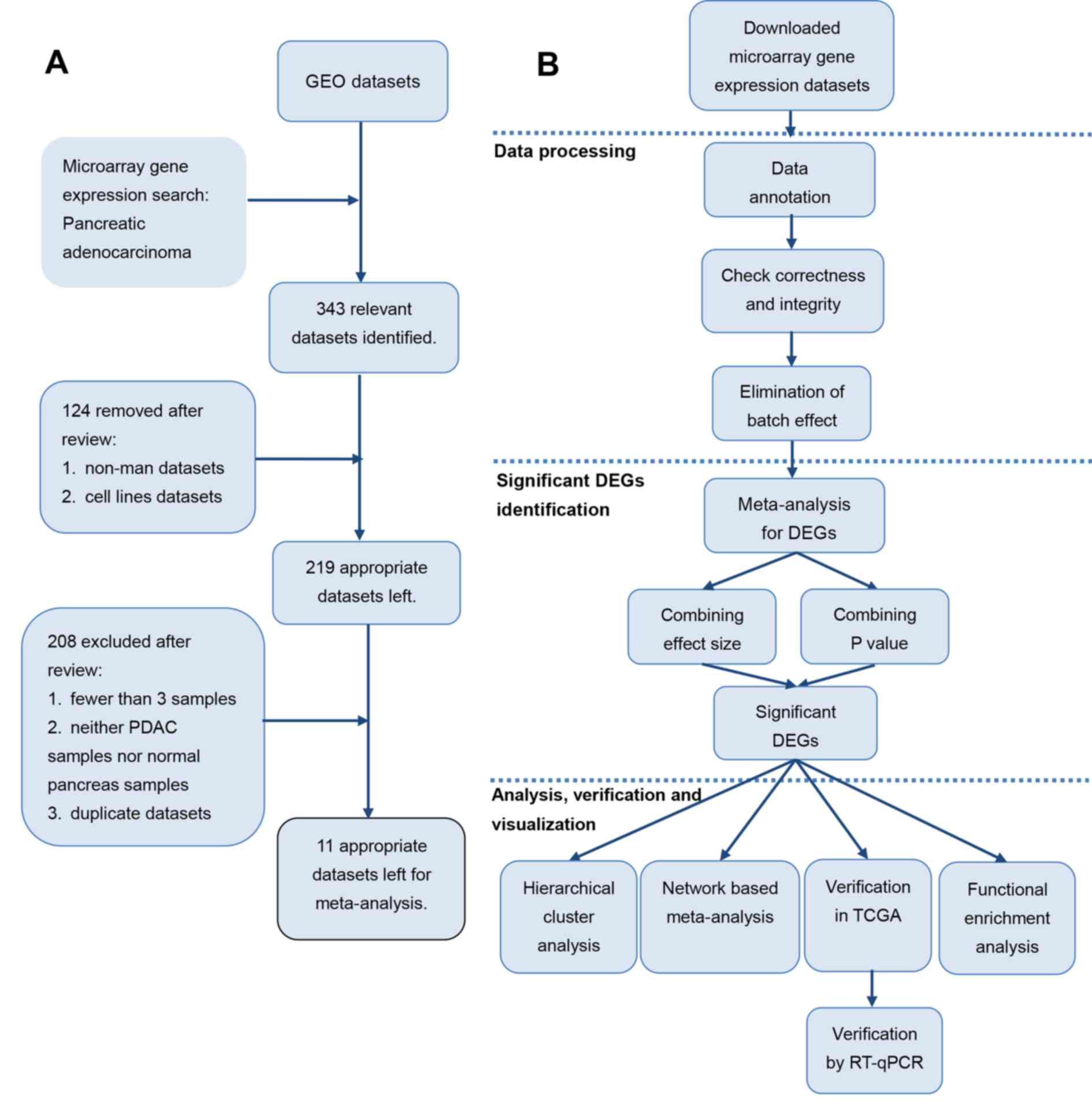
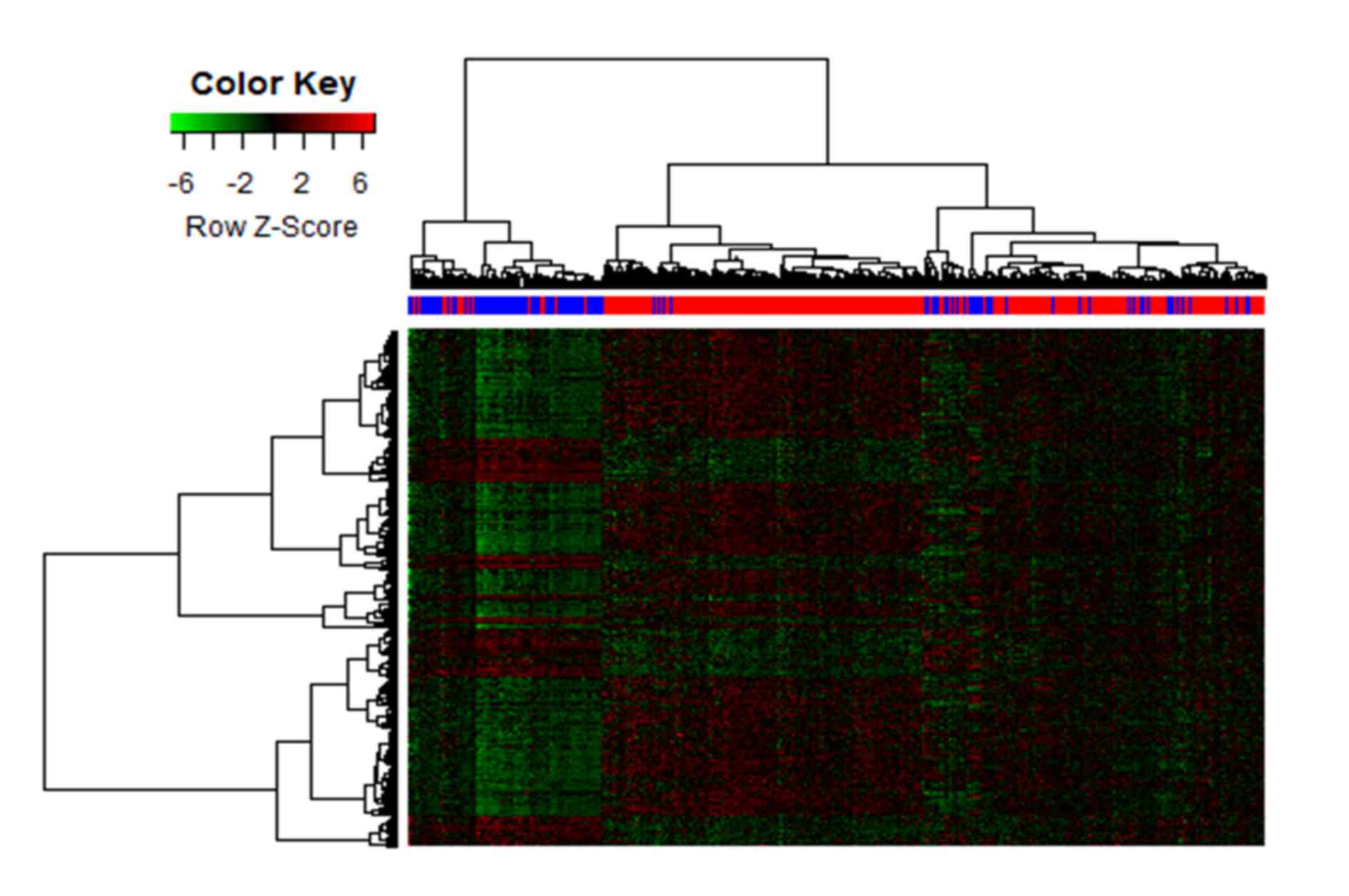
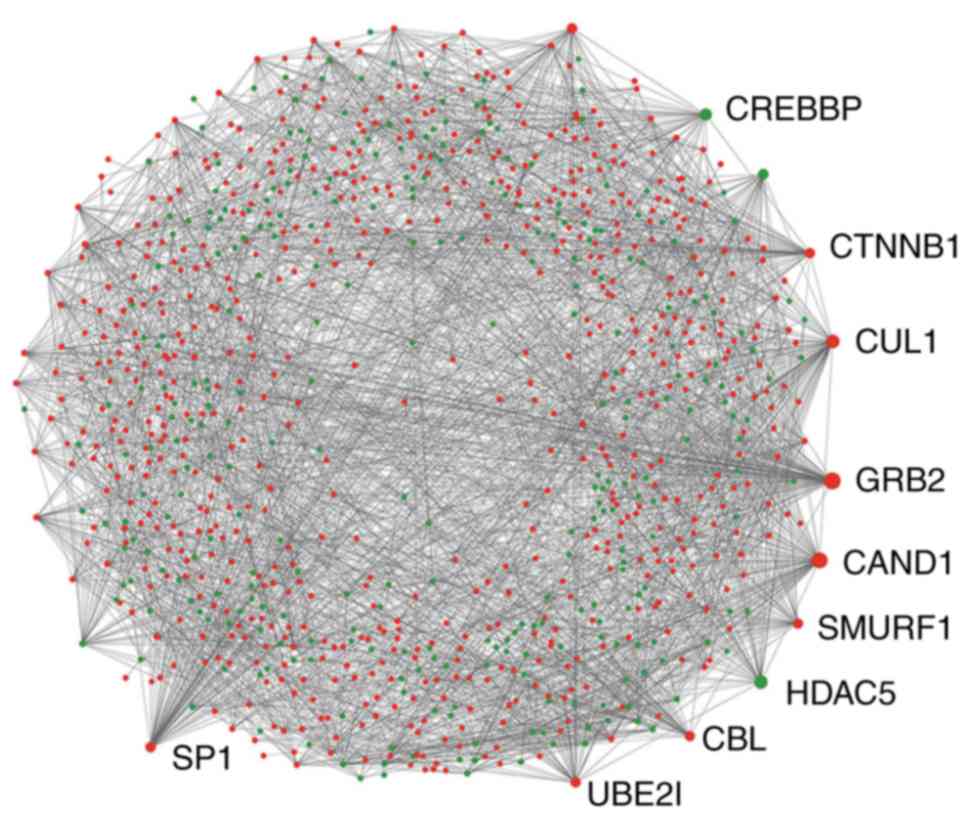
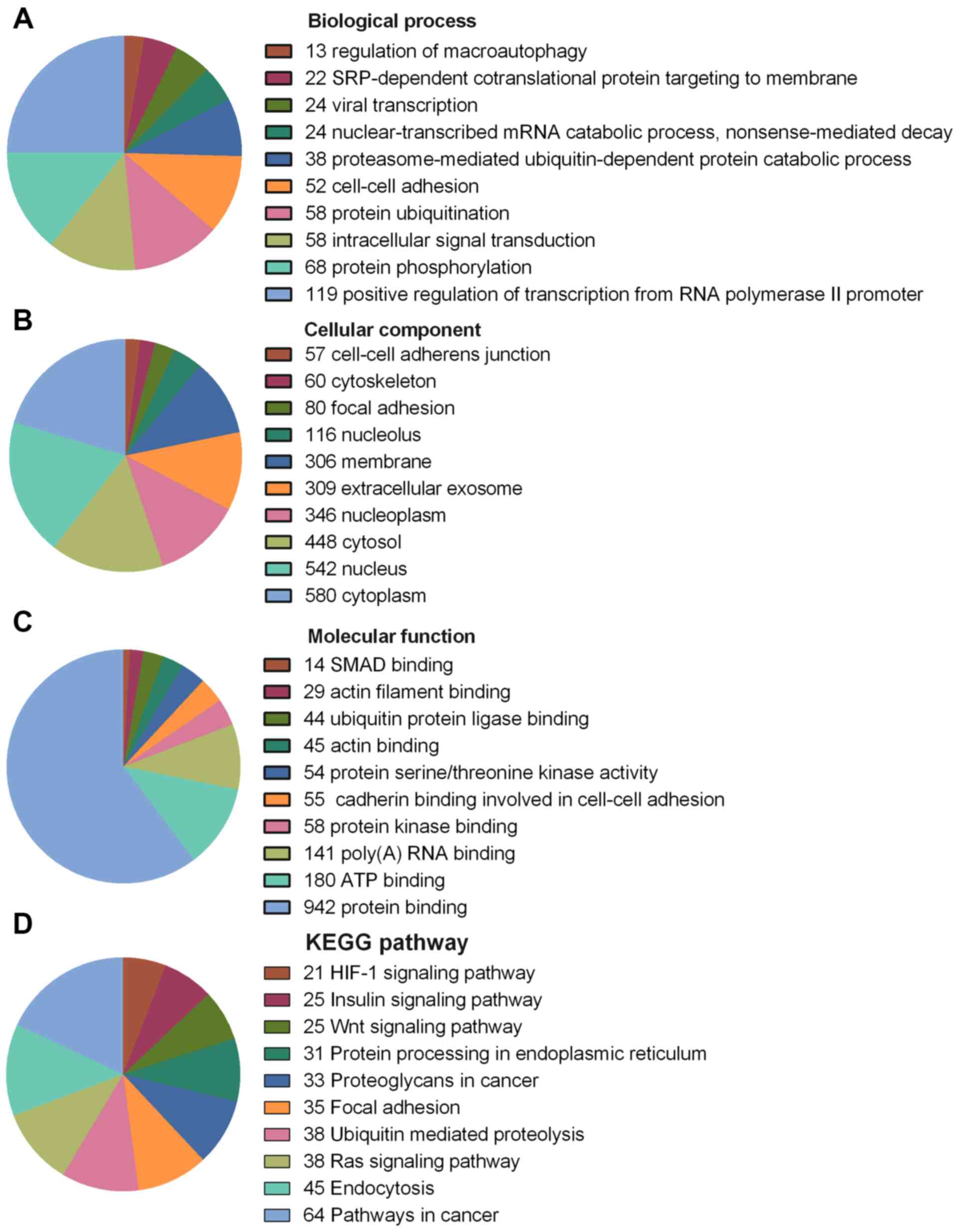
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Crane CH, Varadhachary GR, Yordy JS,
Staerkel GA, Javle MM, Safran H, Haque W, Hobbs BD, Krishnan S,
Fleming JB, et al: Phase II trial of cetuximab, gemcitabine, and
oxaliplatin followed by chemoradiation with cetuximab for locally
advanced (T4) pancreatic adenocarcinoma: Correlation of Smad4(Dpc4)
immunostaining with pattern of disease progression. J Clin Oncol.
29:3037–3043. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hezel AF, Kimmelman AC, Stanger BZ,
Bardeesy N and Depinho RA: Genetics and biology of pancreatic
ductal adenocarcinoma. Genes Dev. 20:1218–1249. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gillen S, Schuster T, Meyer Zum
Büschenfelde C, Friess H and Kleeff J: Preoperative/neoadjuvant
therapy in pancreatic cancer: A systematic review and meta-analysis
of response and resection percentages. PLoS Med. 7:e10002672010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stratton MR, Campbell PJ and Futreal PA:
The cancer genome. Nature. 458:719–724. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jones S, Zhang X, Parsons DW, Lin JC,
Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et
al: Core signaling pathways in human pancreatic cancers revealed by
global genomic analyses. Science. 321:1801–1806. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nagata K, Horinouchi M, Saitou M, Higashi
M, Nomoto M, Goto M and Yonezawa S: Mucin expression profile in
pancreatic cancer and the precursor lesions. J Hepatobiliary
Pancreat Surg. 14:243–254. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sausen M, Phallen J, Adleff V, Jones S,
Leary RJ, Barrett MT, Anagnostou V, Parpart-Li S, Murphy D, Kay Li
Q, et al: Clinical implications of genomic alterations in the
tumour and circulation of pancreatic cancer patients. Nat Commun.
6:76862015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rozenblum E, Schutte M, Goggins M, Hahn
SA, Panzer S, Zahurak M, Goodman SN, Sohn TA, Hruban RH, Yeo CJ and
Kern SE: Tumor-suppressive pathways in pancreatic carcinoma. Cancer
Res. 57:1731–1734. 1997.PubMed/NCBI
|
|
10
|
Bardeesy N, Cheng KH, Berger JH, Chu GC,
Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D and
DePinho RA: Smad4 is dispensable for normal pancreas development
yet critical in progression and tumor biology of pancreas cancer.
Genes Dev. 20:3130–3146. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fong ZV and Winter JM: Biomarkers in
pancreatic cancer: Diagnostic, prognostic, and predictive. Cancer
J. 18:530–538. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Poruk KE, Gay DZ, Brown K, Mulvihill JD,
Boucher KM, Scaife CL, Firpo MA and Mulvihill SJ: The clinical
utility of CA 19-9 in pancreatic adenocarcinoma: Diagnostic and
prognostic updates. Curr Mol Med. 13:340–351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim SM, Kwon IJ, Myoung H, Lee JH and Lee
SK: Identification of human papillomavirus (HPV) subtype in oral
cancer patients through microarray technology. Eur Arch
Otorhinolaryngol. 275:535–543. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gunel T, Hosseini MK, Gumusoglu E,
Kisakesen HI, Benian A and Aydinli K: Expression profiling of
maternal plasma and placenta microRNAs in preeclamptic pregnancies
by microarray technology. Placenta. 52:77–85. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qiao X, Wang H, Wang X, Zhao B and Liu J:
Microarray technology reveals potentially novel genes and pathways
involved in non-functioning pituitary adenomas. Balkan J Med Genet.
19:5–16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li G, Li X, Yang M, Xu L, Deng S and Ran
L: Prediction of biomarkers of oral squamous cell carcinoma using
microarray technology. Sci Rep. 7:421052017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li J, Chen Z, Tian L, Zhou C, He MY, Gao
Y, Wang S, Zhou F, Shi S, Feng X, et al: LncRNA profile study
reveals a three-lncRNA signature associated with the survival of
patients with oesophageal squamous cell carcinoma. Gut.
63:1700–1710. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ramasamy A, Mondry A, Holmes CC and Altman
DG: Key issues in conducting a meta-analysis of gene expression
microarray datasets. PLoS Med. 5:e1842008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rung J and Brazma A: Reuse of public
genome-wide gene expression data. Nat Rev Genet. 14:89–99. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang S, Jin F, Fan W, Liu F, Zou Y, Hu X,
Xu H and Han P: Gene expression meta-analysis in diffuse low-grade
glioma and the corresponding histological subtypes. Sci Rep.
7:117412017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Xia Q and Lin J: Identification
of the potential oncogenes in glioblastoma based on bioinformatic
analysis and elucidation of the underlying mechanisms. Oncol Rep.
40:715–725. 2018.PubMed/NCBI
|
|
22
|
Xiao W, Pacyna-Gengelbach M, Schluns K, An
Q, Gao Y, Cheng S and Petersen I: Differentially expressed genes
associated with human lung cancer. Oncol Rep. 14:229–234.
2005.PubMed/NCBI
|
|
23
|
Tang F, He Z, Lei H, Chen Y, Lu Z, Zeng G
and Wang H: Identification of differentially expressed genes and
biological pathways in bladder cancer. Mol Med Rep. 17:6425–6434.
2018.PubMed/NCBI
|
|
24
|
Peng C, Ma W, Xia W and Zheng W:
Integrated analysis of differentially expressed genes and pathways
in triplenegative breast cancer. Mol Med Rep. 15:1087–1094. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun W, Ma X, Shen J, Yin F, Wang C and Cai
Z: Bioinformatics analysis of differentially expressed pathways
related to the metastatic characteristics of osteosarcoma. Int J
Mol Med. 38:466–474. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hoshida Y, Nijman SM, Kobayashi M, Chan
JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K,
Hashimoto M, et al: Integrative transcriptome analysis reveals
common molecular subclasses of human hepatocellular carcinoma.
Cancer Res. 69:7385–7392. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang Y, Zhang Z, Tang Y, Chen X and Zhou
J: Identification of potential target genes in pancreatic ductal
adenocarcinoma by bioinformatics analysis. Oncol Lett.
16:2453–2461. 2018.PubMed/NCBI
|
|
28
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC Cancer Staging Manual. 8th. Springer
International Publishing; New York, NY: 2017, View Article : Google Scholar
|
|
29
|
Xia J, Fjell CD, Mayer ML, Pena OM,
Wishart DS and Hancock RE: INMEX-a web-based tool for integrative
meta-analysis of expression data. Nucleic Acids Res. 41:W63–W70.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Johnson WE, Li C and Rabinovic A:
Adjusting batch effects in microarray expression data using
empirical bayes methods. Biostatistics. 8:118–127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cochran WG: The combination of estimates
from different experiments. Biometrics. 10:101–129. 1954.
View Article : Google Scholar
|
|
32
|
Marot G, Foulley JL, Mayer CD and
Jaffrézic F: Moderated effect size and P-value combinations for
microarray meta-analyses. Bioinformatics. 25:2692–2699. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Breuer K, Foroushani AK, Laird MR, Chen C,
Sribnaia A, Lo R, Winsor GL, Hancock RE, Brinkman FS and Lynn DJ:
InnateDB: Systems biology of innate immunity and beyond-recent
updates and continuing curation. Nucleic Acids Res. 41:D1228–D1233.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.PubMed/NCBI
|
|
36
|
Pei H, Li L, Fridley BL, Jenkins GD,
Kalari KR, Lingle W, Petersen G, Lou Z and Wang L: FKBP51 affects
cancer cell response to chemotherapy by negatively regulating Akt.
Cancer Cell. 16:259–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang G, Schetter A, He P, Funamizu N,
Gaedcke J, Ghadimi BM, Ried T, Hassan R, Yfantis HG, Lee DH, et al:
DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity
and predicts clinical outcome in pancreatic ductal adenocarcinoma.
PLos One. 7:e315072012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang G, He P, Tan H, Budhu A, Gaedcke J,
Ghadimi BM, Ried T, Yfantis HG, Lee DH, Maitra A, et al:
Integration of metabolomics and transcriptomics revealed a fatty
acid network exerting growth inhibitory effects in human pancreatic
cancer. Clin Cancer Res. 19:4983–4993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Donahue TR, Tran LM, Hill R, Li Y,
Kovochich A, Calvopina JH, Patel SG, Wu N, Hindoyan A, Farrell JJ,
et al: Integrative survival-based molecular profiling of human
pancreatic cancer. Clin Cancer Res. 18:1352–1363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Crnogorac-Jurcevic T, Chelala C, Barry S,
Harada T, Bhakta V, Lattimore S, Jurcevic S, Bronner M, Lemoine NR
and Brentnall TA: Molecular analysis of precursor lesions in
familial pancreatic cancer. PLos One. 8:e548302013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Park M, Kim M, Hwang D, Park M, Kim WK,
Kim SK, Shin J, Park ES, Kang CM, Paik YK and Kim H:
Characterization of gene expression and activated signaling
pathways in solid-pseudopapillary neoplasm of pancreas. Mod Pathol.
27:580–593. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lunardi S, Jamieson NB, Lim SY, Griffiths
KL, Carvalho-Gaspar M, Al-Assar O, Yameen S, Carter RC, McKay CJ,
Spoletini G, et al: IP-10/CXCL10 induction in human pancreatic
cancer stroma influences lymphocytes recruitment and correlates
with poor survival. Oncotarget. 5:11064–11080. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Janky R, Binda MM, Allemeersch J, Van den
Broeck A, Govaere O, Swinnen JV, Roskams T, Aerts S and Topal B:
Prognostic relevance of molecular subtypes and master regulators in
pancreatic ductal adenocarcinoma. BMC Cancer. 16:6322016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jha PK, Vijay A, Sahu A and Ashraf MZ:
Comprehensive gene expression meta-analysis and integrated
bioinformatic approaches reveal shared signatures between
thrombosis and myeloproliferative disorders. Sci Rep. 6:370992016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang X, Ning Y and Guo X: Integrative
meta-analysis of differentially expressed genes in osteoarthritis
using microarray technology. Mol Med Rep. 12:3439–3445. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Han H, Bearss DJ, Browne LW, Calaluce R,
Nagle RB and Von Hoff DD: Identification of differentially
expressed genes in pancreatic cancer cells using cDNA microarray.
Cancer Res. 62:2890–2896. 2002.PubMed/NCBI
|
|
47
|
Lu KH, Patterson AP, Wang L, Marquez RT,
Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I,
et al: Selection of potential markers for epithelial ovarian cancer
with gene expression arrays and recursive descent partition
analysis. Clin Cancer Res. 10:3291–3300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Laughlin KM, Luo D, Liu C, Shaw G,
Warrington KH Jr, Qiu J, Yachnis AT and Harrison JK: Hematopoietic-
and neurologic-expressed sequence 1 expression in the murine GL261
and high-grade human gliomas. Pathol Oncol Res. 15:437–444. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang C, Xu B, Lu S, Zhao Y and Liu P: HN1
contributes to migration, invasion, and tumorigenesis of breast
cancer by enhancing MYC activity. Mol Cancer. 16:902017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Varisli L, Ozturk BE, Akyuz GK and Korkmaz
KS: HN1 negatively influences the β-catenin/E-cadherin interaction,
and contributes to migration in prostate cells. J Cell Biochem.
116:170–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ramirez NE, Zhang Z, Madamanchi A, Boyd
KL, O'Rear LD, Nashabi A, Li Z, Dupont WD, Zijlstra A and Zutter
MM: The α2β1 integrin is a metastasis suppressor in mouse models
and human cancer. J Clin Invest. 121:226–237. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Robertson JH, Yang SY, Winslet MC and
Seifalian AM: Functional blocking of specific integrins inhibit
colonic cancer migration. Clin Exp Metastasis. 26:769–780. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Shimoyama S, Gansauge F, Gansauge S,
Oohara T and Beger HG: Altered expression of extracellular matrix
molecules and their receptors in chronic pancreatitis and
pancreatic adenocarcinoma in comparison with normal pancreas. Int J
Pancreatol. 18:227–234. 1995.PubMed/NCBI
|
|
54
|
Li Y, Wagner ER, Yan Z, Wang Z, Luther G,
Jiang W, Ye J, Wei Q, Wang J, Zhao L, et al: The calcium-binding
protein S100A6 accelerates human osteosarcoma growth by promoting
cell proliferation and inhibiting osteogenic differentiation. Cell
Physiol Biochem. 37:2375–2392. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang XH, Zhang LH, Zhong XY, Xing XF, Liu
YQ, Niu ZJ, Peng Y, Du H, Zhang GG, Hu Y, et al: S100A6
overexpression is associated with poor prognosis and is
epigenetically up-regulated in gastric cancer. Am J Pathol.
177:586–597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Maelandsmo GM, Florenes VA, Mellingsaeter
T, Hovig E, Kerbel RS and Fodstad O: Differential expression
patterns of S100A2, S100A4 and S100A6 during progression of human
malignant melanoma. Int J Cancer. 74:464–469. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
He X, Xu X, Khan AQ and Ling W: High
expression of S100A6 predicts unfavorable prognosis of lung
squamous cell cancer. Med Sci Monit. 23:5011–5017. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ohuchida K, Mizumoto K, Ishikawa N, Fujii
K, Konomi H, Nagai E, Yamaguchi K, Tsuneyoshi M and Tanaka M: The
role of S100A6 in pancreatic cancer development and its clinical
implication as a diagnostic marker and therapeutic target. Clin
Cancer Res. 11:7785–7793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Logsdon CD, Simeone DM, Binkley C,
Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R and Hanash
S: Molecular profiling of pancreatic adenocarcinoma and chronic
pancreatitis identifies multiple genes differentially regulated in
pancreatic cancer. Cancer Res. 63:2649–2657. 2003.PubMed/NCBI
|
|
60
|
Okada Y, Yamazaki H, Sekine-Aizawa Y and
Hirokawa N: The neuron-specific kinesin superfamily protein KIF1A
is a unique monomeric motor for anterograde axonal transport of
synaptic vesicle precursors. Cell. 81:769–780. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
De S, Cipriano R, Jackson MW and Stark GR:
Overexpression of kinesins mediates docetaxel resistance in breast
cancer cells. Cancer Res. 69:8035–8042. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hattori Y, Kanamoto N, Kawano K, Iwakura
H, Sone M, Miura M, Yasoda A, Tamura N, Arai H, Akamizu T, et al:
Molecular characterization of tumors from a transgenic mouse
adrenal tumor model: Comparison with human pheochromocytoma. Int J
Oncol. 37:695–705. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Brait M, Loyo M, Rosenbaum E, Ostrow KL,
Markova A, Papagerakis S, Zahurak M, Goodman SM, Zeiger M,
Sidransky D, et al: Correlation between BRAF mutation and promoter
methylation of TIMP3, RARbeta2 and RASSF1A in thyroid cancer.
Epigenetics. 7:710–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Osipovich AB, Jennings JL, Lin Q, Link AJ
and Ruley HE: Dyggve-Melchior-Clausen syndrome: Chondrodysplasia
resulting from defects in intracellular vesicle traffic. Proc Natl
Acad Sci U S A. 105:16171–16176. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Dimitrov A, Paupe V, Gueudry C, Sibarita
JB, Raposo G, Vielemeyer O, Gilbert T, Csaba Z, Attie-Bitach T,
Cormier-Daire V, et al: The gene responsible for
Dyggve-Melchior-Clausen syndrome encodes a novel peripheral
membrane protein dynamically associated with the Golgi apparatus.
Hum Mol Genet. 18:440–453. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shi Z, Hong Y, Zhang K, Wang J, Zheng L,
Zhang Z, Hu Z, Han X, Han Y, Chen T, et al: BAG-1M co-activates
BACE1 transcription through NF-κB and accelerates Aβ production and
memory deficit in Alzheimer's disease mouse model. Biochim Biophys
Acta Mol Basis Dis. 1863:2398–2407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yi X, Hao Y, Xia N, Wang J, Quintero M, Li
D and Zhou F: Sensitive and continuous screening of inhibitors of
β-site amyloid precursor protein cleaving enzyme 1 (BACE1) at
single SPR chips. Anal Chem. 85:3660–3666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chen Q, Liu X, Xu L, Wang Y, Wang S, Li Q,
Huang Y and Liu T: Long non-coding RNA BACE1-AS is a novel target
for anisomycin-mediated suppression of ovarian cancer stem cell
proliferation and invasion. Oncol Rep. 35:1916–1924. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Dong J, Wang R, Ren G, Li X, Wang J, Sun
Y, Liang J, Nie Y, Wu K, Feng B, et al: HMGA2-FOXL2 axis regulates
metastases and epithelial-to-mesenchymal transition of
chemoresistant gastric cancer. Clin Cancer Res. 23:3461–3473. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yao H, Yang Z, Liu Z, Miao X, Yang L, Li
D, Zou Q and Yuan Y: Glypican-3 and KRT19 are markers associating
with metastasis and poor prognosis of pancreatic ductal
adenocarcinoma. Cancer Biomark. 17:397–404. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Dugnani E, Sordi V, Pellegrini S,
Chimienti R, Marzinotto I, Pasquale V, Liberati D, Balzano G,
Doglioni C, Reni M, et al: Gene expression analysis of embryonic
pancreas development master regulators and terminal cell fate
markers in resected pancreatic cancer: A correlation with clinical
outcome. Pancreatology. 18:945–953. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Li CF, Chuang IC, Liu TT, Chen KC, Chen
YY, Fang FM, Li SH, Chen TJ, Yu SC, Lan J and Huang HY:
Transcriptomic reappraisal identifies MGLL overexpression as an
unfavorable prognosticator in primary gastrointestinal stromal
tumors. Oncotarget. 7:49986–49997. 2016.PubMed/NCBI
|
|
73
|
Caba O, Irigoyen A, Jimenez-Luna C,
Benavides M, Ortuño FM, Gallego J, Rojas I, Guillen-Ponce C, Torres
C, Aranda E and Prados J: Identification of gene expression
profiling associated with erlotinib-related skin toxicity in
pancreatic adenocarcinoma patients. Toxicol Appl Pharmacol.
311:113–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wintgens JP, Wichert SP, Popovic L,
Rossner MJ and Wehr MC: Monitoring activities of receptor tyrosine
kinases using a universal adapter in genetically encoded split TEV
assays. Cell Mol Life Sci. 76:1185–1199. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Dharmawardana PG, Peruzzi B, Giubellino A,
Burke TR Jr and Bottaro DP: Molecular targeting of growth factor
receptor-bound 2 (Grb2) as an anti-cancer strategy. Anticancer
Drugs. 17:13–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Liang C, Xu Y, Ge H, Xing B, Li G, Li G
and Wu J: miR-564 inhibits hepatocellular carcinoma cell
proliferation and invasion by targeting the GRB2-ERK1/2-AKT axis.
Oncotarget. 8:107543–107557. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Birchmeier C, Birchmeier W, Gherardi E and
Vande Woude GF: Met, metastasis, motility and more. Nat Rev Mol
Cell Biol. 4:915–925. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang X, Lu X, Zhang T, Wen C, Shi M, Tang
X, Chen H, Peng C, Li H, Fang Y, et al: mir-329 restricts tumor
growth by targeting grb2 in pancreatic cancer. Oncotarget.
7:21441–21453. 2016.PubMed/NCBI
|
|
79
|
Witt O, Deubzer HE, Milde T and Oehme I:
HDAC family: What are the cancer relevant targets? Cancer Lett.
277:8–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Milde T, Oehme I, Korshunov A,
Kopp-Schneider A, Remke M, Northcott P, Deubzer HE, Lodrini M,
Taylor MD, von Deimling A, et al: HDAC5 and HDAC9 in
medulloblastoma: Novel markers for risk stratification and role in
tumor cell growth. Clin Cancer Res. 16:3240–3252. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li A, Liu Z, Li M, Zhou S, Xu Y, Xiao Y
and Yang W: HDAC5, a potential therapeutic target and prognostic
biomarker, promotes proliferation, invasion and migration in human
breast cancer. Oncotarget. 7:37966–37978. 2016.PubMed/NCBI
|
|
82
|
Klieser E, Urbas R, Stättner S, Primavesi
F, Jäger T, Dinnewitzer A, Mayr C, Kiesslich T, Holzmann K, Di
Fazio P, et al: Comprehensive immunohistochemical analysis of
histone deacetylases in pancreatic neuroendocrine tumors: HDAC5 as
a predictor of poor clinical outcome. Hum Pathol. 65:41–52. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
He P, Liang J, Shao T, Guo Y, Hou Y and Li
Y: HDAC5 promotes colorectal cancer cell proliferation by
up-regulating DLL4 expression. Int J Clin Exp Med. 8:6510–6516.
2015.PubMed/NCBI
|
|
84
|
Ozdağ H, Teschendorff AE, Ahmed AA, Hyland
SJ, Blenkiron C, Bobrow L, Veerakumarasivam A, Burtt G,
Subkhankulova T, Arends MJ, et al: Differential expression of
selected histone modifier genes in human solid cancers. BMC
Genomics. 7:902006. View Article : Google Scholar : PubMed/NCBI
|