Introduction
Gastric cancer, the fourth most common cancer in the
world, is the second leading cause of cancer-associated mortality
(1). The molecular mechanisms of
gastric cancer have been extensively studied; however, the early
diagnosis of gastric cancer remains uncommon (2), and the majority of patients have
already reached an advanced stage at the time of diagnosis
(3). The three major causes of
gastric cancer include genetic predisposition, environmental
factors and Helicobacter pylori (Hp) infection (4). Gastric carcinogenesis is a multi-step
process that is closely associated with Hp (5). Hp colonizes the stomach of >50% of
the world's population, and due to its pathogenic role in the
development of gastric cancer, Hp is classified as a class I
carcinogen (6). Therefore, there
is an urgent need to identify novel bio-markers for the early
diagnosis of gastric cancer, as well as new targets for gastric
cancer therapy.
MicroRNAs (miRNAs/miRs) are a type of endogenous
short non-coding RNA molecules ~22 nucleotides in length, which
directly bind to the 3′-untranslated region (3′-UTR) of multiple
target mRNAs, promoting mRNA degradation and preventing translation
to post-transcriptionally regulate gene expression. Numerous
studies have indicated that abnormal expression of miRNAs is
associated with the occurrence and progression of gastric cancer by
regulating the expression of oncogenes and/or tumor inhibitor genes
(7–12). In recent years, various studies of
the role of miRNAs in gastric cancer, including Hp-positive gastric
cancer, have been conducted. For example, it was reported that
miR-101 functions as a growth-suppressive miRNA in Hp-associated
gastric cancer by targeting suppressor of cytokine signaling 2
(13). miR-203 inhibits
Hp-associated gastric cancer growth by repressing peripheral plasma
membrane protein CASK expression (14). miR-Let-7c is significantly
downregulated in Hp-positive gastric carcinogenesis (15). miR-24-3p regulates the progression
of gastric mucosal lesions and suppresses the proliferation and
invasiveness of N87 cells via peroxiredoxin 6 (16). Furthermore, miR-140, a well-studied
tumor suppressor miRNA (17–19),
has been found to inhibit gastric cancer growth via regulating YES
proto-oncogene 1, Src family tyrosine kinase and transcription
factor SRY-box 4 expression (20,21).
However, the role and mechanism of miR-140 in gastric cancer,
particularly in the presence of Hp, remains largely unclear.
Despite a number of achievements in chemotherapy and
alternative therapeutic agents, there has been no major improvement
in the overall survival in patients with gastric cancer over the
past decade (22). Immunotherapy
is a relatively recent strategy in gastric cancer therapy (23–25).
However, the impact of immunotherapy in gastric cancer is
unsatisfactory. Tumor cells escape T-cell-mediated cellular
cytotoxicity through regulating the programmed cell death
(PD)-1/PD-ligand 1 (PD-L1) pathway (26). Targeting the PD-1/PD-L1 pathway has
emerged as a novel strategy in the treatment for a variety of
malignancies (27–29). A previous study suggested that
miR-140 exerts anti-osteosarcoma efficacy via targeting the immune
checkpoint molecule PD-L1 (30).
Therefore, the present study aimed to investigate the role and the
molecular mechanism of miR-140 in Hp-associated gastric cancer, and
to examine its association with immune function in gastric
cancer.
Materials and methods
Clinical specimens
Gastritis tissue samples (15 Hp-positive, 15
Hp-negative) from 30 gastritis patients (21–53 years old; sex
ratio: 1:1), and 30 gastric cancer tissue samples (15 Hp-positive
and 15 Hp-negative) from 30 gastric cancer patients (with or
without Hp infection; 23–57 years old; sex ratio: 1:1) were
collected from patients who underwent gastroscopy at the hospital
from December 2015 to December 2016. All the patients enrolled in
the present study were ≥18 years old, had no other cancer, and were
not taking nonsteroidal anti-inflammatory drugs or proton pump
inhibitors. In total, two gastric biopsies were collected from the
patient: One biopsy was immediately frozen and stored at −80°C
until total RNA extraction, and the second biopsy was used for Hp
detection. Hp infection was confirmed when a rapid urease test
(31) was positive. The present
study was approved by the Ethical Committee of the First Affiliated
Hospital of General Hospital of People's Liberation Army, and
written informed consent was obtained from each patient.
Cell culture
Gastric cancer cell lines AGS, MGC803 (cat. no.
L4678; Wuhan Miaoling Bioscience & Technology Co., Ltd., Wuhan,
China), SGC7901(cat. no. L4801; Wuhan Miaoling Bioscience &
Technology Co., Ltd.), BGC823 (cat. no. L4448; Wuhan Miaoling
Bioscience & Technology Co., Ltd.) and MKN45 (cat. no. L4679;
Wuhan Miaoling Bioscience & Technology Co., Ltd.), and the
immortalized non-tumorigenic cell line GES-1 were purchased from
American Type Culture Collection (Manassas, VA, USA). Cells were
cultured in Dulbecco's modified Eagle's medium or RPMI-1640 (both
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA USA) containing
10% heat-inactivated fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) and 1% streptomycin-penicillin solution
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C with 5%
CO2.
BGC823 cells were infected with different
multiplicity of infections (MOIs) of Hp (0, 1:1, 1:50 or 1:100;
cat. no. MA135390; Thermo Fisher Scientific, Inc.,).
Cell transfection
miR-140 mimic (5′-UGAGAACUGAAUUCCAUGGGUU-3′) and
mimic control (5′-UUCUCCGAACGUGUCACGUTT-3′) were obtained from
Sangon Biotech Co., Ltd. (Shanghai, China). For cell transfection,
BGC823 cells were seeded in 6-well plates (4×105 per
well). Then 100 nM miR-140 mimic, 100 nM mimic control, 100 nM
miR-140 mimic + 1 µg control-plasmid (cat. no. sc-108083; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA), or 100 nM miR-140 mimic
+ 1 µg PD-L1-plasmid (cat. no. sc-401140-ACT; Santa Cruz
Biotechnology, Inc.) was transfected into BGC823 cells using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Cells
were subjected to further experimentation 24 h after transfection.
Transfection efficiency was detected by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
MTT assay
BGC823 cells (5×103 cells/well) were
cultured in 96-well plates. Cells were transfected with miR-140
mimic, mimic control, miR-140 mimic + control - plasmid, or miR-140
mimic + PD-L1 - plasmid at 37°C for 24 h; following this, MTT
solution (20 µl) was added into each well at 24, 48 and 72 h, and
plates were then incubated at 37°C for another 4 h. Then DMSO (100
µl; Nanjing KeyGen Biotech. Co. Ltd., Nanjing, China) was added to
dissolve the formazan crystals. To determine cell viability, the
absorbance was measured at 450 nm.
RT-qPCR
Total RNA from tissues and cells was isolated using
a RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) following the
manufacturer's protocol. RT was performed to synthesize cDNA using
the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Reaction conditions for RT were: 50°C for 15 min and 85°C
for 2 min. SYBR Premix Ex Taq™ II (TliRNaseH Plus) kit (Takara Bio,
Inc., Otsu, Japan) was used to perform qPCR. Amplification
conditions for qPCR were as follows: 95°C for 5 min, followed by 40
cycles of denaturation at 95°C for 15 sec and annealing/elongation
at 60°C for 30 sec. U6 or GAPDH was used as internal control. The
primer sequences used were listed in Table I. Relative gene quantification was
assessed using the 2−ΔΔCq method (32).
 | Table I.Primer sequences used for
quantitative polymerase chain reaction analysis. |
Table I.
Primer sequences used for
quantitative polymerase chain reaction analysis.
| Primer | Direction | Sequence
(5′-3′) |
|---|
| PD-L1 | F |
GGCATTTGCTGAACGCAT |
|
| R |
CAATTAGTGCAGCCAGGT |
| IFN-γ | F |
CTAATTATTCGGTAACTGACTTGA |
|
| R |
ACAGTTCAGCCATCACTTGGA |
| TNF-α | F |
CCTCTTCTCATTCCTGCTC |
|
| R |
CTTCTCCTCCTTGTTGGG |
| miR-140 | F |
CGCGCCAGTGGTTTTACCCT |
|
| R |
CCAGTGCAGGGTCCGAGGTA |
| U6 | F |
GGAGCGAGATCCCTCCAAAAT |
|
| R |
GGCTGTTGTCATACTTCTCATGG |
| GAPDH | F |
CTTTGGTATCGTGGAAGGACTC |
|
| R |
GTAGAGGCAGGGATGATGTTCT |
Western blot analysis
Proteins were extracted from cells or tissues with
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China). A BCA protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.) was used to evaluate the protein
concentrations. Protein samples (25 µg/lane) were separated by 10%
SDS-PAGE, and then transferred to polyvinylidene difluoride
membranes. Membranes were blocked with 5% non-fat milk for 2 h at
room temperature, and then incubated with primary antibodies
against PD-L1 (cat. no. 13684; 1:1,000; Cell Signaling Technology,
Inc., Danvers, MA, USA), phosphorylated (p)-mammalian target of
rapamycin (mTOR; cat. no. 5536; 1:1,000; Cell Signaling Technology,
Inc.), p-ribosomal protein s6 kinase β-1 (S6K1; cat. no. ab60948;
1:1,000; Abcam, Cambridge, MA, USA) and β-actin (cat no. 4970;
1:2,000; Cell Signaling Technology, Inc.) at 4°C overnight.
Subsequently, the PVDF membranes were incubated with horseradish
peroxidase-conjugated anti-rabbit IgG secondary antibody (cat no.
7074; dilution ratio: 1:5,000; Cell Signaling Technology, Inc.) at
room temperature for 1.5 h. Protein blots were visualized using the
SuperSignal West Femto Maximum Sensitivity Substrate (Pierce;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol.
Luciferase reporter assay
TargetScan 7.1 (www.targetscan.org/vert_71) was used to predict the
target genes of miR-140, and PD-L1 was identified as a potential
target of miR-140. To confirm direct target binding, the wild type
(WT) and mutant (MUT) 3′-UTR of PD-L1 was cloned into a pmiR RB
Report™ dual luciferase reporter gene plasmid vector (Guangzhou
RiboBio Co., Ltd., Guangzhou, China). BGC823 cells were
co-transfected with 100 ng WT-PD-L1 or 100 ng MUT-PD-L1 and 50 nM
miR-140 mimic or its control (50 nM; miR-C) vector using
Lipofectamine® 3000, according to the manufacturer's
protocols. After 48 h, luciferase activity was measured by the
dual-luciferase assay system (Promega Corporation, Madison, WI,
USA) as per the manufacturer's protocol, and normalized to
Renilla luciferase activity.
Animal experiments
A total of 50 male C57BL/6 mice (8 weeks old; ~22 g)
were purchased from the Laboratory Animal Center of the Academy of
Military Medical Sciences (Beijing, China). Mice had free access to
food and water. All mice were fed ad libitum and maintained
under standard conditions at 22–30°C and a 12-h light/dark cycle.
The experimental protocol was approved by the Ethical Committee of
the First Affiliated Hospital of General Hospital of People's
Liberation Army, and all experiments were applied according to the
guidance of the Laboratory Animal Care (NIH publication no. 85Y23,
revised 1996) (33). For
generation of subcutaneous tumors, BGC823 cells (5×106)
were injected subcutaneously into the flanks of mice. When the
tumor size reached approximately 50 mm3, the mice were
divided into five groups: Control, miR-140 mimic (mice were
injected with 50 µl solution containing 7 nmol miR-140 mimic, 3 µl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), and 40 µl of serum-free medium Opti-MEM
(Invitrogen), mimic control [mice were injected with 50 µl solution
containing 7 nmol mimic control, 3 µl Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), and 40 µl of
serum-free medium Opti-MEM (Invitrogen; Thermo Fisher Scientific,
Inc.)], miR-140 mimic + control-plasmid [mice were injected with 50
µl solution containing 7 nmol miR-140 mimic, 1 µg control-plasmid,
3 µl Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), and 40 µl of serum-free medium Opti-MEM
(Invitrogen; Thermo Fisher Scientific, Inc.)], and miR-140 mimic +
PD-L1-plasmid [mice were injected with 50 µl solution containing 7
nmol miR-140 mimic, 1 µg PD-L1-plasmid, 3 µl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), and 40 µl of serum-free medium Opti-MEM
(Invitrogen; Thermo Fisher Scientific, Inc.)]. Injections were
performed once a day for 19 days. Tumors were measured every week.
Tumor volume was calculated using the following formula: Volume =
(length)x(width)2/2.
Ex vivo immune analysis
After 19 days, mice were sacrificed and tumors were
dissected and weighed. Single-cell suspensions were extracted from
the tumors. Briefly, the tumor tissues were cut into small pieces
(1–2 mm), washed three times with PBS buffer, and transferred to a
50 ml centrifuge tube. Depending on the amount of tissue, 5–6 times
of 0.25% trypsin solution was added and the tissues digested at
37°C for 20–40 min. After standing for 2–3 min, the suspension was
transferred to a new centrifuge tube. The suspension was filtered
twice with a 200/300 mesh nylon mesh. The filtered suspension was
centrifuged at 1,200 × g for 10 min at 4°C and the supernatant was
discarded. Then, 5 ml of PBS buffer was added, the cells gently
dispersed, and centrifuged again (1,200 × g for 10 min at 4°C).
CD8+ T cells, T regulatory cells (Tregs), and
myeloid-derived suppressor cells (MDSCs) in the tumor cell
suspension were identified by flow cytometry. Data were analyzed
using FlowJo software v7.6 (FlowJo LLC, Ashland, OR, USA).
ELISA
The serum level of IL-10 was measured using the
ELISA kit (cat no. ab108870; Abcam, Cambridge, MA, USA) in
accordance with the manufacturer's instructions.
Statistical analysis
All the experiments were performed three times.
Statistical analysis was performed with SPSS software version 20.0
(IBM Corp., Armonk, NY, USA). Data were displayed as the mean ±
standard deviation of three experimental repeats. Statistical
comparisons between groups were made by one-way analysis of
variance with Tukey's post-hoc test, or Student's t-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
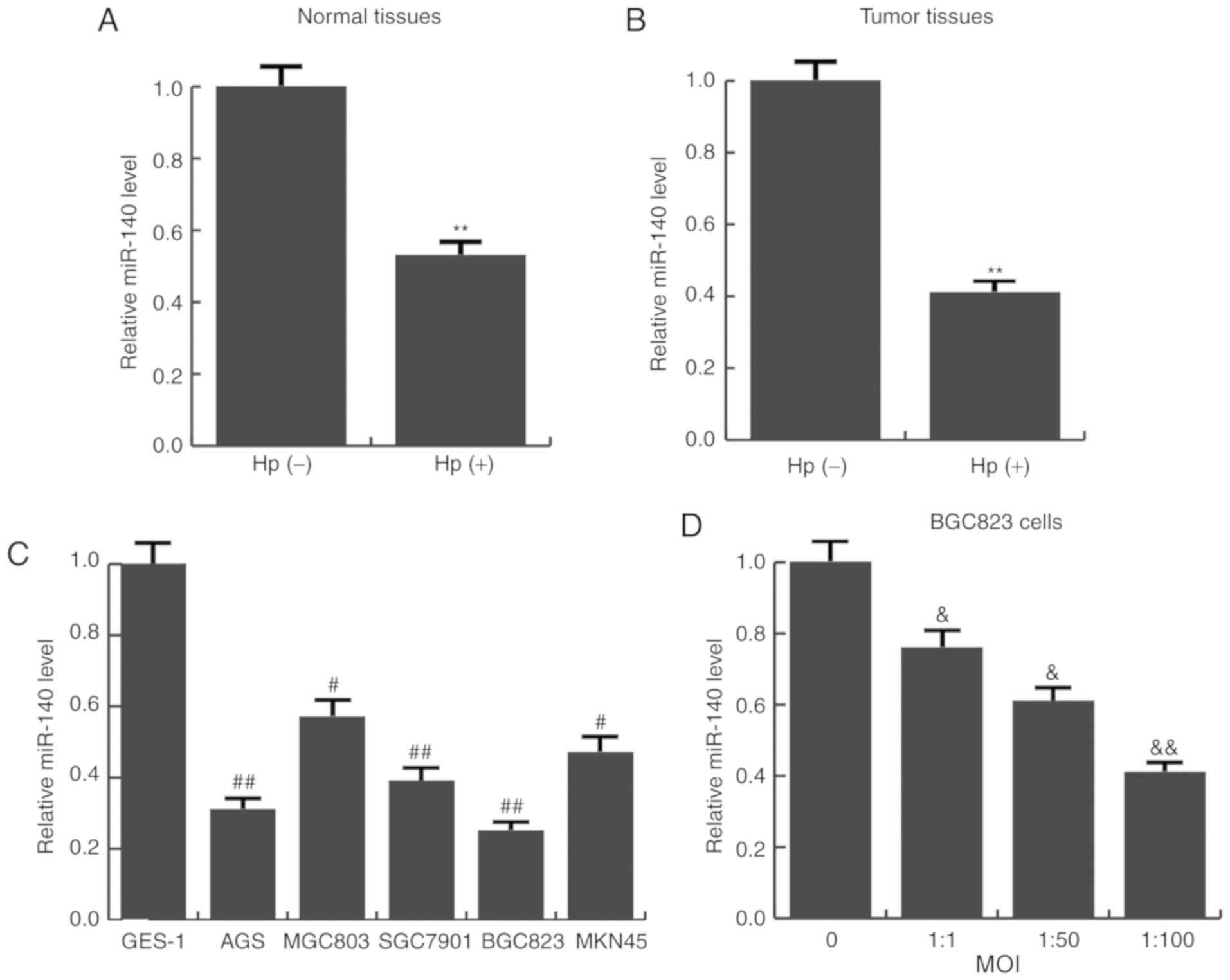
miR-140 is aberrantly downregulated in
Hp-positive tissues and cells
The expression levels of miR-140 in Hp-positive and
Hp-negative gastric cancer tissues, as well as Hp-positive and
Hp-negative normal tissues was detected using RT-qPCR. It was
revealed that the average expression level of miR-140 was
significantly lower in Hp-positive tumor and normal tissues
(Fig. 1A and B). Next, the
expression of miR-140 in various human gastric cancer cell lines
(AGS, MGC803, SGC7901, BGC823 and MKN45) was examined. Consistent
with the tissue results, compared with the immortalized
non-tumorigenic cell line GES-1, miR-140 was significantly
decreased in all 5 GC cell lines examined (Fig. 1C). BGC823 cells had the lowest
miR-140 expression, and were therefore selected for further
functional analysis. BGC823 cells were infected with different MOIs
of Hp (0, 1:1, 1:50, 1:100). The results indicated as the MOI
increased, miR-140 expression gradually reduced (Fig. 1D). These results indicated that
miR-140 expression was significantly downregulated in Hp-infected
conditions and may be associated with gastric cancer
progression.
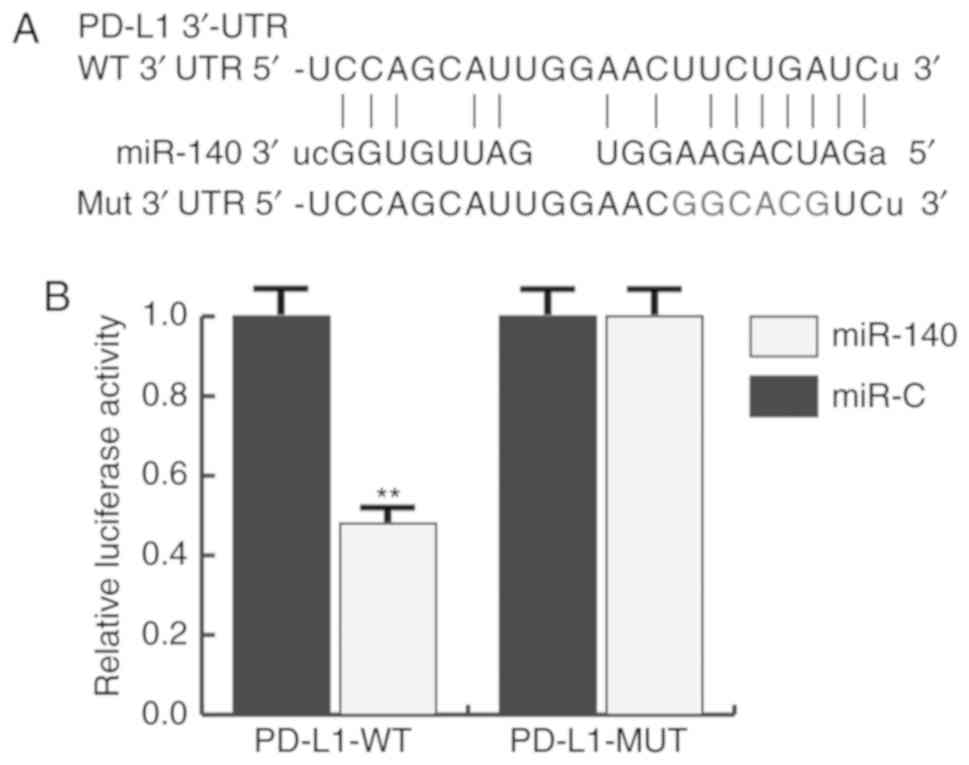
PD-L1 is directly targeted by miR-140
in gastric cancer
TargetScan was used to predict the potential targets
of miR-140, and PD-L1 was identified as a potential target of
miR-140 (Fig. 2A). To investigate
whether PD-L1 was a direct target of miR-140, a luciferase reporter
assay was constructed. The results indicated that miR-140
significantly reduced the luciferase activity of PD-L1-WT in BGC823
cells, but had no effect on the mutant form of PD-L1-MUT (Fig. 2B), indicating that PD-L1 was
directly targeted by miR-140 in gastric cancer cells.
PD-L1 is upregulated in Hp-positive
tumor cells and tissues
The expression level of PD-L1 in Hp-positive and
-negative gastric cancer tissues and cells was subsequently
determined. It was found that the mRNA expression of PD-L1 was
significantly higher in Hp-positive normal (Fig. 3A) and tumor tissues (Fig. 3B). PD-L1 expression was also
detected in five human gastric cancer cell lines, and the results
revealed that compared with GES-1 cells, PD-L1 mRNA (Fig. 3C) and protein (Fig. 3D) expression was significantly
increased in all the gastric cancer cells. In addition, as cells
were transfected with a higher MOI of Hp, PD-L1 mRNA (Fig. 3E) and protein (Fig. 3F) expression was gradually
increased.
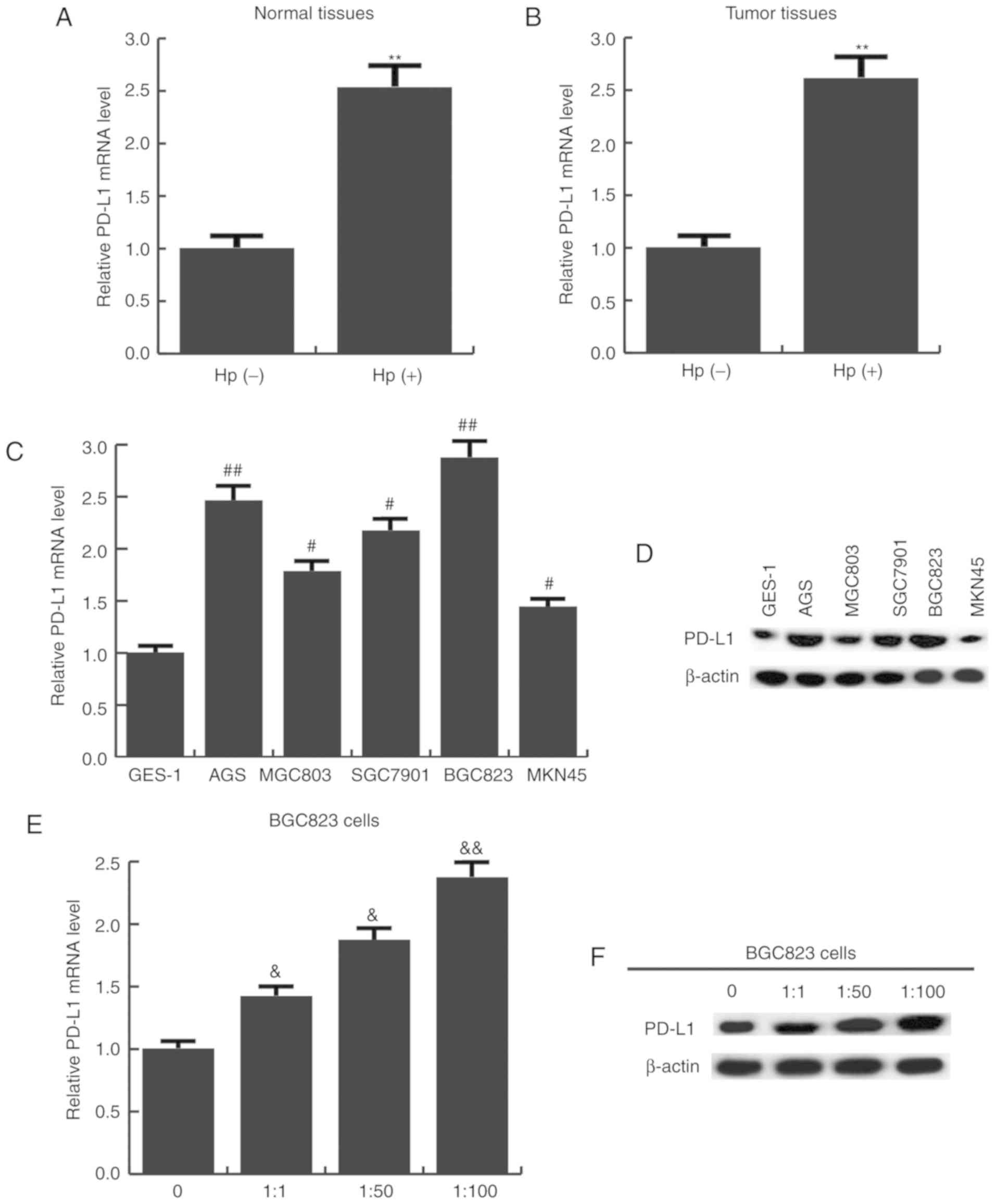 | Figure 3.PD-L1 expression in upregulated in
Hp-positive gastric cancer tissues and cell lines. (A) PD-L1 mRNA
expression in Hp-positive and negative normal tissues, as well as
(B) Hp-positive and negative gastric cancer tissues. **P<0.01 vs
Hp-negative group. (C) PD-L1 mRNA and (D) protein expression in
give gastric cancer cell lines and immortalized GES-1 cells.
#P<0.05, ##P<0.01 vs. GES-1 cells. (E)
PD-L1 mRNA and (F) protein expression in BGC823 cells infected with
different MOIs of Hp (0, 1:1, 1:50, 1:100).
&P<0.05, &&P<0.01 vs. 0 MOI
group. Data are presented as the mean ± standard deviation of three
independent experiments. miR, microRNA; PD-L1, programmed cell
death ligand 1; Hp, Helicobacter pylori; MOI, multiplicity
of infection. |
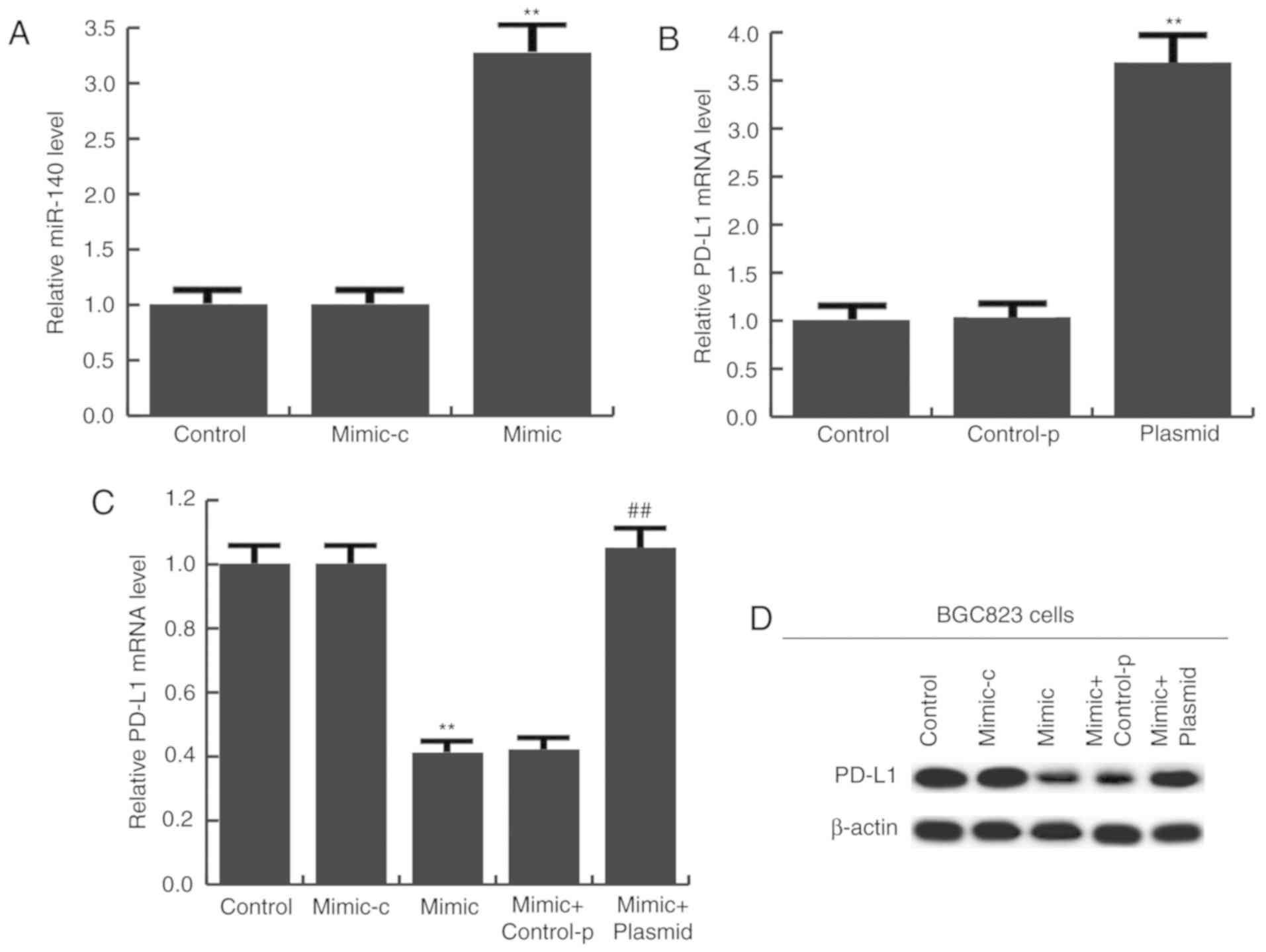
miR-140 has a tumor-suppressive role
in gastric cancer
To determine the potential role of miR-140 in
gastric cancer, BGC823 cells were transfected with miR-140 mimic,
mimic-c, miR-140 mimic + control-plasmid, or miR-140 mimic +
PD-L1-plasmid for 24 h. Transfection efficiency was initially
measured using RT-qPCR. The results confirmed that compared with
the control group, miR-140 expression was significantly increased
in the miR-140 mimic group (Fig.
4A), and PD-L1-plasmid significantly enhanced PD-L1 expression
(Fig. 4B). mRNA (Fig. 4C) and protein (Fig. 4D) expression of PD-L1 was detected
and the results indicated that miR-140 mimic notably reduced PD-L1
expression, and this reduction was inhibited by PD-L1-plasmid
co-transfection (Fig. 4C and
D).
Next, the proliferation of BGC823 cells was detected
by a MTT assay. It was demonstrated that compared with the control
group, miR-140 mimic significantly inhibited BGC823 cell
proliferation, and this inhibition was prevented by PD-L1-plasmid
co-transfection (Fig. 5A).
Consistent with the data obtained from in vitro experiments,
results from the in vivo experiments showed that tumor
volume (Fig. 5B) and mean tumor
weight (Fig. 5C) was significantly
reduced in the miR-140 mimic group, compared with the control
group, and PD-L1-plasmid co-transfection significantly increased
these parameters.
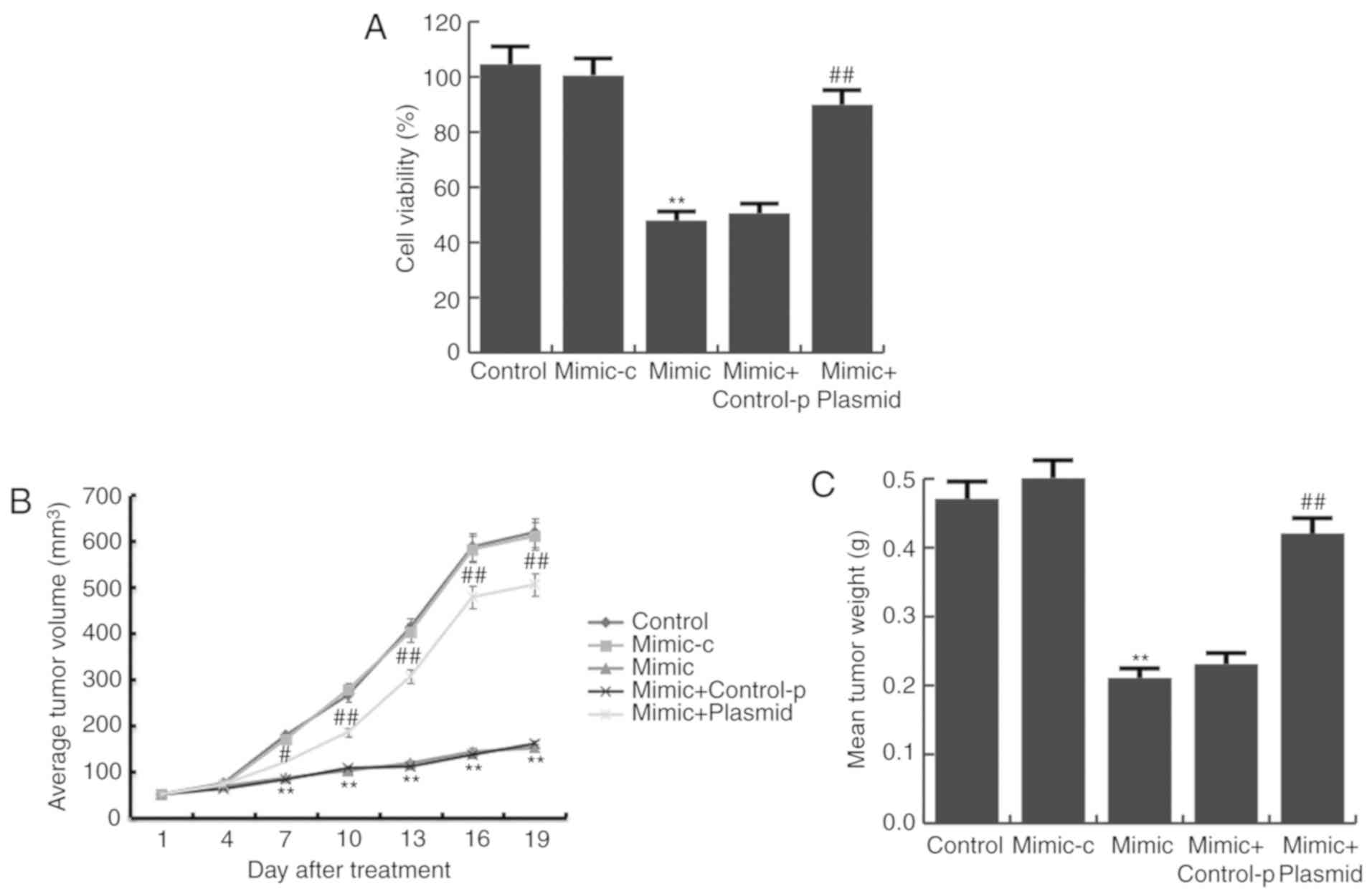 | Figure 5.miR-140 mimic reduces gastric cancer
cell viability. (A) Following transfection with miR-140 mimic,
mimic-c, miR-140 mimic + control-p, or miR-140 mimic +
PD-L1-plasmid for 24 h, BGC823 cell viability was assessed with MTT
assays. (B) Tumor size at different time points following the
intratumoral injection of miR-140 mimic, mimic control, miR-140
mimic + control-p, or miR-140 mimic + PD-L1-plasmid. (C) Mean
weights of the xenograft tumors. Data are presented as the mean ±
standard deviation of three independent experiments. **P<0.01
vs. control; #P<0.05, ##P<0.01 vs.
mimic. miR, microRNA; PD-L1, programmed cell death ligand 1;
mimic-c, miR-140 mimic control; control-p, control plasmid. |
miR-140 enhances antitumor immunity in
gastric cancer
Studies have indicated that PD-L1/PD-1 signaling
inhibition may prevent immune suppression and enhance antitumor
response (26). Thus, it was
investigated whether miR-140 affected the immune response in
gastric cancer. As presented in Fig.
6A, compared with the control group, the CD8+ T cell
population in the miR-140 mimic group was significantly increased,
whereas the MDSC and Tregs cell populations were significantly
reduced. These alterations were prevented by PD-L1 overexpression.
To further characterize the immune responses induced by miR-140,
the mRNA expression of IFN-γ and TNF-α was measured in tumor
tissues, as well as serum IL-10 expression. As expected, miR-140
mimic significantly increased IFN-γ (Fig. 6B) and TNF-α (Fig. 6C) mRNA expression and decreased
serum IL-10 expression (Fig. 6D),
compared with the control group. These alterations were eliminated
by PD-L1 overexpression.
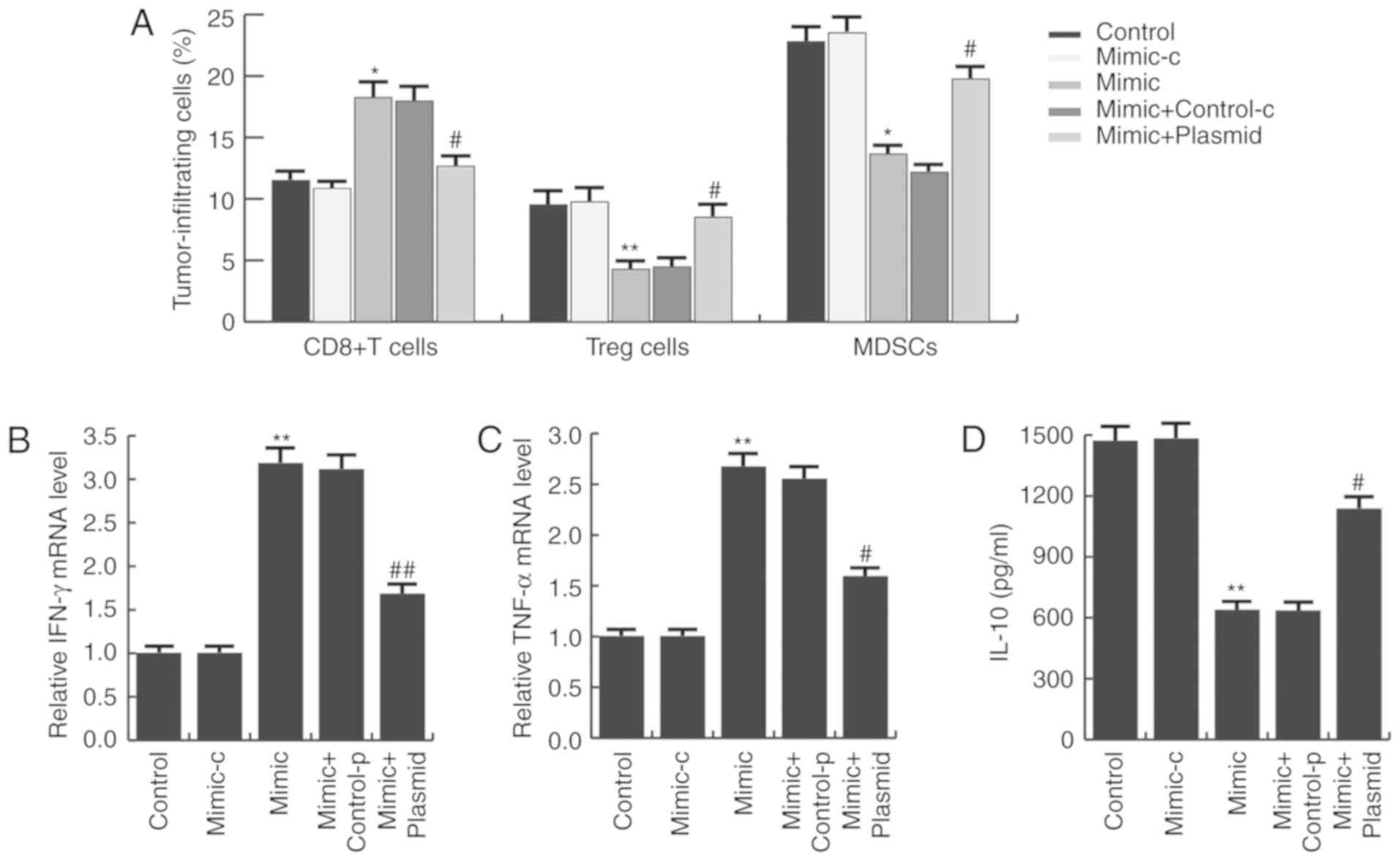 | Figure 6.miR-140 enhances antitumor immunity
in gastric cancer. (A) Ex vivo analysis of the
CD8+ T cell, Treg cell and MDSC populations in the
tumors of each group. Cells were isolated and analyzed by flow
cytometry. (B) Relative mRNA expression of IFN-γ and (C) TNF-α. (D)
Serum IL-10 expression in each group was analyzed by ELISA. Data
are presented as the mean ± standard deviation of three independent
experiments. *P<0.05, **P<0.01 vs. control.
#P<0.05, ##P<0.01 vs. mimic. miR,
microRNA; Treg, T regulatory; MDSC, myeloid-derived suppressor
cell; IFN-γ, interferon γ; TNF-α, tumor necrosis factor-α; IL-10,
interleukin-10; mimic-c, miR-140 mimic control; control-p, control
plasmid. |
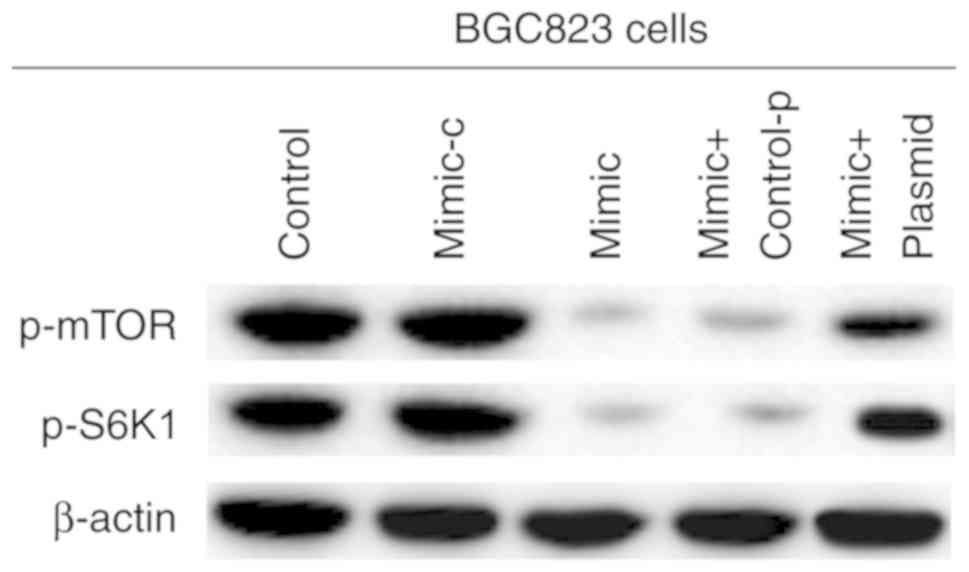
miR-140 suppresses mTOR signaling in
gastric cancer
Previous studies have reported that cell-intrinsic
PD-L1/PD-1 signaling promotes tumor growth by activating downstream
mTOR signaling (34). Therefore,
the effects of miR-140 on mTOR signaling in gastric cancer were
examined. As presented in Fig. 7,
compared with the control group, p-mTOR and p-S6K1 expression was
significantly decreased, and PD-L1 overexpression prevented this
decrease. These data indicated that miR-140 suppressed mTOR
signaling in gastric cancer.
Discussion
To the best of our knowledge, the present study was
the first to identify that miR-140 directly regulated PD-L1
expression in gastric cancer. Upregulation of miR-140 inhibited
gastric cancer growth by targeting PD-L1 in vitro and in
vivo, and was associated with an enhanced antitumor immunity.
Therefore, miR-140 may represent a novel and promising therapeutic
target for gastric cancer treatment through immune checkpoint
inhibition.
In recent years, miRNA-based immunotherapies have
emerged as novel treatments for malignant tumors (35). Tumor cells may escape
T-cell-mediated cellular cytotoxicity by exploiting the PD-1/PD-L1
pathway (26). PD-1 and PD-L1,
once bound, transmit negative regulatory signals to T cells to
induce a resting state, which decreases lymph node CD8+
T cell proliferation and cancer cell recognition. In addition, T
cell apoptosis is induced to effectively reduce the body's immune
response, leading to the unrestrained growth of cancer cells.
Therefore, targeting the PD-1/PD-L1 pathway is a promising strategy
for the treatment of malignant tumors. Evidence has indicated that
miRNAs regulate PD-L1 expression in cancer (36). For example, downregulation of
miR-140-3p is closely associated with overexpression of PDL-1 in
many cancers, including breast and lung cancer (37). In malignant pleural mesothelioma,
tumor suppressor gene miRNAs (miR-15b, miR-16, miR-193a-3p,
miR-195, and miR-200c) downregulate the expression of PD-L1
(38). In pancreatic cancer,
miR-142-5p can inhibit PD-L1 expression and enhance antitumor
immunity (39). The miR-25-93-106b
cluster has been reported to regulate tumor metastasis and immune
evasion by regulating stromal cell-derived factor 1 and PD-L1
expression (40). miR-424 reduces
ovarian cancer chemoresistance by inhibiting PD-L1 expression
(41). miR-140, a well-studied
miRNA in cancer, has been identified as a tumor suppressor in
various cancer types, including gastric cancer (17–21).
In addition, miR-140 has been demonstrated to suppress osteosarcoma
tumor growth by enhancing antitumor immune response, by regulating
PD-L1 (30). However, the role and
mechanism of miR-140 in gastric cancer, particularly in
Hp-associated gastric cancer, remained largely unclear. Therefore,
the present study was conducted.
First, the present study detected the expression of
miR-140 in Hp-positive and -negative gastric cancer tissues,
Hp-positive and -negative normal tissues, and five human gastric
cancer cell lines. The results indicated that miR-140 expression
was significantly downregulated in Hp-positive conditions and was
associated with gastric cancer progression. It was then confirmed
that PD-L1 was a direct target of miR-140 in gastric cancer cells,
and was upregulated in Hp-positive tumor cells and tissues. Further
analysis indicated that miR-140 suppressed gastric cancer growth
in vitro and in vivo, and enhanced antitumor immunity
in gastric cancer. Previous studies have reported that
cell-intrinsic PD-L1/PD-1 signaling promotes tumor growth by
activating downstream mTOR signaling (34). Consistent with the results of
previous research (30,34), the present study demonstrated that
miR-140 suppressed mTOR signaling in gastric cancer.
Taken together, the current study revealed that
miR-140 was significantly reduced in Hp-positive gastric cancer,
and exerted a tumor suppressive effect by targeting immune
checkpoint molecule PD-L1. Thus, miR-140 may be a novel and
promising therapeutic target for the treatment of gastric cancer,
particularly in Hp-positive gastric cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fund of
Development of Health Service of Beijing (grant no.
2011-5002-02).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MZ performed study design, data collection,
statistical analysis, data interpretation, manuscript preparation
and literature search. QL performed study design, statistical
analysis and data interpretation. WL performed data collection. HZ
performed study design, data collection and statistical analysis.
XZ performed data collection and statistical analysis. JL performed
study design, data interpretation and manuscript preparation.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of the First Affiliated Hospital of General Hospital of
People's Liberation Army, and written informed consent was obtained
from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Muhammad JS, Sugiyama T and Zaidi SF:
Gastric pathophysiological ins and outs of helicobacter
pylori: A review. J Pak Med Assoc. 63:1528–1533.
2013.PubMed/NCBI
|
|
2
|
Ali Z, Deng Y and Ma C: Progress of
research in gastric cancer. J Nanosci Nanotechnol. 12:8241–8248.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lordick F, Allum W, Carneiro F, Mitry E,
Tabernero J, Tan P, Van Cutsem E, van de Velde C and Cervantes A:
Unmet needs and challenges in gastric cancer: The way forward.
Cancer Treat Rev. 40:692–700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Graham DY: History of Helicobacter
pylori, duodenal ulcer, gastric ulcer and gastric cancer. World
J Gastroenterol. 20:5191–5204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Falt P, Hanousek M, Kundrátová E and Urban
O: Precancerous conditions and lesions of the stomach. Klin Onkol.
26 (Suppl):S22–S28. 2013.(In Czech). View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiao D, Zhang H, He L, Peng X, Wang Y, Xue
G, Su P and Zhang J: High natural variability bacteria
identification and typing: Helicobacter pylori analysis
based on peptide mass fingerprinting. J Proteomics. 98:112–122.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katada T, Ishiguro H, Kuwabara Y, Kimura
M, Mitui A, Mori Y, Ogawa R, Harata K and Fujii Y: MicroRNA
expression profile in undifferentiated gastric cancer. Int J Oncol.
34:537–542. 2009.PubMed/NCBI
|
|
8
|
Bou Kheir T, Futoma-Kazmierczak E,
Jacobsen A, Krogh A, Bardram L, Hother C, Grønbæk K, Federspiel B,
Lund AH and Friis-Hansen L: miR-449 inhibits cell proliferation and
is down-regulated in gastric cancer. Mol Cancer. 10:292011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chiang Y, Zhou X, Wang Z, Song Y, Liu Z,
Zhao F, Zhu J and Xu H: Expression levels of microRNA-192 and −215
in gastric carcinoma. Pathol Oncol Res. 18:585–591. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H, Cheng Y, Jia C, Yu S, Xiao Y and
Chen J: MicroRNA-29s could target AKT2 to inhibit gastric cancer
cells invasion ability. Med Oncol. 32:3422015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang R, Li F, Wang W, Wang X, Li S and
Liu J: The effect of antisense inhibitor of miRNA 106b~25 on the
proliferation, invasion, migration, and apoptosis of gastric cancer
cell. Tumour Biol. 37:10507–10515. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiang XJ, Deng J, Liu YW, Wan LY, Feng M,
Chen J and Xiong JP: MiR-1271 Inhibits Cell Proliferation, Invasion
and EMT in Gastric Cancer by Targeting FOXQ1. Cell Physiol Biochem.
36:1382–1394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou X, Xia Y, Li L and Zhang G: MiR-101
inhibits cell growth and tumorigenesis of Helicobacter
pylori related gastric cancer by repression of SOCS2. Cancer
Biol Ther. 16:160–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou X, Xu G, Yin C, Jin W and Zhang G:
Down-regulation of miR-203 induced by Helicobacter pylori
infection promotes the proliferation and invasion of gastric cancer
by targeting CASK. Oncotarget. 5:11631–11640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fassan M, Saraggi D, Balsamo L, Cascione
L, Castoro C, Coati I, De Bernard M, Farinati F, Guzzardo V, Valeri
N, et al: Let-7c down-regulation in Helicobacter
pylori-related gastric carcinogenesis. Oncotarget. 7:4915–4924.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Q, Wang N, Wei H, Li C, Wu J and Yang
G: miR-24-3p Regulates Progression of Gastric Mucosal Lesions and
Suppresses Proliferation and Invasiveness of N87 Via Peroxiredoxin
6. Dig Dis Sci. 61:3486–3497. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan X, Zhu Z, Xu S, Yang LN, Liao XH,
Zheng M, Yang D, Wang J, Chen D, Wang L, et al: MicroRNA-140-5p
inhibits hepatocellular carcinoma by directly targeting the unique
isomerase Pin1 to block multiple cancer-driving pathways. Sci Rep.
7:459152017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu L, Lu Y, Han X, Zhao W, Li J, Mao J,
Wang B, Shen J, Fan S, Wang L, et al: microRNA −140-5p inhibits
colorectal cancer invasion and metastasis by targeting ADAMTS5 and
IGFBP5. Stem Cell Res Ther. 7:1802016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Su Y, Xiong J, Hu J, Wei X, Zhang X and
Rao L: MicroRNA-140-5p targets insulin like growth factor 2 mRNA
binding protein 1 (IGF2BP1) to suppress cervical cancer growth and
metastasis. Oncotarget. 7:68397–68411. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y,
Zhang T, Khaliq J and Li Y: miR-140-5p suppresses the
proliferation, migration and invasion of gastric cancer by
regulating YES1. Mol Cancer. 16:1392017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zou J and Xu Y: MicroRNA-140 Inhibits Cell
Proliferation in Gastric Cancer Cell Line HGC-27 by Suppressing
SOX4. Med Sci Monit. 22:2243–2252. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song Z, Wu Y, Yang J, Yang D and Fang X:
Progress in the treatment of advanced gastric cancer. Tumour Biol.
39:10104283177146262017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang K, Peng Z, Huang X, Qiao Z, Wang X,
Wang N, Xi H, Cui J, Gao Y, Huang X, et al: Phase II Trial of
Adjuvant Immunotherapy with Autologous Tumor-derived Gp96
Vaccination in Patients with Gastric Cancer. J Cancer. 8:1826–1832.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mimura K, Teh JL, Okayama H, Shiraishi K,
Kua LF, Koh V, Smoot DT, Ashktorab H, Oike T, Suzuki Y, et al:
PD-L1 expression is mainly regulated by interferon gamma associated
with JAK-STAT pathway in gastric cancer. Cancer Sci. 109:43–53.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dos Santos Fernandes G, da Motta Girardi
D, Dib Batista Bugiato Faria L, Giacomini Bernardes JP and de
Almeida Coudry R: Impressive response to immunotherapy in a
metastatic gastric cancer patient: Could somatic copy number
alterations help patient selection? J Immunother Cancer. 5:842017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goodman A, Patel SP and Kurzrock R:
PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat Rev
Clin Oncol. 14:203–220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Salmaninejad A, Valilou SF, Shabgah AG,
Aslani S, Alimardani M, Pasdar A and Sahebkar A: PD-1/PD-L1
pathway: Basic biology and role in cancer immunotherapy. J Cell
Physiol. Feb 19–2019.(Epub ahead of print). View Article : Google Scholar
|
|
28
|
Liu Y, Wu L, Tong R, Yang F, Yin L, Li M,
You L, Xue J and Lu Y: PD-1/PD-L1 Inhibitors in Cervical Cancer.
Front Pharmacol. 10:652019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pawłowska A, Suszczyk D, Okła K,
Barczyński B, Kotarski J and Wertel I: Immunotherapies based on
PD-1/PD-L1 pathway inhibitors in ovarian cancer treatment. Clin Exp
Immunol. 195:334–344. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ji X, Wang E and Tian F: MicroRNA-140
suppresses osteosarcoma tumor growth by enhancing anti-tumor immune
response and blocking mTOR signaling. Biochem Biophys Res Commun.
495:1342–1348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McNicholl AG, Ducons J, Barrio J, Bujanda
L, Forné-Bardera M, Aparcero R, Ponce J, Rivera R, Dedeu-Cuso JM,
Garcia-Iglesias P, et al: Helicobacter pylori Study Group of
the Asociación Española de Gastroenterología (AEG): Accuracy of the
Ultra-Rapid Urease Test for diagnosis of Helicobacter pylori
infection. Gastroenterol Hepatol. 40:651–657. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bayne K: Revised Guide for the Care and
Use of Laboratory Animals available. American Physiological
Society. Physiologist. 39(199): 208–211. 1996.
|
|
34
|
Kleffel S, Posch C, Barthel SR, Mueller H,
Schlapbach C, Guenova E, Elco CP, Lee N, Juneja VR, Zhan Q, et al:
Melanoma cell-intrinsic PD-1 receptor functions promote tumor
growth. Cell. 162:1242–1256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cortez MA, Anfossi S, Ramapriyan R, Menon
H, Atalar SC, Aliru M, Welsh J and Calin GA: Role of miRNAs in
immune responses and immunotherapy in cancer. Genes Chromosomes
Cancer. 58:244–253. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Q, Lin W, Tang X, Li S, Guo L, Lin Y
and Kwok HF: The Roles of microRNAs in Regulating the Expression of
PD-1/PD-L1 Immune Checkpoint. Int J Mol Sci. 18:182017. View Article : Google Scholar
|
|
37
|
Kapodistrias N, Bobori C and
Theocharopoulou G: MiR-140-3p Downregulation in Association with
PDL-1 Overexpression in Many Cancers: A Review from the Literature
Using Predictive Bioinformatics Tools. Adv Exp Med Biol.
988:225–233. 2017.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kao SC, Cheng YY, Williams M, Kirschner
MB, Madore J, Lum T, Sarun KH, Linton A, McCaughan B, Klebe S, et
al: Tumor Suppressor microRNAs Contribute to the Regulation of
PD-L1 Expression in Malignant Pleural Mesothelioma. J Thorac Oncol.
12:1421–1433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jia L, Xi Q, Wang H, Zhang Z, Liu H, Cheng
Y, Guo X, Zhang J, Zhang Q, Zhang L, et al: miR-142-5p regulates
tumor cell PD-L1 expression and enhances anti-tumor immunity.
Biochem Biophys Res Commun. 488:425–431. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cioffi M, Trabulo SM, Vallespinos M, Raj
D, Kheir TB, Lin ML, Begum J, Baker AM, Amgheib A, Saif J, et al:
The miR-25-93-106b cluster regulates tumor metastasis and immune
evasion via modulation of CXCL12 and PD-L1. Oncotarget.
8:21609–21625. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang
T, Song W, Chen Y, OuYang J, Chen J, et al: miR-424(322) reverses
chemoresistance via T-cell immune response activation by blocking
the PD-L1 immune checkpoint. Nat Commun. 7:114062016. View Article : Google Scholar : PubMed/NCBI
|