Introduction
Gliomas are the most common primary brain tumors.
They arise from cancerous brain and spinal cord glial cells
(1,2). Glioblastoma multiforme (GBM) is a
World Health Organization grade IV glioma (3). It accounts for 56.1% of all gliomas
and has a five-year survival rate of only 5.5% (4). Despite a number of treatment methods,
including chemotherapeutic, radiological and surgical
interventions, there has been no significant change in the
therapeutic effect. Novel therapeutic approaches that are more
effective are needed and require more knowledge of the pathogenesis
of GBM.
Long non-coding RNA (lncRNA) is a type of RNA that
contains over 200 nucleotides and that does not encode protein
(5,6). LncRNAs serve important roles in human
diseases, especially involving tumors (7,8). In
gliomas, lncRNAs are involved in the regulation of various
biological processes (9). For
example, lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1)
is highly expressed in GBM tissues and perturbed expression of
NEAT1 suppresses cell proliferation and invasion (10). High expression of lncRNA H19
promotes cell migration and enhances angiogenesis (11,12).
The expression of HOX transcript antisense RNA lncRNA is increased
in GBM and is significantly associated with high grade brain tumors
(13). Additionally, as
competitive endogenous RNAs (ceRNAs), lncRNAs and messenger RNAs
(mRNAs) can regulate one other by competing for the shared
microRNAs (miRNAs/miRs) (14,15).
LncHERG is an lncRNA that acts as a competing RNA of miR-940. When
lncHERG is highly expressed, cell multiplication, invasion and
migration are inhibited in GBM (16).
In this study, the gene expression profile in GBM
was analyzed using a microarray dataset and RNA-sequencing
(RNA-Seq) datasets. Key lncRNAs in GBM were screened. Based on the
ceRNA hypothesis (15), the
lncRNA-miRNA-mRNA network was constructed and Gene Ontology (GO)
and Kyoto Encyclopedia of Gene and Genomes (KEGG) pathway analysis
of mRNAs in the network were performed. Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
cell proliferation assay were used to explore the functional role
of the key lncRNAs in GBM.
Materials and methods
Data, sample and cell information
Gene expression profile datasets GSE2223 (17,18)
and GSE59612 (19) were obtained
from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) (20). Gene expression profile of GBM
determined by RNA-Seq of the Cancer Genome Atlas (TCGA-GBM) was
extracted from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). The GSE2223
dataset included microarray data of 54 samples. Of these, four GBM
adjacent normal brain tissues and 27 GBM tissues were included in
the present study. A total of 17 GBM adjacent normal brain tissues
and 39 GBM tissues of 92 samples in the GSE59612 dataset were
selected in the present study. For the TCGA dataset, five GBM
adjacent normal brain tissues and 156 GBM tissues were
included.
A total of 10 GBM tissues were collected from GBM
patients who were treated surgically at the People's Hospital of
Zhangjiajie (Zhangjiajie, China) between April 2015 and March 2017.
The 10 GBM adjacent normal brain tissues were obtained from
patients with head trauma who underwent surgical treatment in the
same hospital. The age of the participants was 30–60 years old,
with a mean age of 49.6±6.7 years, and the male:female ratio was
8:6. All samples were collected immediately following surgical
resection and frozen rapidly in liquid nitrogen at −70°C. The
experimental study was approved by the hospital's ethics committee
and written informed consent was obtained from all
participants.
The SHG-44 and U251 GBM cell lines were obtained
from the American Type Culture Collection (ATCC; Manassas, VA, USA)
and maintained in RPMI 1640 medium (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing 10% fetal bovine serum (GE Healthcare
Life Sciences, Logan, UT, USA), penicillin (100 units/ml), and
streptomycin (100 ug/ml) and incubated in a 5% CO2
incubator at 37°C.
Small interfering si-NC, (si)-DLEU1 and si-tumor
necrosis factor receptor-associated factor 4 (TRAF4) was purchased
from Shanghai GenePharma Co., Ltd. (Shanghai, China). The si-NC,
si-DLEU1, si-TRAF4 and the empty vectors (10 nM) were transfected
into SHG-44 and U251 cell line using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Silencing was
confirmed 12 h after transfection, by reverse
transcription-polymerase chain reaction (RT-PCR) and western blot
analysis. The specific primer sequences are presented in Table I.
 | Table I.Sequences for si-NC, si-DLEU1#1,
si-DLEU1#2, si-TRAF4#1 and si-TRAF4#2. |
Table I.
Sequences for si-NC, si-DLEU1#1,
si-DLEU1#2, si-TRAF4#1 and si-TRAF4#2.
| Name | Guide | Passenger |
|---|
| si-NC |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
5′-ACGUGACACGUUCGGAGAATT-3′ |
| si-DLEU1#1 |
5′-UUUUUUGUGCAGUUUCAGCAA-3′ |
5′GCUGAAACUGCACAAAAAAUC-3′ |
| si-DLEU1#2 |
5′-UUCCUUUUUGAUAGUAUUCAA-3′ |
5′GAAUACUAUCAAAAAGGAAAA-3′ |
| si-TRAF4#1 |
5′-UUAAUAAAUACAAUUCCGGAU-3′ |
5′CCGGAAUUGUAUUUAUUAAUU-3′ |
| si-TRAF4#2 |
5′-UUUCAUAGGUGAAACGUGGAU-3′ |
5′CCACGUUUCACCUAUGAAACA-3′ |
Data analyses
Differentially expressed genes (DEGs) including
differentially expressed mRNAs (DEmRNAs) and differentially
expressed lncRNAs (DElncRNAs) between GBM and GBM adjacent normal
brain tissues of the GSE2223 and GSE59612 datasets with normalized
expression were analyzed using the Limma package (version 3.34.6;
http://www.bioconductor.org/packages/release/bioc/html/limma.html).
DEGs between GBM and GBM adjacent normal brain tissues of the
TCGA-GBM were screened using the DESeq package (version 3.34.6,
http://www.bioconductor.org/packages/release/bioc/html/DESeq.html).
Count data were used to normalize data in the DESeq analysis. The
fold-change (FC) of the DEGs, log2FC and false discovery
rate (FDR) were obtained following testing. The cutoff thresholds
were set to a |log2FC|≥1 and FDR <0.05. The
overlapping genes of the three datasets and genes displaying
consistent regulation were included. Following annotation with
Blast2GO5.0 (https://www.blast2go.com/), the lncRNA gene-type DEGs
were selected as candidate lncRNAs.
Identification of cancer-associated
lncRNAs
Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia.cancer-pku.cn/) was used
to analyze expression levels, survival analysis and correlation of
genes between tumors, and corresponding normal tissues. The
expression levels of candidate lncRNAs between cancer and normal
control were analyzed on the GEPIA website, which demonstrated 31
tumors types which have been tested. The parameters for expression
levels screening are P-value Cutoff (0.01) and log2FC Cutoff
(1) on this site.
Construction of lncRNA-miRNA-mRNA
network and correlation analysis of DElncRNAs and DEmRNAs
The integrated miRNA-DEmRNA and miRNA-DElncRNA pairs
were simultaneously predicted by miRanda (http://www.microrna.org/microrna/home.do) and
RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/),
respectively. The parameters were set to energy <-20 and score
>150 in miRanda and energy <-25 in RNAhybrid. Only the
overlap results of miRanda and RNAhybrid were selected as the
miRNA-DEmRNA and miRNA-DElncRNA pairs. The DElncRNA-miRNA-DEmRNA
networks were constructed by miRNA-bridges using Cytoscape software
(version 3.4.0) (21,22). Correlation analysis of DElncRNAs
and DEmRNAs was performed using the GEPIA website.
GO and KEGG pathway analysis
To investigate the underlying functional role of
lncRNAs, GO biological processes and KEGG pathway analysis were
performed for the mRNAs in lncRNA-miRNA-mRNA interactions with
DAVID 6.8 software (https://david.ncifcrf.gov/summary.jsp).
RT-PCR
Total RNA from GBM and normal brain samples was
isolated using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. After
DNase digestion, RNA quantification and purity were measured by the
ratio of 260/280 nm. RNA integrity was measured by 1.2% agarose gel
electrophoresis. The reverse transcriptase reaction kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to
reverse-transcribe RNA samples at 50°C for 60 min. RT-PCR was
performed in triplicate according to the manufacturer's protocol of
SYBR Green PCR Master Mix and reactions were carried using in a PCR
Thermal Cycler (Takara Bio, Inc., Otsu, Japan). The conditions
were: Initial denaturation at 95°C for 10 min, followed by 40
cycles of denaturation at 95°C for 5 sec, and annealing and
extension at 55–58°C for 30 sec. The relative expression levels of
genes were calculated as relative quantification, calculated as
2−∆∆Cq (18).
Western blotting
Total proteins were extracted from cells using Radio
Immunoprecipitation Assay Lysis Buffer (Beyotime Institute of
Biotechnology, Shanghai, China). The proteins were quantified using
the bicinchoninic acid protein assay kit (Shanghai Solarbio
Bioscience & Technology Co., Ltd., Shanghai, China). Cell
lysates were separated by 10% SDS-PAGE, transferred to
polyvinylidene fluoride membranes, and membranes were blocked at
room temperature with 5% skimmed milk in TSB Tween-20 (0.05% v/v;
TBS-T) for 1 h and incubated with specific antibodies at 4°C
overnight. The primary antibody was a 1:1,000 dilution of rabbit
anti-TRAF4 (cat. no. Ab108991; Abcam, Cambridge, UK) or mouse
anti-GAPDH (cat. no. AG019; Beyotime Institute of Biotechnology).
The membranes were washed three times for 10 min every time with
TBS-T, followed by incubation with secondary antibodies goat
anti-mouse immunoglobulin (Ig)G (1:1,000; cat. no. A0216; Beyotime
Institute of Biotechnology) and goat anti-rabbit IgG (1:2,000; cat.
no. Ab6721; Abcam) for 1 h at room temperature. The intensities of
the immunoreactivity were detected with an enhanced
chemiluminescence kit (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The images were developed on X-ray film. The experiments were
repeated ≥3 times.
Cell proliferation assay
A MTT kit (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) was used to analyze cell proliferation according to the
manufacturer's protocol. All the cells were cultured in 96-well
plates in the dark, the formazan was dissolved with dimethyl
sulfoxide (DMSO) and the absorbance value at 570 nm was detected
every 24 h.
Statistical analyses
SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA)
was used to analyze statistical significance. All the experiments
were independently performed three times. The data are expressed as
the mean ± standard deviation. The difference between the groups
was analyzed with an analysis of variance (ANOVA) or Student's t
test. Post hoc tests were performed using a Tukey test following
the ANOVA. * and ** refer to the statistically significant
difference of expression (P<0.05) and extremely significant
difference of expression (P<0.01), respectively. P<0.005 was
considered to indicate a statistically significant difference.
Results
Analysis of DEGs
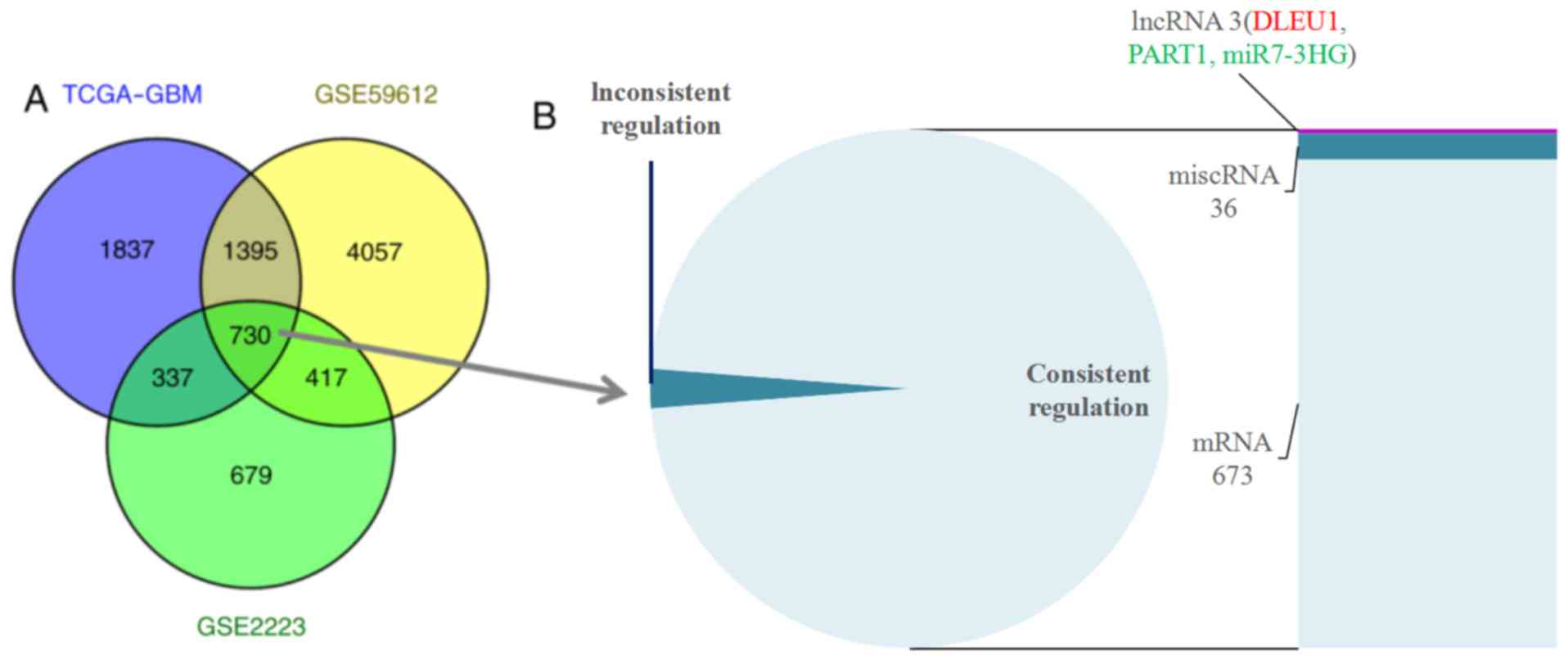
A total of 730 overlapping DEGs were screened from
the GSE2223, GSE59612 and TCGA-GBM datasets (Fig. 1A). Among these, 712 DEGs displaying
consistent regulation in all the three datasets were selected for
further study. The annotation analysis revealed three lncRNAs, 36
miscellaneous RNAs (miscRNAs) and 673 mRNAs (Fig. 1B). LncRNA DLEU1 was upregulated,
while prostate androgen-regulated transcript 1 (PART1) and miR7-3HG
were downregulated.
Identification of cancer-associated
lncRNAs in GBM
Among the DElncRNAs, the expression of DLEU1 was
upregulated in GBM and a number of other tumor types, including
cervical squamous cell carcinoma and endocervical adenocarcinoma,
colon adenocarcinoma, lymphoid neoplasm diffuse large B-cell
lymphoma, brain lower grade glioma, lung squamous cell carcinoma,
ovarian serous cystadenocarcinoma, pancreatic adenocarcinoma,
rectum adenocarcinoma, stomach adenocarcinoma, thymoma, and uterine
carcinosarcoma (Fig. 2). The
results highlighted the important role of DLEU1 in
carcinogenesis.
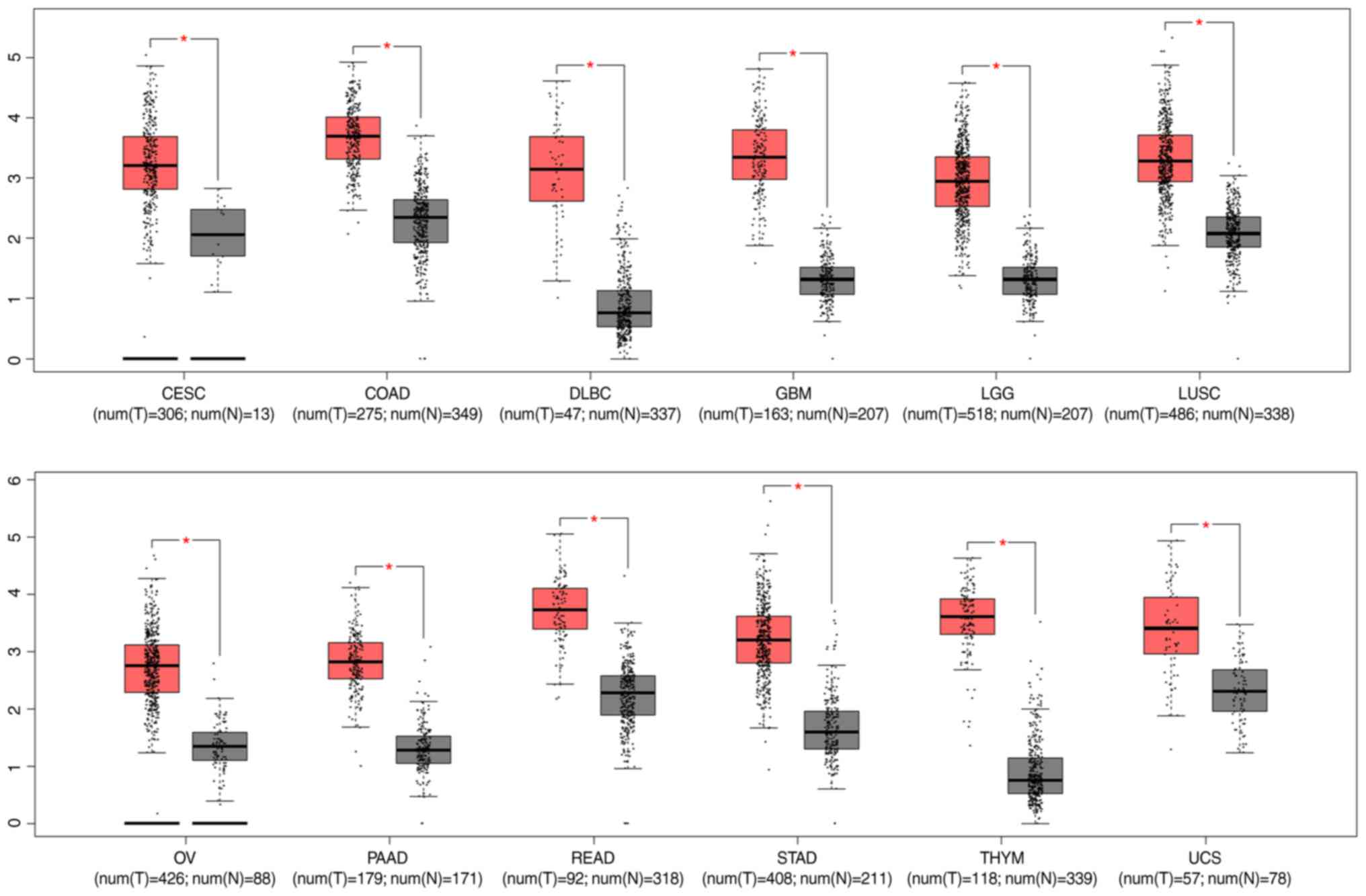 | Figure 2.Expression level of lncRNA DLEU1 in
multiple types of cancer. *P<0.05. CESC, cervical squamous cell
carcinoma and endocervical adenocarcinoma; COAD, colon
adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell
lymphoma; GBM, glioblastoma multiforme; LGG, brain lower grade
glioma; LUSC, lung squamous cell carcinoma; OV, ovarian serous
cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; READ, rectum
adenocarcinoma; STAD, stomach adenocarcinoma; THYM, thymoma; UCS,
uterine carcinosarcoma; num, number; T, tumour; N, normal. |
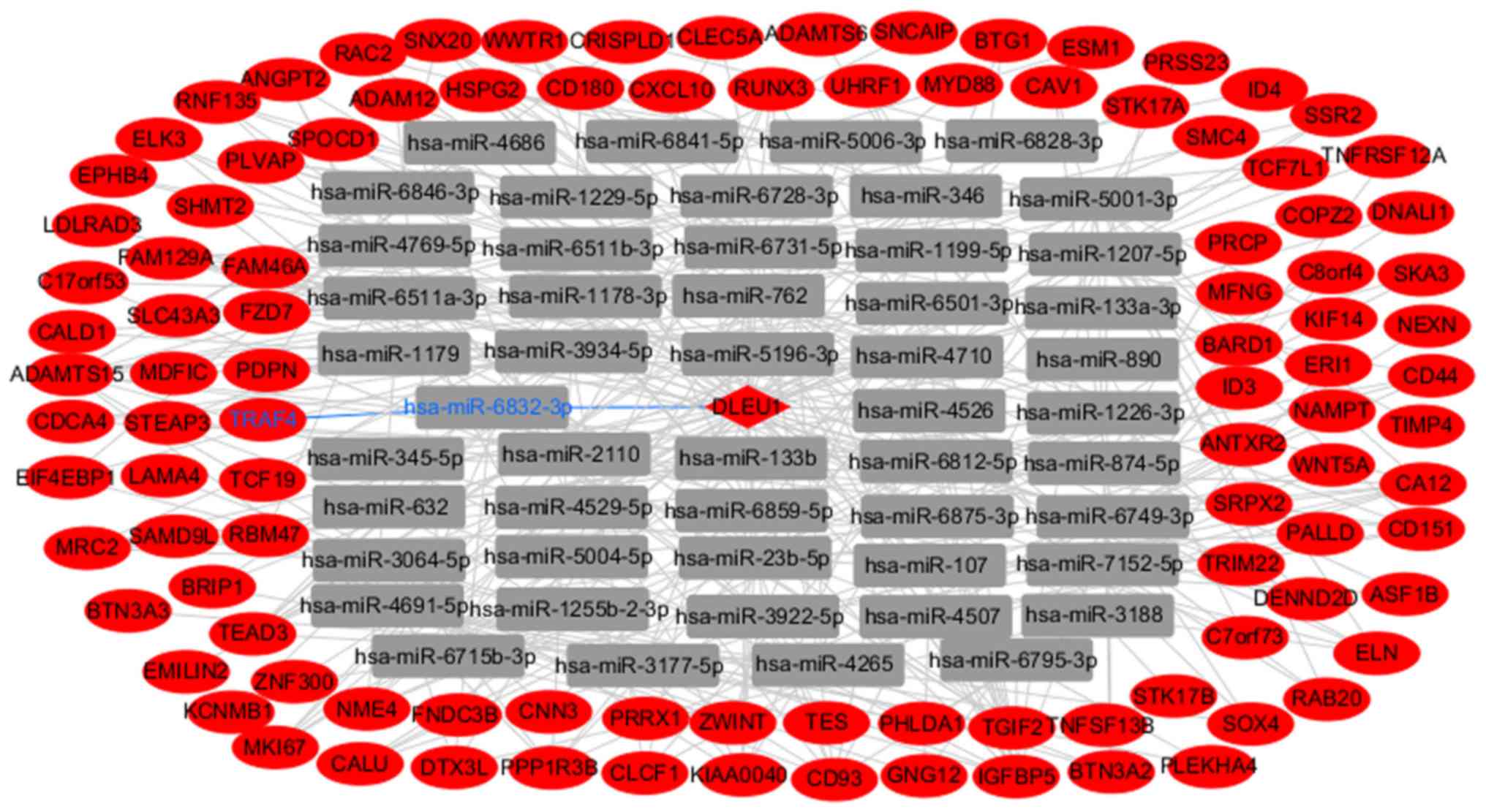
LncRNA-miRNA-mRNA network
The integrated DLEU1-miRNA-DEmRNA interactions were
identified with the miRanda and RNAhybrid methods. The network was
constructed using Cytoscape software (Fig. 3). In the lncRNA DLEU1-mediated
ceRNA network, DLEU1 interacted with as many as 315 miRNAs and 105
DEmRNAs. Among the miRNAs, miR-107, miR-1179, miR-133a, miR-133b
and miR-346 have been reported to be downregulated, and are
involved in the progression of in GBM.
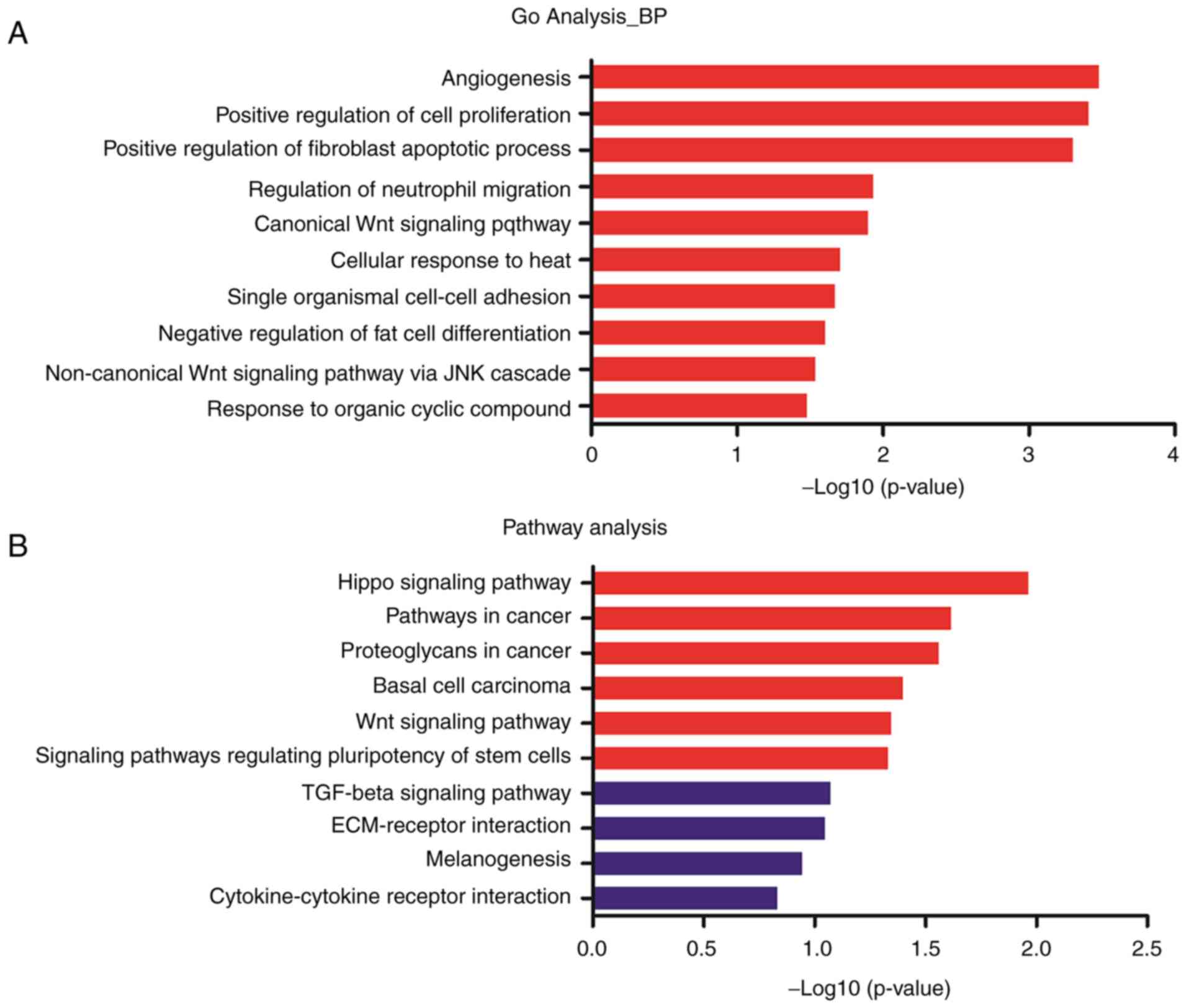
GO and pathway analysis
To investigate the functional role of genes in the
DLEU1-miRNA-DEmRNA network, GO and pathway analysis of mRNAs were
performed with DAVID 6.8 software (23,24).
The top 10 GO terms and pathway terms are presented in Fig. 4. The mRNAs mainly enriched in the
tumorigenesis associated GO terms were angiogenesis, positive
regulation of cell proliferation, positive regulation of fibroblast
apoptotic process, regulation of neutrophil migration and others.
The pathway analysis revealed mRNAs primarily enriched in important
pathways associated with tumorigenesis (Hippo signaling pathway,
pathways in cancer and Wnt signaling pathway). The genes involved
in the significant GO terms and pathway terms are presented in
Table II. Wnt family member
(WNT)5A, frizzled class receptor 7 (FZD7), transcription factor 7
like 1 (TCF7L1), WW domain containing transcription regulator 1
(WWTR1) and cluster of differentiation (CD)44 were enriched in at
least four significant GO or pathway terms.
 | Table II.Significant GO and pathway analysis
of differentially expressed mRNAs in the DLEU1-miRNA-mRNAs
network. |
Table II.
Significant GO and pathway analysis
of differentially expressed mRNAs in the DLEU1-miRNA-mRNAs
network.
| Terms | Genes | P-value |
|---|
| GO |
|
|
|
Angiogenesis | CAV1, SRPX2,
TNFRSF12A, HSPG2, ESM1, ELK3, ANGPT2, EPHB4 | 0.0003296 |
|
Positive regulation of cell
proliferation | KIF14, NAMPT,
SHMT2, TNFSF13B, RAC2, CLCF1, SOX4, ID4, ESM1, WWTR1,
CXCL10 | 0.0003889 |
|
Positive regulation of
fibroblast apoptotic process | BTG1, STK17B,
STK17A | 0.0004981 |
|
Regulation of neutrophil
migration | MYD88, RAC2 | 0.0116385 |
|
Canonical Wnt signaling
pathway | WNT5A, SOX4,
TCF7L1, FZD7 | 0.0126456 |
|
Cellular response to heat | MKI67, C8ORF4,
CXCL10 | 0.0196642 |
| Single
organismal cell-cell adhesion | SRPX2, CD44,
CD93, PDPN | 0.0212821 |
|
Negative regulation of fat
cell differentiation | WNT5A, ID4,
WWTR1 | 0.0249487 |
|
Non-canonical Wnt signaling
pathway via JNK cascade | WNT5A,
FZD7 | 0.0288453 |
|
Response to organic cyclic
compound | NAMPT, MKI67,
ANGPT2 | 0.0331943 |
| Mammary
gland involution | CAV1, IGFBP5 | 0.0345148 |
| Signal
transduction | NAMPT, PDPN, MRC2,
GNG12, ELK3, CXCL10, MYD88, RAC2, TNFSF13B, CLEC5A, ANGPT2, TRAF4,
IGFBP5 | 0.0375587 |
|
Negative regulation of protein
export from nucleus | SOX4, BARD1 | 0.0401515 |
| Kidney
morphogenesis | SOX4,
WWTR1 | 0.0457556 |
| T cell
mediated immunity | BTN3A3, BTN3A2 | 0.0457556 |
|
Cartilage development | WNT5A, CD44,
PRRX1 | 0.0465266 |
|
Positive regulation of protein
binding | WNT5A, MFNG,
CAV1 | 0.0493935 |
| Pathway |
|
|
| Hippo
signaling pathway | WNT5A,
TEAD3, WWTR1, TCF7L1, FZD7 | 0.010874 |
|
Pathways in cancer | WNT5A,
LAMA4, RAC2, GNG12, TCF7L1, FZD7, TRAF4 | 0.024194 |
|
Proteoglycans in cancer | WNT5A, CAV1,
CD44, HSPG2, FZD7 | 0.027611 |
| Basal
cell carcinoma | WNT5A, TCF7L1,
FZD7 | 0.040012 |
| Wnt
signaling pathway | WNT5A, RAC2,
TCF7L1, FZD7 | 0.045011 |
|
Signaling pathways regulating
pluripotency of stem cells | WNT5A, ID4,
ID3, FZD7 | 0.046643 |
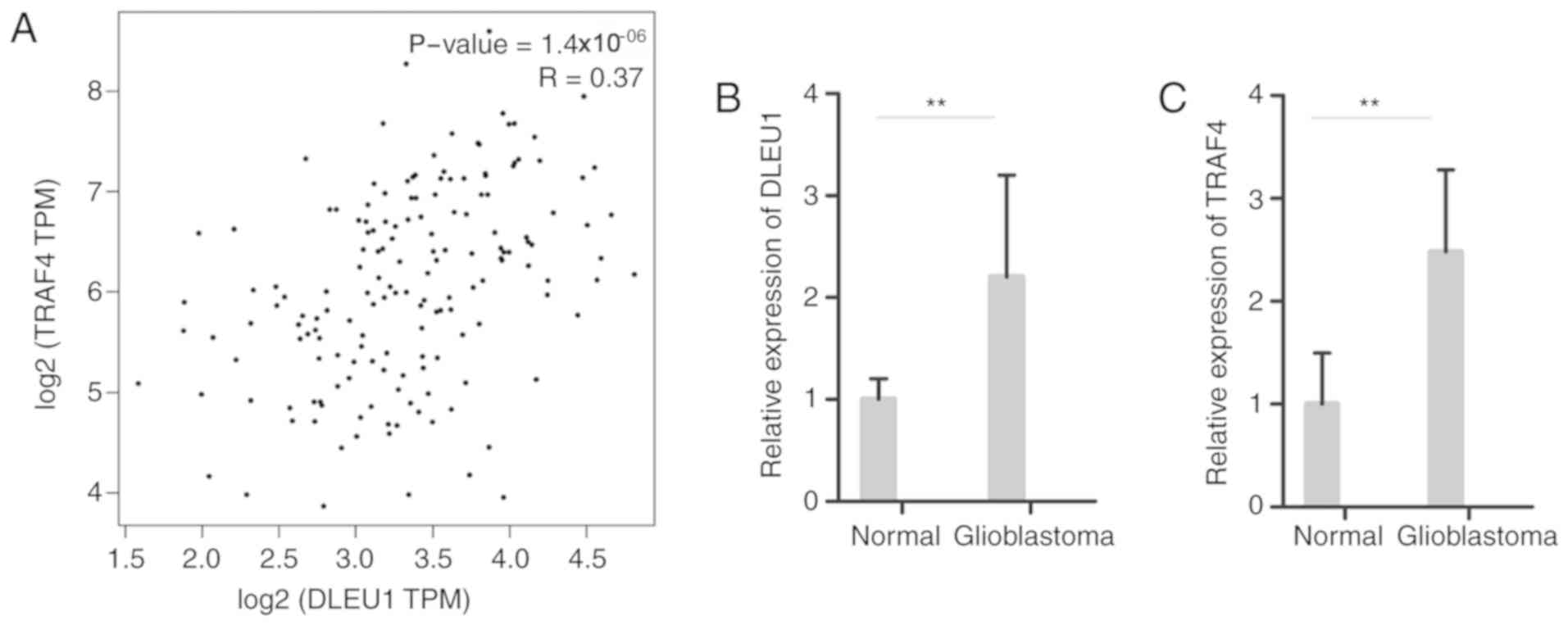
DLEU1 is positively associated with
TRAF4
TRAF4 serves an important role in other cancers
according to previous studies (25–28).
Querying GEPIA revealed that the expression of TRAF4 in the
DLEU1-miRNA-DEmRNA network was positively associated with DLEU1
(Fig. 5A). To validate the
prediction, the relative expression levels of DLEU1 and DLEU1 were
analyzed by RT-PCR in 10 and 10 GBM adjacent normal brain tissues.
The expression levels of DLEU1 and TRAF4 were both significantly
increased in GBM compared with normal control (P<0.01; Fig. 5B and C).
Silencing DLEU1 downregulates TRAF4
and inhibits cell proliferation
The effects of silencing lncRNA were studied.
Following silencing DLEU1 (Fig.
6A), cell viability decreased significantly in SHG-44 and U251
cell (P<0.01; Fig. 6B).
However, there was no significant difference between the negative
control and blank control concerning cell viability. Compared with
the negative control transfected with the empty vector, the
expression of TRAF4 was downregulated in SHG-44 and U251 cell
transfected with si-DLEU1 at transcription (Fig. 6C) and the protein level (Fig. 6D).
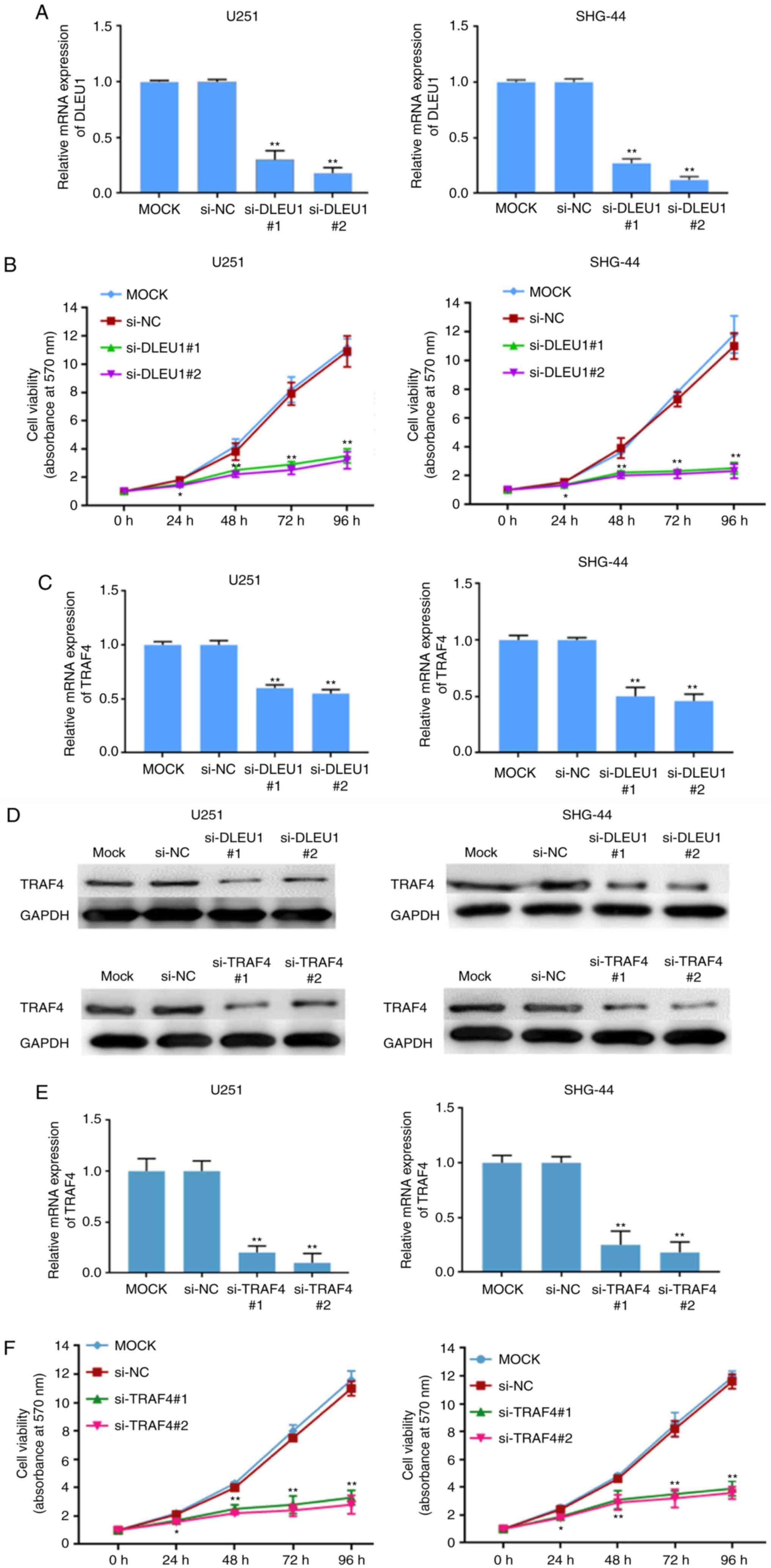 | Figure 6.Silencing of lncRNA DLEU1
downregulates TRAF4 and inhibits cell proliferation. Silencing of
lncRNA DLEU1 inhibited cell proliferation by MTT assays in (A)
SHG-44 and (B) U251 cell. Silencing of lncRNA DLEU1 downregulates
TRAF4 in SHG-44 and U251 cell transfected with si-DLEU1 at (C) the
transcription and the (D) protein level. Silencing of TRAF4
inhibited cell proliferation as measured by MTT assays in (E)
SHG-44 cells and (F) U251 cells. MOCK, SHG-44 cells and U251 cells
transfected empty vector with no any other sequence; NC, SHG-44
cells and U251 cells transfected with non-targeting sequence in
humans; si-DLEU1, SHG-44 cells and U251 cells transfected with
si-DLEU1; si-TRAF4, SHG-44 cells and U251 cells transfected with
si-TRAF4. *P<0.05 and **P<0.01, n=3. Si, small interfering;
lnc, long non-coding; NC, negative control; TRAF4, TNF receptor
associated factor 4; DLEU1, deleted in lymphocytic leukemia 1. |
Silencing TRAF4 inhibits cell
proliferation
Similarly, the proliferation of cells was studied by
silencing TRAF. Compared with the negative control transfected with
the empty vector. After silencing TRAF4 (Fig. 6E), cell viability decreased
significantly in SHG-44 and U251 cells (P<0.01; Fig. 6F). However, there was no
significant difference between the negative control and blank
control concerning cell viability.
Discussion
In the present study, microarray data profiling and
RNA-Seq profiles of GBM were integrated and re-analyzed. A total of
712 DEGs (673 DEmRNAs, 36 miscellaneous RNA and 3 DElncRNAs) were
identified between GBM, and GBM adjacent normal brain tissues.
Among the three lncRNAs (DLEU1, PART1 and miR7-3HG), DLEU1 was more
expressed in a number of types of cancer, including GBM. The
DLEU1-miRNA-DEmRNAs network was constructed. Several miRNAs in the
DLEU1-miRNA-mRNAs network have been reported in the progression of
tumorigenesis in GBM. For example, miR-107 is downregulated in
glioma tissues and cell lines (29). Decreased expression of miRNA-107
promotes cell growth and invasion, inhibits cell apoptosis, and
predicts a poor prognosis (30–32).
miR-1179 and miR-346 (33) are
downregulated in glioma tissues and cell lines, and their high
expression inhibits cell proliferation. In GBM (34), miR-133a (35,36),
miR-133b (37,38) and miR-346 are downregulated in
glioma tissues and cell lines, and high expression inhibits cell
proliferation, migration, and invasion. The regulation of the
association of these miRNAs in the authors' prediction was in
concordance with these previous studies. Therefore, the analysis
and prediction were credible.
DEmRNAs in the DLEU1-miRNA-mRNAs network were mainly
enriched in the pathway terms of the Hippo signaling pathway,
pathways in cancer and Wnt signaling pathway. The Hippo signaling
pathway is reportedly a major signaling pathway that regulates cell
proliferation and growth, and is important role in the
tumorigenesis of GBM (39). CD44
was reported to be upregulated in GBM and protected cancer cells by
attenuating the activation of the Hippo signaling pathway (40). Suppression of the activity of the
Hippo signaling pathway using Amlexanox was reported to inhibit GBM
cell proliferation and induce GBM cell apoptosis (41). The Wnt signaling pathway affects
tumor initiation, cell migration and invasion of GBM (42). Multiple factors, including small
molecules, including SEN461 (43),
small non-coding RNAs, such as miRNA34a (44) and miRNA-577 (45), and mRNAs, such as zinc finger E-box
binding homeobox 1 (46), WNT3A
(47), and homeobox A13 (48), influence GBM progression via the
Wnt signaling pathway. Presently, six DEmRNAs [WNT5A, TEA domain
family members (TEAD)3, WWTR1, TCF7L1, FZD7 and Rac family small
GTPase 2] in the DLEU1-miRNA-mRNA network were demonstrated to be
enriched in the Hippo and Wnt signaling pathways. WNT5A induces GBM
cell migration (49). WNT5A is
increased in GBM tissues and overexpression of WNT5A promotes the
differentiation, proliferation, migration, and invasive growth of
GBM cells (49–51). WWTR1, also known as TAZ, is an
important regulator of the Hippo signaling pathway. The interaction
of WWTR1 with TEAD drives mesenchymal differentiation of malignant
glioma and influences tumor progression (52). As a direct target, WWTR1 is
upregulated in GBM and promotes cell proliferation (53). Finally, the high expression of FZD7
has been detected in GBM and is associated with poor survival
(54,55).
TRAF4 is overexpressed in tissues or cells in
osteosarcoma (25), colon cancer
(26), oral squamous cell
carcinoma (27) and breast cancer
(28). Presently, the knockdown of
TRAF4 inhibited cell proliferation, migration and invasion, and
induced apoptosis. A prior study reported the significantly high
expression of DLEU1 in epithelial carcinoma, with associated
promotion of cell multiplication, migration, and invasion and
suppression of cell apoptosis (56). Other authors reported the
intensified expression of DLEU1 in gastric cancer and the
association with lymph node metastasis, tumor size, and advanced
stage of pathology. Silencing of DLEU1 inhibited cell proliferation
(57). In accordance with these
findings, the expression level of DLEU1 and TRAF4 was confirmed to
be high in GBM tissues by RT-PCR. The expression of DLEU1 was
positively associated with TRAF4. RNA interference with DLEU1
downregulated TRAF and inhibited GBM cell proliferation. When TRAF4
knockdown occurs, cell proliferation is inhibited, with similar
results to DLEU1. All results indicate that DLEU1 may affect the
tumorigenesis of GBM by regulating the expression of TRAF4.
In conclusion, DLEU1 was intensified in GBM tissues
and silencing of DLEU1 inhibited cell proliferation. Furthermore,
miRNAs and mRNAs in the DLEU1-mediated ceRNA network are involved
in cell differentiation, proliferation, migration, and invasive
growth of GBM cells. These collective observations support the idea
that DLEU1 may serve a pivotal role in the tumorigenesis of GBM.
This study identifies a novel lncRNA that may act as a ceRNA in
GBM. Further studies are needed to understand the molecular role of
DLEU1 in GBM progression.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Natural Science
Foundation of Hunan Province (grant no. C2013-225).
Availability of data materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GH and JW were responsible for the concept and
design of the present study. JW and XQ acquired the data. JW and DP
performed data analysis and experiments. JW drafted the article.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The experimental study was approved by the ethics
committee of People's Hospital of Zhangjiajie and written informed
consent was obtained from all participants.
Patient consent for publication
Written informed consent was obtained from all
participants.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kohler BA, Ward E, McCarthy BJ, Schymura
MJ, Ries LA, Eheman C, Jemal A, Anderson RN, Ajani UA and Edwards
BK: Annual report to the nation on the status of cancer, 1975–2007,
featuring tumors of the brain and other nervous system. J Natl
Cancer Inst. 103:714–736. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ohgaki H and Kleihues P: Epidemiology and
etiology of gliomas. Acta Neuropathol. 109:93–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ostrom QT, Gittleman H, Liao P,
Vecchione-Koval T, Wolinsky Y, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and other central nervous
system tumors diagnosed in the United States in 2010–2014. Neuro
Oncol. 19:v1–v88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lipovich L. Johnson R and Lin CY: MacroRNA
underdogs in a microRNA world: Evolutionary, regulatory, and
biomedical significance of mammalian long non-protein-coding RNA.
Biochim Biophys Acta. 1799:597–615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li M, Deng H, Peng H and Wang Q:
Functional nanoparticles in targeting glioma diagnosis and
therapies. J Nanosci Nanotechnol. 14:415–432. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi Y, Wang Y, Luan W, Wang P, Tao T,
Zhang J, Qian J, Liu N and You Y: Long non-coding RNA H19 promotes
glioma cell invasion by deriving miR-675. PLoS One. 9:e862952014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai
Y, Wu D, Wang Y, Zhuang Z and Xia H: Increased level of H19 long
noncoding RNA promotes invasion, angiogenesis, and stemness of
glioblastoma cells. J Neurosurg. 2016:129–136. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tan SK, Pastori C, Penas C, Komotar RJ,
Ivan ME, Wahlestedt C and Ayad NG: Serum long noncoding RNA HOTAIR
as a novel diagnostic and prognostic biomarker in glioblastoma
multiforme. Mol Cancer. 17:742018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi J, Wang YJ, Sun CR, Qin B, Zhang Y and
Chen G: Long noncoding RNA lncHERG promotes cell proliferation,
migration and invasion in glioblastoma. Oncotarget.
8:108031–108041. 2007.
|
|
17
|
Bredel M, Bredel C, Juric D, Duran GE, Yu
RX, Harsh GR, Vogel H, Recht LD, Scheck AC and Sikic BI: Tumor
necrosis factor-alpha-induced protein 3 as a putative regulator of
nuclear factor-kappaB-mediated resistance to O6-alkylating agents
in human glioblastomas. J Clin Oncol. 24:274–287. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bredel M, Bredel C, Juric D, Harsh GR,
Vogel H, Recht LD and Sikic BI: Functional network analysis reveals
extended gliomagenesis pathway maps and three novel MYC-interacting
genes in human gliomas. Cancer Res. 65:8679–8689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gill BJ, Pisapia DJ, Malone HR, Goldstein
H, Lei L, Sonabend A, Yun J, Samanamud J, Sims JS, Banu M, et al:
MRI-localized biopsies reveal subtype-specific differences in
molecular and cellular composition at the margins of glioblastoma.
Proc Natl Acad Sci USA. 111:12550–12555. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bindea G, Galon J and Mlecnik B: CluePedia
Cytoscape plugin: Pathway insights using integrated experimental
and in silico data. Bioinformatics. 29:661–663. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yao W, Wang X, Cai Q, Gao S, Wang J and
Zhang P: Knockdown of TRAF4 expression suppresses osteosarcoma cell
growth in vitro and in vivo. Int J Mol Med. 34:1655–1660. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang K, Wang F and Han JJ: TRAF4 promotes
the growth and invasion of colon cancer through the Wnt/β-catenin
pathway. Int J Clin Exp Pathol. 8:1419–1426. 2015.PubMed/NCBI
|
|
27
|
Yang J, Wei D, Wang W, Shen B, Xu S and
Cao Y: TRAF4 enhances oral squamous cell carcinoma cell growth,
invasion and migration by Wnt-β-catenin signaling pathway. Int J
Clin Exp Pathol. 8:11837–11846. 2015.PubMed/NCBI
|
|
28
|
Zhu L, Zhang S, Huan X, Mei Y and Yang H:
Down-regulation of TRAF4 targeting RSK4 inhibits proliferation,
invasion and metastasis in breast cancer xenografts. Biochem
Biophys Res Commun. 500:810–816. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen L, Zhang R, Li P, Liu Y, Qin K, Fa
ZQ, Liu YJ, Ke YQ and Jiang XD: P53-induced microRNA-107 inhibits
proliferation of glioma cells and down-regulates the expression of
CDK6 and Notch-2. Neurosci Lett. 534:327–332. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen L, Chen XR, Chen FF, Liu Y, Li P,
Zhang R, Yan K, Yi YJ, Xu ZM and Jiang XD: MicroRNA-107 inhibits
U87 glioma stem cells growth and invasion. Cell Mol Neurobiol.
33:651–657. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He J, Zhang W, Zhou Q, Zhao T, Song Y,
Chai L and Li Y: Low-expression of microRNA-107 inhibits cell
apoptosis in glioma by upregulation of SALL4. Int J Biochem Cell
Biol. 45:1962–1973. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ji Y, Wei Y, Wang J, Ao Q, Gong K and Zuo
H: Decreased expression of microRNA-107 predicts poorer prognosis
in glioma. Tumour Biol. 36:4461–4466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wolter M, Werner T, Malzkorn B and
Reifenberger G: Role of microRNAs located on chromosome arm 10q in
malignant gliomas. Brain Pathol. 26:344–358. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu X, Cai N, Zhi T, Bao Z, Wang D, Liu Y,
Jiang K, Fan L, Ji J and Liu N: MicroRNA-1179 inhibits glioblastoma
cell proliferation and cell cycle progression via directly
targeting E2F transcription factor 5. Am J Cancer Res. 7:1680–1692.
2017.PubMed/NCBI
|
|
35
|
Wang J, Li J, Guo F and Yan Y:
MicroRNA-133a inhibits the malignant behavior of glioma via
downregulation of matrix metallopeptidase 9. Mol Med Rep.
13:3220–3226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sakr M, Takino T, Sabit H, Nakada M, Li Z
and Sato H: miR-150-5p and miR-133a suppress glioma cell
proliferation and migration through targeting membrane-type-1
matrix metalloproteinase. Gene. 587:155–162. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang J, Li Y and Jiang C: MiR-133b
contributes to arsenic-induced apoptosis in U251 glioma cells by
targeting the hERG channel. J Mol Neurosci. 55:985–994. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li C, Liu Z, Yang K, Chen X, Zeng Y, Liu
J, Li Z and Liu Y: miR-133b inhibits glioma cell proliferation and
invasion by targeting Sirt1. Oncotarget. 7:36247–36254.
2016.PubMed/NCBI
|
|
39
|
Artinian N, Cloninger C, Holmes B,
Benavides-Serrato A, Bashir T and Gera J: Phosphorylation of the
hippo pathway component AMOTL2 by the mTORC2 kinase promotes YAP
signaling, resulting in enhanced glioblastoma growth and
invasiveness. J Biol Chem. 290:19387–19401. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu Y, Stamenkovic I and Yu Q: CD44
attenuates activation of the hippo signaling pathway and is a prime
therapeutic target for glioblastoma. Cancer Res. 70:2455–2464.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu Y, Lu J, Zhang Z, Zhu L, Dong S, Guo
G, Li R, Nan Y, Yu K, Zhong Y and Huang Q: Amlexanox, a selective
inhibitor of IKBKE, generates anti-tumoral effects by disrupting
the Hippo pathway in human glioblastoma cell lines. Cell Death Dis.
8:e30222017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee Y, Lee JK, Ahn SH, Lee J and Nam DH:
WNT signaling in glioblastoma and therapeutic opportunities. Lab
Invest. 96:137–150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
De Robertis A, Valensin S, Rossi M, Tunici
P, Verani M, De Rosa A, Giordano C, Varrone M, Nencini A, Pratelli
C, et al: Identification and characterization of a small-molecule
inhibitor of Wnt signaling in glioblastoma cells. Mol Cancer Ther.
12:1180–1189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rathod SS, Rani SB, Khan M, Muzumdar D and
Shiras A: Tumor suppressive miRNA-34a suppresses cell proliferation
and tumor growth of glioma stem cells by targeting Akt and Wnt
signaling pathways. FEBS Open Bio. 4:485–495. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang W, Shen C, Li C, Yang G, Liu H, Chen
X, Zhu D, Zou H, Zhen Y, Zhang D and Zhao S: miR-577 inhibits
glioblastoma tumor growth via the Wnt signaling pathway. Mol
Carcinog. 55:575–585. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kahlert UD, Maciaczyk D, Doostkam S, Orr
BA, Simons B, Bogiel T, Reithmeier T, Prinz M, Schubert J,
Niedermann G, et al: Activation of canonical WNT/β-catenin
signaling enhances in vitro motility of glioblastoma cells by
activation of ZEB1 and other activators of
epithelial-to-mesenchymal transition. Cancer Lett. 325:42–53. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kaur N, Chettiar S, Rathod S, Rath P,
Muzumdar D, Shaikh ML and Shiras A: Wnt3a mediated activation of
Wnt/β-catenin signaling promotes tumor progression in glioblastoma.
Mol Cell Neurosci. 54:44–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Duan R, Han L, Wang Q, Wei J, Chen L,
Zhang J, Kang C and Wang L: HOXA13 is a potential GBM diagnostic
marker and promotes glioma invasion by activating the Wnt and TGF-β
pathways. Oncotarget. 6:27778–27793. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kamino M, Kishida M, Kibe T, Ikoma K,
Iijima M, Hirano H, Tokudome M, Chen L, Koriyama C, Yamada K, et
al: Wnt-5a signaling is correlated with infiltrative activity in
human glioma by inducing cellular migration and MMP-2. Cancer Sci.
102:540–548. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yu JM, Jun ES and Jung JS, Suh SY, Han JY,
Kim JY, Kim KW and Jung JS: Role of Wnt5a in the proliferation of
human glioblastoma cells. Cancer Lett. 257:172–181. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hu B, Wang Q, Wang YA, Hua S, Sauvé CG,
Ong D, Lan ZD, Chang Q, Ho YW, Monasterio MM, et al: Epigenetic
Activation of WNT5A drives glioblastoma stem cell differentiation
and invasive growth. Cell. 167:1281–1295.e18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bhat KP, Salazar KL, Balasubramaniyan V,
Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL,
Kim SH, et al: The transcriptional coactivator TAZ regulates
mesenchymal differentiation in malignant glioma. Genes Dev.
25:2594–2609. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yuan J, Xiao G, Peng G, Liu D, Wang Z,
Liao Y, Liu Q, Wu M and Yuan X: MiRNA-125a-5p inhibits glioblastoma
cell proliferation and promotes cell differentiation by targeting
TAZ. Biochem Biophys Res Commun. 457:171–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Schiffgens S, Wilkens L, Brandes AA, Meier
T, Franceschi E, Ermani M, Hartmann C, Sandalcioglu IE and Dumitru
CA: Sex-specific clinicopathological significance of novel
(Frizzled-7) and established (MGMT, IDH1) biomarkers in
glioblastoma. Oncotarget. 7:55169–55180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wald JH, Hatakeyama J, Printsev I, Cuevas
A, Fry WHD, Saldana MJ, VanderVorst K, Rowson-Hodel A, Angelastro
JM, Sweeney C and Carraway KL Rd: Suppression of planar cell
polarity signaling and migration in glioblastoma by Nrdp1-mediated
Dvl polyubiquitination. Oncogene. 36:5158–5167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang LL, Sun KX, Wu DD, Xiu YL, Chen X,
Chen S, Zong ZH, Sang XB, Liu Y and Zhao Y: DLEU1 contributes to
ovarian carcinoma tumourigenesis and development by interacting
with miR-490-3p and altering CDK1 expression. J Cell Mol Med.
21:3055–3065. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li X, Li Z, Liu Z, Xiao J, Yu S and Song
Y: Long non-coding RNA DLEU1 predicts poor prognosis of gastric
cancer and contributes to cell proliferation by epigenetically
suppressing KLF2. Cancer Gene Ther. 25:58–67. 2018. View Article : Google Scholar : PubMed/NCBI
|