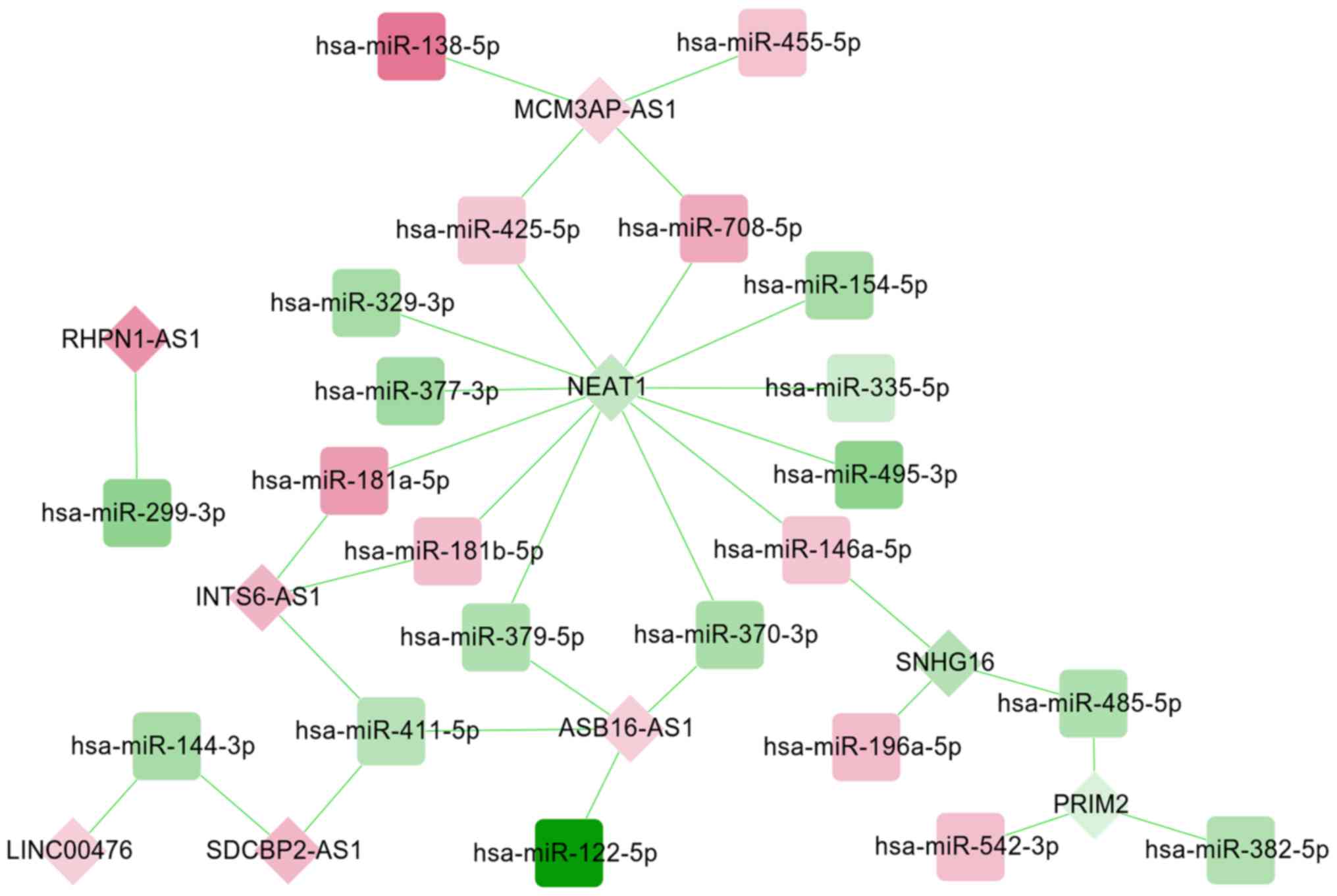
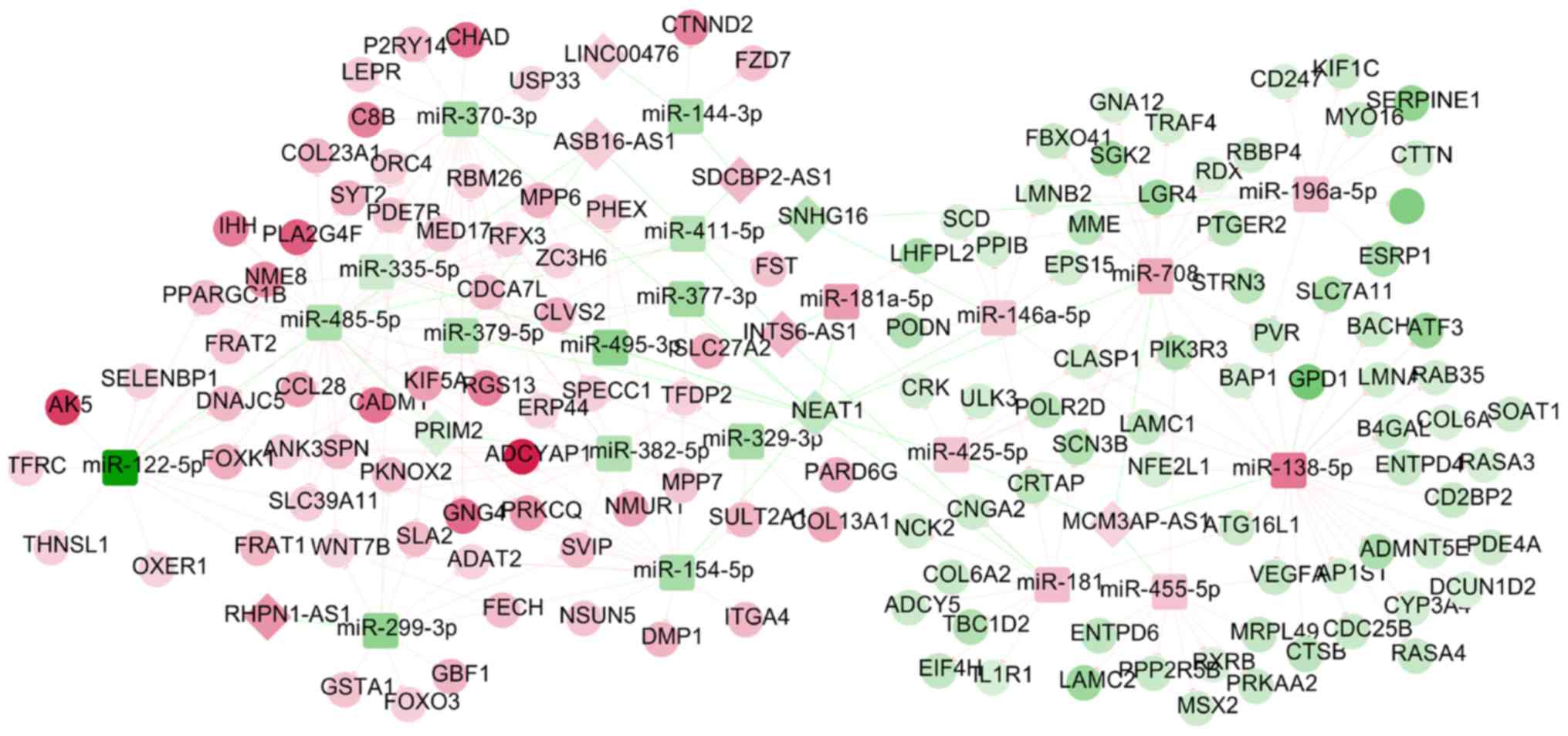
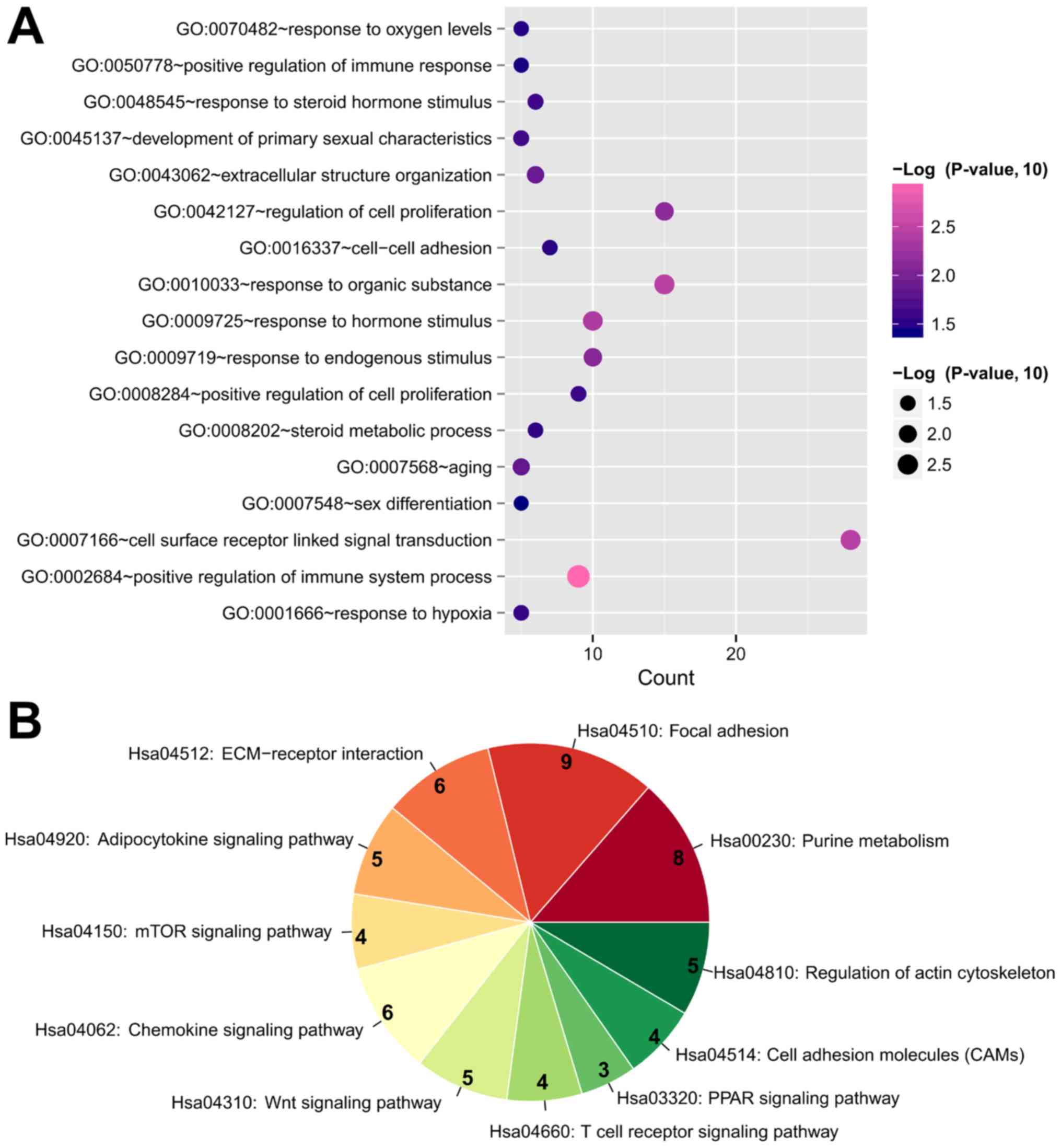
|
1
|
Lang N, Yuan HS, Wang HL, Liao J, Li M,
Guo FX, Shi S and Chen ZQ: Epidemiological survey of ossification
of the ligamentum flavum in thoracic spine: CT imaging observation
of 993 cases. Eur Spine J. 22:857–862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mori K, Imai S, Kasahara T, Nishizawa K,
Mimura T and Matsusue Y: Prevalence, distribution and morphology of
thoracic ossification of the posterior longitudinal ligament in
Japanese: Results of CT-based cross-sectional study. Spine (Phila
Pa 1976). 65:394–399. 2014. View Article : Google Scholar
|
|
3
|
Moon BJ, Kuh SU, Kim S, Kim KS, Yong EC
and Dong KC: Prevalence, distribution and significance of
incidental thoracic ossification of the ligamentum flavum in Korean
patients with back or leg pain: MR-based cross sectional study. J
Korean Neurosurg Soc. 58:112–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ono K, Yonenobu K, Miyamoto S and Okada K:
Pathology of ossification of the posterior longitudinal ligament
and ligamentum flavum. Clin Orthop Relat Res. 359:18–26. 1999.
View Article : Google Scholar
|
|
5
|
Wang H, Wei F, Long H, Han G, Sribastav
SS, Li Z, Huang Y, Zhu R and Liang C: Surgical outcome of thoracic
myelopathy caused by ossification of ligamentum flavum. J Clin
Neurosci. 45:83–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yabe Y, Honda M, Hagiwara Y, Tohjo Y,
Nakajima S, Ando A, Sonofuchi K and Itoi E: Thoracic radiculopathy
caused by ossification of the ligamentum flavum. Ups J Med Sci.
118:54–58. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhong ZM, Wu Q, Meng TT, Zhu YJ, Qu DB,
Wang JX, Jiang JM, Lu KW, Zheng S and Zhu SY: Clinical outcomes
after decompressive laminectomy for symptomatic ossification of
ligamentum flavum at the thoracic spine. J Clin Neurosci. 28:77–81.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ren L, Hu H, Sun X, Li F, Zhou JJ and Wang
YM: The roles of inflammatory cytokines in the pathogenesis of
ossification of ligamentum flavum. Am J Transl Res. 5:582–585.
2013.PubMed/NCBI
|
|
9
|
Ning S, Chen Z, Fan D, Sun C, Zhang C,
Zeng Y, Li W, Hou X, Qu X and Ma Y: Genetic differences in
osteogenic differentiation potency in the thoracic ossification of
the ligamentum flavum under cyclic mechanical stress. Int J Mol
Med. 39:135–143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qu X, Chen Z, Fan D, Sun C, Zeng Y, Hou X
and Ning S: Notch signaling pathways in human thoracic ossification
of the ligamentum flavum. J Orthop Res. 34:1481–1491. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hou XF, Fan DW, Sun CG and Chen ZQ:
Recombinant human bone morphogenetic protein-2-induced ossification
of the ligamentum flavum in rats and the associated global
modification of histone H3. J Neurosurg Spine. 21:334–341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhong ZM, Chen JT, Zhang Y, Zha DS, Lin
ZS, Zhao CY, Xu JC, Li T and Xu Z: Growth/differentiation Factor-5
induces osteogenic differentiation of human ligamentum flavum cells
through activation of ERK1/2 and p38 MAPK. Cell Physiol Biochem.
26:179–186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang C, Chen Z, Meng X, Li M, Zhang L and
Huang A: The involvement and possible mechanism of pro-inflammatory
tumor necrosis factor alpha (TNF-α) in thoracic ossification of the
ligamentum flavum. PLoS One. 12:e01789862017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Park JO, Lee BH, Kang YM, Kim TH, Yoon JY,
Kim H, Kwon UH, Lee KI, Lee HM and Moon SH: Inflammatory cytokines
induce fibrosis and ossification of human ligamentum flavum cells.
J Spinal Disord Tech. 26:E6–E12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang B, Chen Z, Meng X, Li M, Yang X and
Zhang C: iTRAQ quantitative proteomic study in patients with
thoracic ossification of the ligamentum flavum. Biochem Biophys Res
Commun. 487:834–839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hausser J and Zavolan M: Identification
and consequences of miRNA-target interactions-beyond repression of
gene expression. Nat Rev Genet. 15:599–612. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yin J, Zhuang G, Zhu Y, Hu X, Zhao H,
Zhang R, Guo H, Fan X and Cao Y: MiR-615-3p inhibits the osteogenic
differentiation of human lumbar ligamentum flavum cells via
suppression of osteogenic regulators GDF5 and FOXO1. Cell Biol Int.
41:779–786. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu X, Chen Z, Fan D, Sun C and Yan Z:
MiR-132-3p regulates the osteogenic differentiation of thoracic
ligamentum flavum cells by inhibiting multiple osteogenesis-related
genes. Int J Mol Sci. 17(pii): E13702016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qu X, Chen Z, Fan D, Sun C, Yan Z, Guo Z,
Qi Q and Li W: MiR-199b-5p inhibits osteogenic differentiation in
ligamentum flavum cells by targeting JAG1 and modulating the Notch
signalling pathway. J Cell Mol Med. 21:1159–1170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yayama T, Mori K, Okumura N, Nishizawa K,
Kumagai K, Nakamura A and Imai S: Wnt signaling pathway correlates
with ossification of the spinal ligament: A microRNA array and
immunohistochemical study. J Orthop Sci. 23:26–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: ceRNA hypothesis: The Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han Y, Hong Y, Li L, Li T, Zhang Z, Wang
J, Xia H, Tang Y, Shi Z, Han X, et al: A Transcriptome-level study
identifies changing expression profiles for ossification of the
ligamentum flavum of the spine. Mol Ther Nucleic Acids. 12:872–883.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Larkin MA, Blackshields G, Brown NP,
Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm
A, Lopez R, et al: Clustal W and Clustal X version 2.0.
Bioinformatics. 23:2947–2948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dominguez E, Mauborgne A, Mallet J,
Desclaux M and Pohl M: SOCS3-mediated blockade of JAK/STAT3
signaling pathway reveals its major contribution to spinal cord
neuroinflammation and mechanical allodynia after peripheral nerve
injury. J Neurosci. 30:5754–5766. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fukushima A, Kajiya H, Izumi T, Shigeyama
C, Okabe K and Anan H: Pro-inflammatory cytokines induce suppressor
of cytokine signaling-3 in human periodontal ligament cells. J
Endod. 36:1004–1008. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen H, Cai W, Chu ESH, Tang J, Wong CC,
Wong SH, Sun W, Liang Q, Fang J, Sun Z and Yu J: Hepatic
cyclooxygenase-2 overexpression induced spontaneous hepatocellular
carcinoma formation in mice. Oncogene. 36:4415–4426. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li D, Hao X and Song Y: Identification of
the Key MicroRNAs and the miRNA-mRNA regulatory pathways in
prostate cancer by bioinformatics methods. Biomed Res Int.
2018:62041282018.PubMed/NCBI
|
|
36
|
Pal J, Patil V, Mondal B, Shukla S, Hegde
AS, Arivazhagan A, Santosh V and Somasundaram K: Epigenetically
silenced GNG4 inhibits SDF1α/CXCR4 signaling in mesenchymal
glioblastoma. Genes Cancer. 7:136–147. 2016.PubMed/NCBI
|
|
37
|
Liang L, Zeng JH, Qin XG, Chen JQ, Luo DZ
and Chen G: Distinguishable prognostic signatures of left- and
right-sided colon cancer: A study based on sequencing data. Cell
Physiol Biochem. 48:475–490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liao J, Chen Z, He Q, Liu Y and Wang J:
Differential gene expression analysis and network construction of
recurrent cardiovascular events. Mol Med Rep. 13:1746–1764. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Q, Shen Y, Jiang Y, Zhao S, Zhou D
and Xu N: Overexpression of miR-182 inhibits ossification of
ligamentum flavum cells by targeting NAMPT. Exp Cell Res.
367:119–131. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cao Y, Wu TD, Wu H, Lang Y, Li DZ, Ni SF,
Lu HB and Hu JZ: Synchrotron radiation micro-CT as a novel tool to
evaluate the effect of agomir-210 in a rat spinal cord injury
model. Brain Res. 1655:55–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ren D, Lin B, Zhang X, Peng Y, Ye Z, Ma Y,
Liang Y, Cao L, Li X, Li R, et al: Maintenance of cancer stemness
by miR-196b-5p contributes to chemoresistance of colorectal cancer
cells via activating STAT3 signaling pathway. Oncotarget.
8:49807–49823. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Y, Mao G, Lv Y, Huang Q and Wang G:
MicroRNA-181b stimulates inflammation via the nuclear factor-κB
signaling pathway in vitro. Exp Ther Med. 10:1584–1590. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yamamoto K, Seike M, Takeuchi S, Soeno C,
Miyanaga A, Noro R, Minegishi Y, Kubota K and Gemma A: MiR-379/411
cluster regulates IL-18 and contributes to drug resistance in
malignant pleural mesothelioma. Oncol Rep. 32:2365–2372. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao C, Wang S, Zhao Y, Du F, Wang W, Lv P
and Qi L: Long noncoding RNA NEAT1 modulates cell proliferation and
apoptosis by regulating miR-23a-3p/SMC1A in acute myeloid leukemia.
J Cell Physiol. 234:6161–6172. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Meng XH, Chen XD, Greenbaum J, Zeng Q, You
SL, Xiao HM, Tan LJ and Deng HW: Integration of summary data from
GWAS and eQTL studies identified novel causal BMD genes with
functional predictions. Bone. 113:41–48. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Feng F, Chen A, Huang J, Xia Q, Chen Y and
Jin X: Long noncoding RNA SNHG16 contributes to the development of
bladder cancer via regulating miR-98/STAT3/Wnt/β-catenin pathway
axis. J Cell Biochem. 119:9408–9418. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lu YF, Cai XL, Li ZZ, Lv J, Xiang YA, Chen
JJ, Chen WJ, Sun WY, Liu XM and Chen JB: LncRNA SNHG16 functions as
an oncogene by sponging miR-4518 and up-regulating PRMT5 expression
in Glioma. Cell Physiol Biochem. 45:1975–1985. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yan J, Guo D, Yang S, Sun H, Wu B and Zhou
D: Inhibition of miR-222-3p activity promoted osteogenic
differentiation of hBMSCs by regulating Smad5-RUNX2 signal axis.
Biochem Biophys Res Commun. 470:498–503. 2016. View Article : Google Scholar : PubMed/NCBI
|