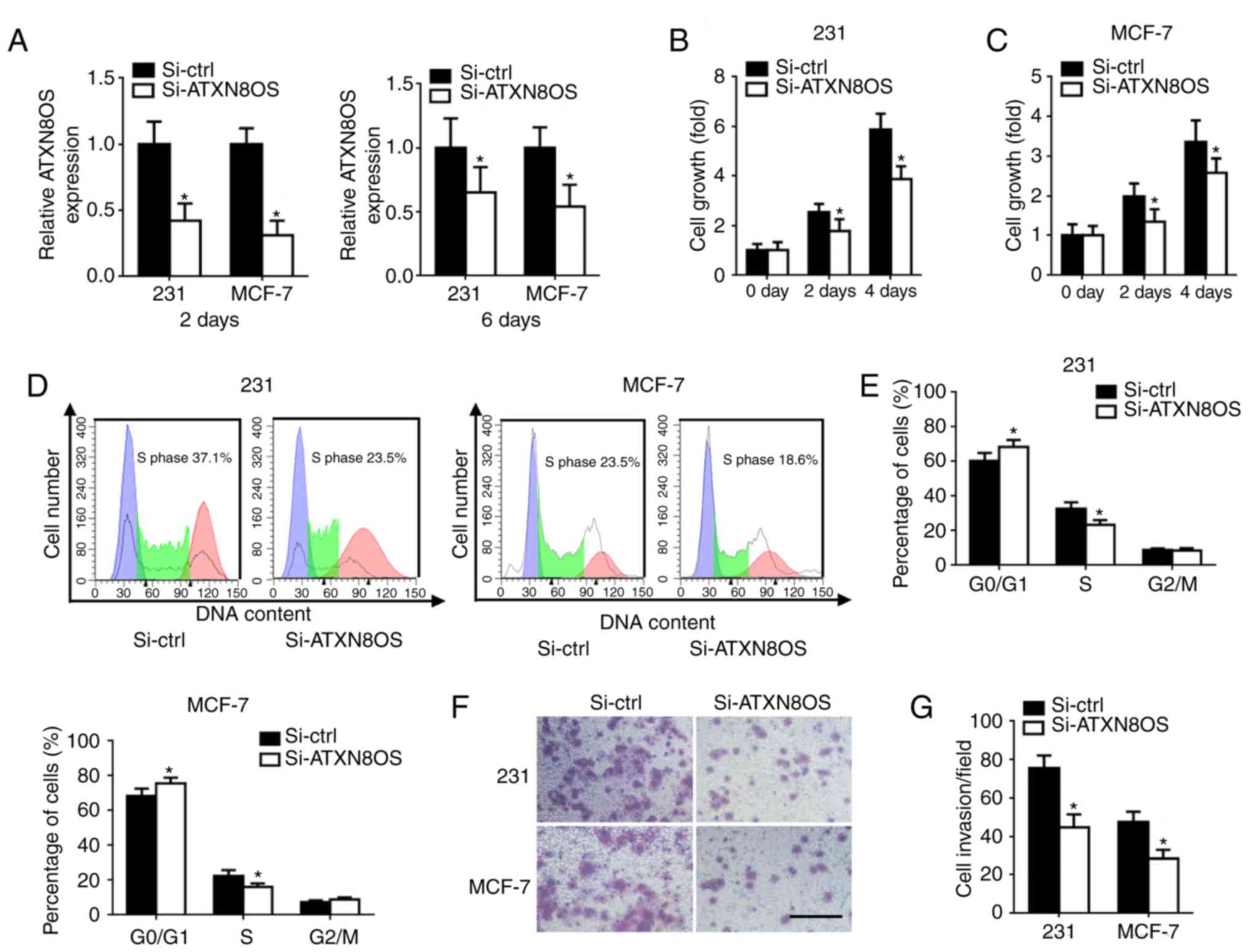
Down-regulation of ATXN8OS inhibits BC
cell proliferation and invasion
To investigate the function of ATXN8OS in BC cells,
MDA-MB-231 and MCF7 BC cells were transfected with si-ATXN8OS.
RT-qPCR showed an efficient knockdown of ATXN8OS in the cell lines
(Fig. 2A). The CCK-8 assay was
used to measure the effect of ATXN8OS on the proliferation of human
BC cells. Cell viability was reduced significantly in
ATXN8OS-knockdown BC cells compared with the control knockdown
cells (Fig. 2B and C). Flow
cytometry analysis showed that transfection with si-ATXN8OS induced
G0/G1 arrest and decreased the S phase
population compared with the control knockdown (Fig. 2D and E). A Transwell assay was
performed to determine whether ATXN8OS regulated BC cell invasion.
Knockdown of ATXN8OS in cells reduced the number of invasive cells
compared with the control knockdown (Fig. 2F and G).
ATXN8OS directly inhibits miR-204
expression by targeting its 3′ untranslated region (UTR)
To explore the molecular mechanism of ATXN8OS in BC,
StarBase V3.0 was used to predict the potential targets of ATXN8OS.
The analysis revealed that ATXN8OS contains complementary binding
sites to the 3′UTR of miR-204. RT-qPCR showed that the expression
level of miR-204 was negatively correlated with the expression of
ATXN8OS (P=0.0017; Fig. 3A). To
identify the relationship between ATXN8OS and miR-204, the
expression levels of miR-204 and its downstream genes in si-ATXN8OS
BC cells were determined. The results demonstrated that the
expression of miR-204 was significantly upregulated after knockdown
of ATXN8OS in human MDA-MB-231 and MCF-7 BC cells (Fig. 3B). The expression of downstream
targets of miR-204, including JAK2, FOXA1, ANGPT1 and TGFβR2, was
significantly decreased after the inhibition of ATXN8OS in both of
the human BC cell lines tested (Fig.
3C-E). To determine whether ATXN8OS suppresses miR-204 by
directly binding miR-204 in BC cells, Wt ATXN8OS and Mut ATXN8OS
sequences containing the target sites of miR-204 were cloned into a
luciferase reporter system (Fig.
3F). The luciferase activity of Wt ATXN8OS was reduced in the
BC cells containing the miR-204 mimic; however, the luciferase
activity of Wt ATXN8OS was increased in the BC cells containing the
miR-204 inhibitor (Fig. 3G).
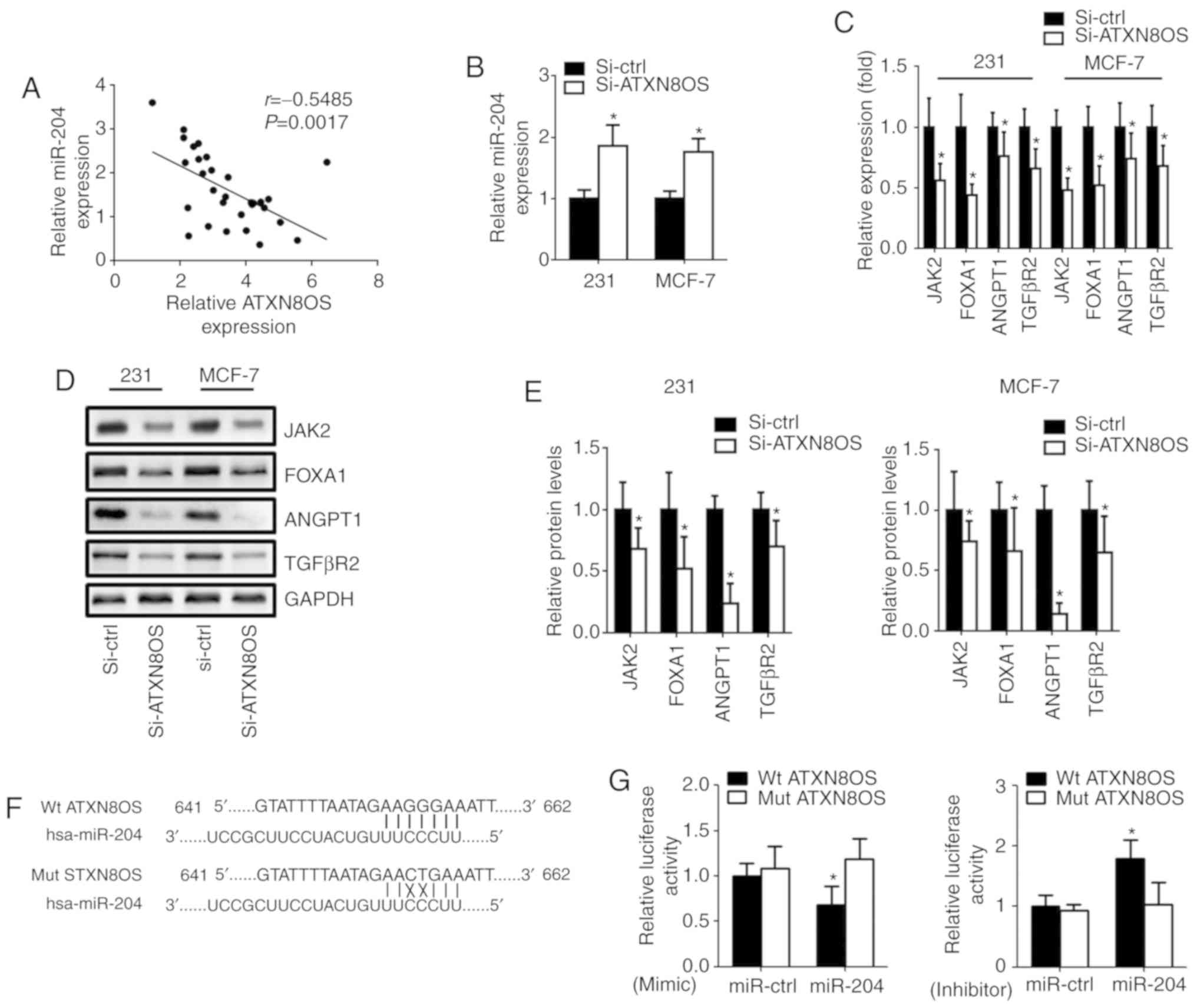 | Figure 3.ATXN8OS targets miR-204. (A) An
inverse relationship was found between ATXN8OS and miR-204 in BC
tissues. (B) Up-regulation of miR-204 was found in BC cells
subjected to knockdown of ATXN8OS. (C) Reverse
transcription-quantitative PCR showed reduced expression of JAK2,
FOXA1, ANGPT1 and TGFβR2 in 231 and MCF-7 cells 48 h
post-transfection. (D) The protein levels of JAK2, FOXA1, ANGPT1
and TGFβR2 were determined by western blot analysis. GAPDH was used
as an internal control. (E) Relative protein expression was
calculated based on the densitometric analysis of band intensities.
(F) Putative ATXN8OS target sequences in miR-204 are displayed. (G)
Dual-luciferase reporter system analysis was conducted in 231 cells
co-transfected with Wt- or Mut-ATXN8OS and miR-204 or miR-con
mimics/inhibitors. *P<0.05 vs. respective control. n=5. ATXN8OS,
ataxin 8 opposite strand; BC, breast cancer; miR-204, microRNA-204;
miR-con, microRNA control; si-ATXN8OS, small interfering RNA
targeting ATXN8OS; si-ctrl, control small interfering RNA; 231,
MDA-MB-231; JAK2, tyrosine protein kinase JAK2; FOXA1, forkhead box
A1; ANGPT1, angiopoietin-1; TGFβR2, TGF-β receptor type 2; Wt, wild
type; Mut, mutant. |
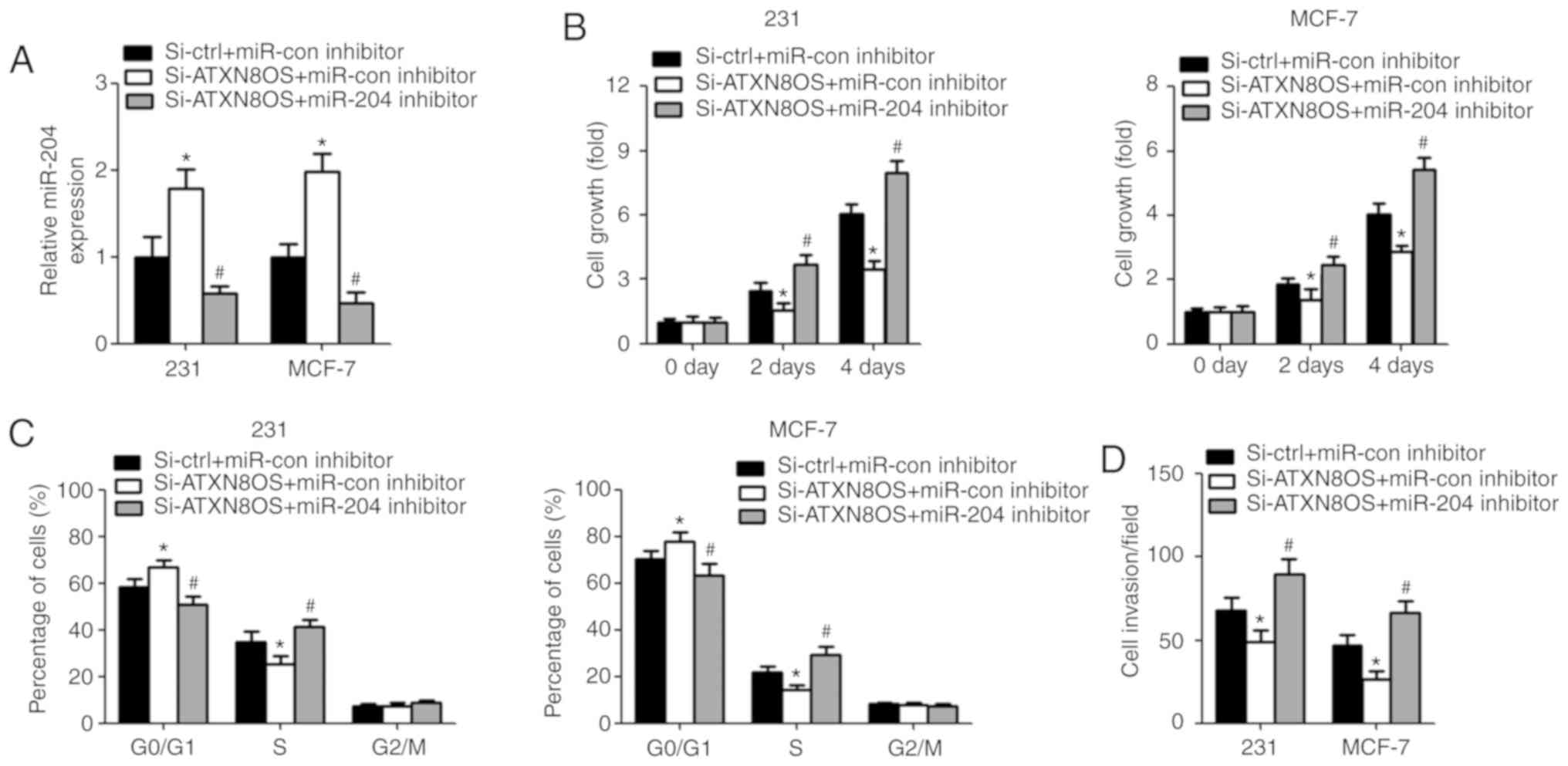
Inhibition of miR-204 reverses
ATXN8OS-induced effects on BC
To confirm whether ATXN8OS exerts its biological
function by targeting miR-204, BC cells were co-transfected with
si-ATXN8O and the miR-204 inhibitor. RT-qPCR showed a reduced
expression of miR-204 in BC cells after transfection with the
miR-204 inhibitor (Fig. 4A).
Proliferation and cell cycle distribution were analysed using the
CCK-8 assay and flow cytometry, respectively. The results showed
that cell growth was increased in the si-ATXN8OS cells
co-transfected with the miR-204 inhibitor (Fig. 4B). The inhibition of miR-204
resulted in fewer BC cells in the G0/G1 phase
and an increased percentage of cells in the S phase (Fig. 4C). Invasion was increased in cells
co-transfected with si-ATXN8OS and the miR-204 inhibitor compared
with cells co-transfected with si-ATXN8OS and the miR-con inhibitor
(Fig. 4D).
Discussion
The identification of novel molecular targets for BC
is becoming increasingly important, with the aim of improving the
diagnosis, therapeutic strategies and clinical follow-up of BC
(26–28). Accumulating evidence supports the
role of lncRNAs functioning as ceRNAs in the occurrence and
progression of a number of human cancer types (29,30).
Despite rapid improvements in the detection, diagnosis, treatment
and prediction of the prognosis of BC, the recurrence and mortality
rates of BC remain some of the biggest challenges for patients with
BC. Therefore, it is important to determine the exact molecular
mechanism of action involved in the initiation and progression of
BC, and to explore possible prognostic markers. Increasing
experimental evidence indicates that lncRNAs can exert important
functions in many biological processes and are closely associated
with the development and prognosis of cancer (31). Consequently, elucidation of the
relationship between lncRNAs and their downstream targets would
shed light on the diagnosis and treatment of patients with BC.
The results of the present study found that ATXN8OS
was up-regulated in human BC tissues compared with normal human
tissues; an increased expression of ATXN8OS was also found in the
BC cell lines tested. These data indicated that ATXN8OS might
promote the occurrence of carcinomas. In addition, the expression
of ATXN8OS was found to be related to tumour size, TNM stage and
lymphatic metastasis, suggesting that ATXN8OS may be involved in
the development of BC. Conversely, it was found that the knockdown
of ATXN8OS suppressed proliferation, increased the percentage of
cells in the G0/G1 phase of the cell cycle
and decreased cell invasion in BC cell lines. Therefore, ATXN8OS
may be a novel factor involved in the progression of BC, by
arresting the cell cycle in the G0/G1 phase.
In addition, the overall survival rate was low in patients with
ATXN8OS overexpression according to the 60-month follow-up survival
survey, indicating a strong correlation between ATXN8OS
overexpression and poor prognostic outcomes of BC. This result
suggested that ATXN8OS may be an important prognostic marker in
forecasting the prognosis of BC. There are three sub-types of BC
cell lines, two of which were used in the present study. The
luminal-like MCF7 cell line and the basal-like MDA-MB-231 cell line
were used; in addition, the normal breast epithelium cell line
MCF10A was used. The other sub-type of BC cell lines, the Her-2
elevated type, including SKBR3, will be investigated in future
studies.
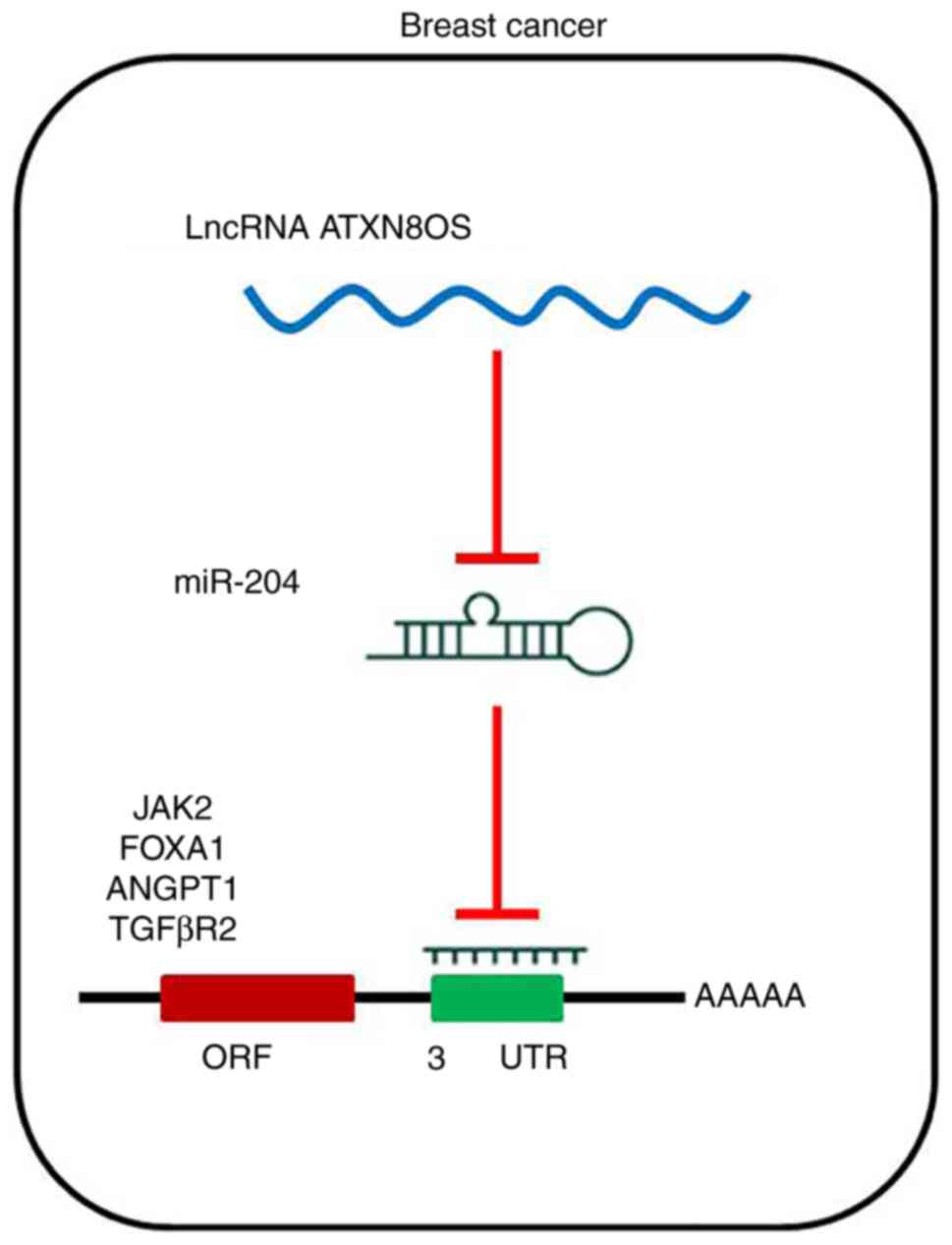
miR-204 was identified as a predicted target gene of
ATXN8OS, and their expression was found to be inversely correlated
in BC tissues and cell lines. miR-204 has been identified as an
anti-oncogene and is reported to be down-regulated in diverse human
malignancies, including intrahepatic cholangiocarcinoma, glioma,
non-small cell lung cancer, endometrial cancer, gastric cancer,
head and neck squamous cell carcinoma, and thyroid cancer (32–35).
Previous studies revealed that JAK2 and FOXA1 are direct targets of
miR-204 through binding sites in their 3′UTRs in BC (2,36).
Another previous study showed that miR-204 functions by
down-regulating the expression of ANGPT1 and TGFβR2 by targeting
binding sites in their 3′UTRs in BC (37). Based on these previous studies, it
is proposed that the expression of JAK2, FOXA1, ANGPT1 and TGFβR2
may be inhibited in si-ATXN8OS cells if miR-204 is a direct
downstream target of ATXN8OS. The reduced expression of
transcriptional control genes downstream of miR-204 observed in the
present study suggested that miR-204 is a direct downstream target
of ATXN8OS in BC cell lines (Fig.
5). More experiments need to be performed to understand the
binding interaction between miR-204 and the lncRNA ATXN8OS. The
inhibition of miR-204 expression reversed the enhancing effects of
ATXN8OS on proliferation, cell cycle and invasion of BC cells in
vitro.
In conclusion, the results of the present study
showed that ATXN8OS was up-regulated in the tissues of patients
with BC and in BC cell lines, and that its aberrant overexpression
was significantly correlated with poor prognostic outcomes and
lower overall survival rates. The results of the present study
revealed the potential molecular mechanism by which ATXN8OS exerts
its stimulating functions on proliferation and invasion in BC
cells, by sequestering miR-204. The findings of the present study
indicated that ATXN8OS may be an oncogenic factor that promotes the
development and progression of BC, and might be a potential
biomarker for the clinical diagnosis and treatment of patients with
BC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Fujian Province (grant no. 2016J05196),
Military Medical Scientific Youth Cultivation Project (grant no.
15QNP205) and Fujian Provincial Science and Technology Major
Project (grant no. 2012YZ0001-1).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JT contributed to the conception and design of the
study. ZD, LL, HC, WW and LZ performed the experiments. SY and JC
analysed the data and JT contributed to manuscript drafting.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients, and the study protocol (no. 20171026) was approved by the
Ethics Committee of 900th Hospital of the Joint Logistics Support
Force.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tian T, Wang M, Lin S, Guo Y, Dai Z, Liu
K, Yang P, Dai C, Zhu Y, Zheng Y, et al: The impact of lncRNA
dysregulation on clinicopathology and survival of breast cancer: A
systematic review and meta-analysis. Mol Ther Nucleic Acids.
12:359–369. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang X, Qiu W, Zhang G, Xu S, Gao Q and
Yang Z: MicroRNA-204 targets JAK2 in breast cancer and induces cell
apoptosis through the STAT3/BCl-2/survivin pathway. Int J Clin Exp
Pathol. 8:5017–5025. 2015.PubMed/NCBI
|
|
3
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feng W, Wang C, Liang C, Yang H, Chen D,
Yu X, Zhao W, Geng D, Li S, Chen Z and Sun M: The dysregulated
expression of KCNQ1OT1 and its interaction with downstream factors
miR-145/CCNE2 in breast cancer cells. Cell Physiol Biochem.
49:432–446. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bertero T, Cottrill KA, Lu Y, Haeger CM,
Dieffenbach P, Annis S, Hale A, Bhat B, Kaimal V, Zhang YY, et al:
Matrix remodeling promotes pulmonary hypertension through feedback
mechanoactivation of the YAP/TAZ-miR-130/301 circuit. Cell Rep.
13:1016–1032. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cai H, Xu J, Han Y, Lu Z, Han T, Ding Y
and Ma L: Integrated miRNA-risk gene-pathway pair network analysis
provides prognostic biomarkers for gastric cancer. Onco Targets
Ther. 9:2975–2986. 2016.PubMed/NCBI
|
|
8
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ,
Lee KH, Yeh SD, Hong TM and Chen YL: MicroRNA-320 suppresses the
stem cell-like characteristics of prostate cancer cells by
downregulating the Wnt/beta-catenin signaling pathway.
Carcinogenesis. 34:530–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang T, Zou P, Wang T, Xiang J, Cheng J,
Chen D and Zhou J: Down-regulation of miR-320 associated with
cancer progression and cell apoptosis via targeting Mcl-1 in
cervical cancer. Tumour Biol. 37:8931–8940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gooding AJ, Zhang B, Jahanbani FK, Gilmore
HL, Chang JC, Valadkhan S and Schiemann WP: The lncRNA BORG Drives
Breast Cancer Metastasis and Disease Recurrence. Sci Rep.
7:126982017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ransohoff JD, Wei Y and Khavari PA: The
functions and unique features of long intergenic non-coding RNA.
Nat Rev Mol Cell Biol. 19:143–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fang Y, Wang J, Wu F, Song Y, Zhao S and
Zhang Q: Long non-coding RNA HOXA-AS2 promotes proliferation and
invasion of breast cancer by acting as a miR-520c-3p sponge.
Oncotarget. 8:46090–46103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Zhou J, Wang Z, Wang P and Li S:
Upregulation of SOX2 activated LncRNA PVT1 expression promotes
breast cancer cell growth and invasion. Biochem Biophys Res Commun.
493:429–436. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cho JY: Molecular diagnosis for
personalized target therapy in gastric cancer. J Gastric Cancer.
13:129–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang S, Lan F and Xia Y: lncRA ANCR
inhibits non-small cell lung cancer Cell migration and invasion by
inactivating TGF-β pathway. Med Sci Monit. 24:6002–6009. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dong H, Wang W, Mo S, Chen R, Zou K, Han
J, Zhang F and Hu J: SP1-induced lncRNA AGAP2-AS1 expression
promotes chemoresistance of breast cancer by epigenetic regulation
of MyD88. J Exp Clin Cancer Res. 37:2022018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen IC, Lin HY, Hsiao YC, Chen CM, Wu YR,
Shiau HC, Shen YF, Huang KS, Su MT, Hsieh-Li HM and Lee-Chen GJ:
Internal ribosome entry segment activity of ATXN8 opposite strand
RNA. PLoS One. 8:e738852013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koob MD, Moseley ML, Schut LJ, Benzow KA,
Bird TD, Day JW and Ranum LP: An untranslated CTG expansion causes
a novel form of spinocerebellar ataxia (SCA8). Nat Genet.
21:379–384. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fan CN, Ma L and Liu N: Systematic
analysis of lncRNA-miRNA-mRNA competing endogenous RNA network
identifies four-lncRNA signature as a prognostic biomarker for
breast cancer. J Transl Med. 16:2642018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schnipper LE, Davidson NE, Wollins DS,
Tyne C, Blayney DW, Blum D, Dicker AP, Ganz PA, Hoverman JR,
Langdon R, et al: American Society of clinical oncology statement:
A conceptual framework to assess the value of cancer treatment
options. J Clin Oncol. 33:2563–2577. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang D, Zhu C, Zhang Y, Zheng Y, Ma F, Su
L and Shao G: MicroRNA-30e-3p inhibits cell invasion and migration
in clear cell renal cell carcinoma by targeting Snail1. Oncol Lett.
13:2053–2058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Campos-Parra AD, López-Urrutia E, Orozco
Moreno LT, López-Camarillo C, Meza-Menchaca T, Figueroa González G,
Bustamante Montes LP and Pérez-Plasencia C: Long non-coding RNAs as
new master regulators of resistance to systemic treatments in
breast cancer. Int J Mol Sci. 19:E27112018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang XJ, Xia Y, He GF, Zheng LL, Cai YP,
Yin Y and Wu Q: MALAT1 promotes angiogenesis of breast cancer.
Oncol Rep. 40:2683–2689. 2018.PubMed/NCBI
|
|
28
|
Wang G, Chen X, Liang Y, Wang W, Fang Y
and Shen K: Long noncoding RNA signature and disease outcome in
estrogen receptor-positive breast cancer patients treated with
tamoxifen. J Breast Cancer. 21:277–287. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J, Yuan L, Zhang X, Hamblin MH, Zhu
T, Meng F, Li Y, Chen YE and Yin KJ: Altered long non-coding RNA
transcriptomic profiles in brain microvascular endothelium after
cerebral ischemia. Exp Neurol. 277:162–170. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Troy A and Sharpless NE: Genetic ‘lnc’-age
of noncoding RNAs to human disease. J Clin Invest. 122:3837–3840.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Courboulin A, Paulin R, Giguère NJ,
Saksouk N, Perreault T, Meloche J, Paquet ER, Biardel S, Provencher
S, Côté J, et al: Role for miR-204 in human pulmonary arterial
hypertension. J Exp Med. 208:535–548. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sacconi A, Biagioni F, Canu V, Mori F, Di
Benedetto A, Lorenzon L, Ercolani C, Di Agostino S, Cambria AM,
Germoni S, et al: miR-204 targets Bcl-2 expression and enhances
responsiveness of gastric cancer. Cell Death Dis. 3:e4232012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H,
Li W, Hu B, Cheng SY and Li M: Loss of miR-204 expression enhances
glioma migration and stem cell-like phenotype. Cancer Res.
73:990–999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chung TK, Lau TS, Cheung TH, Yim SF, Lo
KW, Siu NS, Chan LK, Yu MY, Kwong J, Doran G, et al: Dysregulation
of microRNA-204 mediates migration and invasion of endometrial
cancer by regulating FOXC1. Int J Cancer. 130:1036–1045. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shen SQ, Huang LS, Xiao XL, Zhu XF, Xiong
DD, Cao XM, Wei KL, Chen G and Feng ZB: miR-204 regulates the
biological behavior of breast cancer MCF-7 cells by directly
targeting FOXA1. Oncol Rep. 38:368–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Flores-Pérez A, Marchat LA,
Rodríguez-Cuevas S, Bautista-Piña V, Hidalgo-Miranda A, Ocampo EA,
Martínez MS, Palma-Flores C, Fonseca-Sánchez MA, Astudillo-de la
Vega H, et al: Dual targeting of ANGPT1 and TGFBR2 genes by miR-204
controls angiogenesis in breast cancer. Sci Rep. 6:345042016.
View Article : Google Scholar : PubMed/NCBI
|