Introduction
Lung cancer, one of the most prevalent human
malignancies, is the leading cause of cancer-associated mortality
globally (1). In total, ~1.82
million novel lung cancer cases and 1.59 million mortalities occur
per year globally, according to the data from GLOBOCAN 2012
(2). According to histological
analysis, lung cancer may be divided into two predominant
categories, namely small cell lung cancer and non-small cell lung
cancer (NSCLC) (3). NSCLC accounts
for ~85% of all lung cancer cases (4). NSCLC includes three major subtypes,
namely squamous-cell carcinoma, adenocarcinoma and large-cell
carcinoma (4). Currently, surgery,
radiotherapy and chemotherapy are the primary therapeutic
techniques for patients with NSCLC (5). Despite advances in diagnosis and
therapy in previous years, their clinical efficiency in patients
with NSCLC remains unsatisfactory with a 5-year survival rate of
only 15% (6,7). The late disease presentation, tumor
heterogeneities within histological subtypes, cancer metastasis,
high recurrence rates and the poor understanding of NSCLC
pathogenesis are responsible for the poor prognosis of patients
with NSCLC (8,9). Therefore, the elucidation of the
mechanisms underlying NSCLC genesis and development will help
identify promising diagnostic indicators and therapeutic targets
for the treatment of the malignancy.
MicroRNAs (miRNAs/miRs), which are 18–22 nucleotides
in length, are an abundant class of endogenous, noncoding short RNA
molecules (10). miRNAs may
negatively regulate gene expression through the recognition and
preferential binding to the 3′-untranslated regions (3′-UTRs) of
their targets, which thus result in translation suppression and/or
mRNA cleavage (11). miRNAs may
modulate over one half of all gene expression of human proteins and
may participate in the modulation of numerous cellular biological
behaviors, including cell proliferation, survival, apoptosis,
differentiation, metabolism and metastasis (12). Numerous deregulated miRNAs have
been identified in various disorders, including human malignancies
(13–15). The aberrant expression of miRNAs is
strongly associated with the tumorigenesis and tumor development
(16–18). miRNAs may function as
tumor-suppressing or tumor-promoting miRNAs in different types of
human cancer, depending on the biological functions of their target
genes (19). As a consequence,
research on cancer-associated miRNAs in NSCLC may benefit the
identification of effective therapeutic methods for patients with
NSCLC.
miR-208a is reportedly expressed abnormally in
numerous human malignancies (20–22).
The expression level and potential functions of miR-208a in NSCLC,
however, remain unknown. Thus, the aims of the present study were
to measure the expression level, clinical significance and detailed
functions of miR-208a in NSCLC and the associated regulatory
mechanism.
Materials and methods
Tissue specimens and ethical
statement
A total of 52 primary NSCLC tissues (21
adenocarcinoma, 26 squamous cell carcinoma and 5 large cell
neuroendocrine carcinoma) and adjacent non-cancerous tissues were
obtained from Qilu Hospital of Shandong University (Shandong,
China) between March 2014 and June 2016. All patients were treated
with surgical resection. The present study included 28 males and 24
females, with an age range of 47–73 years (median age, 62 years).
All participants had not undergone preoperative chemotherapy or
radiotherapy. Patients who had been treated with preoperative
chemotherapy or radiotherapy were excluded from this research.
Tumor-Node-Metastasis (TNM) stage (23) was used for staging. Fresh surgical
tissue samples were immediately frozen in liquid nitrogen and
stored at −80°C until used. The Ethics Committee of the Qilu
Hospital of Shandong University ethically approved the present
study. In addition, all patients provided written informed consent
prior to the surgery.
Cell lines and culture conditions
In total, five NSCLC cell lines, including SK-MES-1,
NCI-H522, NCI-H460, SPC-A1 and A-549, in addition to a
non-tumorigenic bronchial epithelium BEAS-2B cell line were ordered
from the Shanghai Institute of Biochemistry and Cell Biology
(Shanghai, China).
LHC-9 medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) was utilized for the culture of the
BEAS-2B cell line. Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS was used
to culture all the aforementioned NSCLC cell lines. All cells were
grown at 37°C in a humidified condition supplied with 5%
CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) was applied for the isolation of total RNA from
tissue specimens or cultured cells as aforementioned. For the
quantification of miR-208a, complementary DNA (cDNA) was prepared
from total RNA with the TaqMan MicroRNA Reverse Transcription kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The temperature protocol for reverse
transcription was: 16°C for 30 min, 42°C for 30 min and 85°C for 5
min. Subsequently, cDNA was amplified by qPCR using the TaqMan
MicroRNA PCR kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The cycling
conditions for qPCR were as follows: 50°C for 2 min, 95°C for 10
min; 40 cycles of denaturation at 95°C for 15 sec; and
annealing/extension at 60°C for 60 sec. For the detection of Src
kinase signaling inhibitor 1 (SRCIN1) mRNA expression, reverse
transcription was performed using the PrimeScript RT Reagent kit
followed by qPCR using the SYBR Premix Ex Taq™ kit (both from
Takara Biotechnology Co., Ltd., Dalian, China) according to the
manufacturer's protocols. The temperature protocol for reverse
transcription was: 37°C for 15 min and 85°C for 5 sec. The
thermocycling conditions for qPCR were as follows: 95°C for 10 min,
followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The
expression levels of miR-208a and SRCIN1 mRNA were normalized to
that of U6 snRNA and GAPDH, respectively. RT-qPCR was performed
three times using an ABI7500 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The 2−ΔΔCq
method (23) was used to analyze
the relative miR-208a and SRCIN1 mRNA expression. The primers were
designed as follows: miR-208a,
5′-GTCATCTAGAAAGCTTGATGCAGGAAAGAGCTTTGG-3′ (forward) and
5′-TGACAGATCTCAGCTGACATCCTCTAGGCTGGGGTT-3′ (reverse); U6,
5′-GTGCTCGCTTCGGCAGCACATAT-3′ (forward) and
5′-AAAATATGGAACGCTTCACGAA-3′ (reverse); SRCIN1,
5′-GAACGGCTGCGCTATCTCAA-3′ (forward) and
5′-GGATCTTCTCCACCGATTTCTCC-3′ (reverse); and GAPDH,
5′-GCTGGCGCTGAGTACGTCGTGGAGT-3′ (forward) and
5′-CACAGTCTTCTGGGTGGCAGTGATGG-3′ (reverse).
RNA oligonucleotide and cell
transfection
The miR-208a inhibitor used to knockdown endogenous
miR-208a expression was chemically synthesized by Shanghai
GenePharma Co. Ltd. (Shanghai, China). The negative control (NC)
miRNA inhibitor functioned as the control for miR-208a. Small
interfering (si)RNA against the expression of SRCIN1 and its NC
siRNA were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou,
China). The SRCIN1 siRNA sequence was 5′-AAGCTGTGTCTGTTGAGGCTG-3′
and the NC siRNA sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. For cell
transfection, H460 and A549 cells were plated into 6-well plates at
a density of 6×105 cells per well. The transfection
experiments were mediated using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) based on the manufacturer's
protocol. The quantity of siRNA transfected was 100 pmol. After 8 h
incubation at 37°C, transfected cells were washed with PBS (Gibco;
Thermo Fisher Scientific, Inc.) and the culture medium was replaced
with fresh DMEM containing 10% FBS.
Cell Counting Kit-8 (CCK-8) assay
A total of 24 h after transfection, the transfected
H460 and A549 cells were harvested and subsequently inoculated into
the 96-well plates at an initial density of 3,000 cells/well. The
cells were then incubated at 37°C in a humidified incubator with 5%
CO2 for 0, 24, 48 and 72 h. A CCK-8 assay was applied to
measure cellular proliferation at each time point. The cells were
then incubated with a total of 10 µl CCK-8 assay solution (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) at 37°C for
additional 2 h. The optical density at 450 nm wavelength was read
with a microplate reader (BioTek Instruments, Inc., Winooski, VT,
USA).
Matrigel invasion assay
A Matrigel invasion assay was adopted to evaluate
the cellular invasive ability using a Matrigel pre-coated 24-well
Boyden chamber (BD Biosciences, Franklin Lakes, NJ, USA). At 48 h
post-transfection, the H460 and A549 cells were harvested, washed
with phosphate buffer solution and suspended in FBS-free DMEM. A
total of 5×104 cells were seeded on the upper chambers
and allowed to invade the reverse side of the chamber under
chemoattractant conditions with 10% FBS medium in the lower
chambers. After 24 h of culture at 37°C, the non-invaded cells that
remained on the upper surface of the upper chambers were removed
gently using a cotton swab. The invaded cells were fixed using 100%
methanol at room temperature for 30 min and stained with 0.1%
crystal violet at room temperature for 30 min. Finally, the
invasive ability was assessed by counting the number of invaded
cells in five randomly selected visual fields under an IX71
inverted light microscope (×200 magnification; Olympus Corporation,
Tokyo, Japan).
miR-208a target prediction and
luciferase reporter assay
The following online miRNA target prediction
algorithms, TargetScan (24)
(http://www.targetscan.org/index.html)
and miRanda (25) (http://www.microrna.org/microrna/), were utilized
to identify the putative target genes of miR-208a. Luciferase
plasmids were chemically synthesized by Shanghai GenePharma Co.,
Ltd. The 3′-UTR segments of the SRCIN1 gene containing the
wild-type or mutant miR-208a binding sites were inserted into
pMIR-GLO™ luciferase vector to generate the wild-type luciferase
plasmid (pMIR-SRCIN1-Wt-3′-UTR) or mutant luciferase plasmid
(pMIR-SRCIN1-Mut-3′-UTR). H460 and A549 cells were inoculated into
the 24-well plates at a density of 60 to 70% confluence. miR-208a
inhibitor or NC inhibitor were cotransfected with
pMIR-SRCIN1-Wt-3′-UTR or pMIR-SRCIN1-Mut-3′-UTR into the cells
using Lipofectamine™ 2000 reagent. Subsequent to incubation at 37°C
for 48 h, transfected cells were harvested and subjected to the
quantification of luciferase activity using a dual-luciferase
reporter assay system (Promega Corporation, Madison, WI, USA).
Luciferase activity was normalized relative to that of the Renilla
luciferase activity.
Western blot analysis
Total protein was isolated from tissue samples or
cultured H460 and A549 cells using radioimmunoprecipitation assay
lysis buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
supplemented with protease inhibitors (Roche Diagnostics, Basel,
Switzerland). A bicinchoninic acid kit (Beyotime Institute of
Biotechnology, Shanghai, China) was adopted to measure the
concentration of the total protein extracts, according to the
manufacturer's protocol. Equal amounts of proteins (30 µg) were
loaded, separated by 10% SDS-PAGE and then transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). Following blocking in 5% non-fat milk at room temperature for
2 h, the membranes were incubated overnight at 4°C with primary
antibodies against SRCIN1 (cat. no. 3757; Cell Signaling
Technology, Inc., Danvers, MA, USA), extracellular regulated kinase
(ERK; cat. no. sc-514302; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA), phosphorylated (p-)ERK (cat. no. sc-7383; Santa Cruz
Biotechnology, Inc.) or GAPDH (cat. no. sc-51907; Santa Cruz
Biotechnology, Inc.). All primary antibodies were used at a
dilution of 1:1,000. Subsequently, the membranes were rinsed with
Tris-buffered saline containing 0.1% Tween-20 (TBST) for three
times and probed using a goat anti-mouse IgG-HRP secondary antibody
conjugated with horseradish peroxidase (1:5,000 dilution; cat. no.
sc-2005; Santa Cruz Biotechnology, Inc.) at room temperature for 1
h. An enhanced chemiluminescence immunoblot detection system
(Pierce; Thermo Fisher Scientific, Inc.) was applied to visualize
the protein signals. Relative protein expression was analyzed using
Quantity One software (version 4.62; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) and presented as the density ratio compared with
GAPDH.
Statistical analysis
All data were presented as the mean ± standard
deviation and analyzed using SPSS software version 18.0 (SPSS,
Inc., Chicago, IL, USA). A Student's t-test and one-way analysis of
variance followed by a Tukey or Dunnett's test were used to compare
the differences between two groups and multiple groups,
respectively. Spearman's correlation analysis was performed to
examine the association between miR-208a and SRCIN1 in NSCLC
tissues. P<0.05 was considered to indicate a statistically
significant difference.
Results
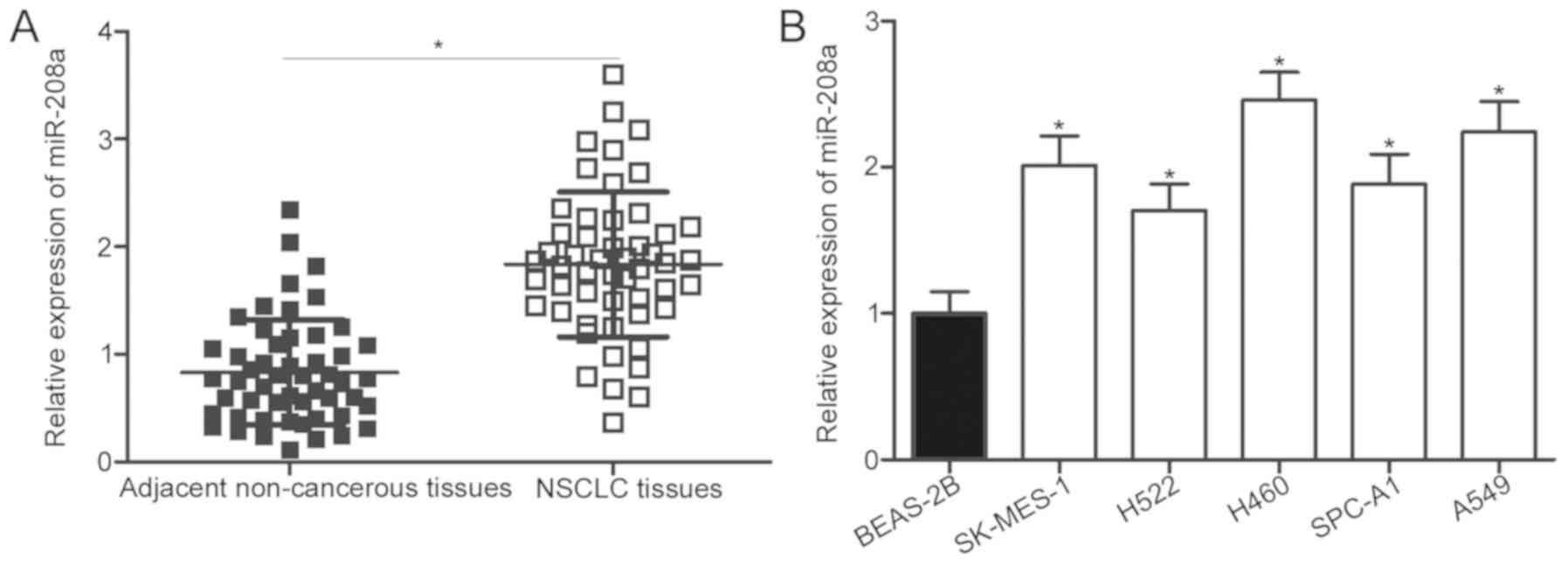
miR-208a is overexpressed in NSCLC
tissue specimens and cell lines
To investigate the expression profile of miR-208a in
NSCLC, RT-qPCR was utilized for the determination of miRNA
expression levels in NSCLC tissues and adjacent non-cancerous
tissues. The expression levels of miR-208a were significantly
upregulated in NSCLC tissues compared with that in adjacent
non-cancerous tissues (P<0.05; Fig.
1A). Subsequently, miR-208a expression was detected in five
human NSCLC cell lines, including SK-MES-1, H522, H460, SPC-A1 and
A-549. Fig. 1B reveals that the
miR-208a expression levels were significantly higher in all NSCLC
cell lines compared with that in the non-tumorigenic bronchial
epithelium BEAS-2B cell line (P<0.05). To investigate the
clinical value of miR-208a dysregulation in patients with NSCLC,
all patients with NSCLC were divided into miR-208a high/low
expression groups based on median expression of miR-208a (1.83). As
presented in Table I, high
miR-208a expression was significantly associated with
Tumor-Node-Metastasis (TNM) stage (P=0.026) and lymph node
metastasis (P=0.012). These results suggest that miR-208a may serve
an important function in the development of NSCLC.
 | Table I.Association between miR-208a
expression and the clinicopathological features of patients with
non-small cell lung cancer. |
Table I.
Association between miR-208a
expression and the clinicopathological features of patients with
non-small cell lung cancer.
|
|
| miR-208a expression
group |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Cases | High | Low | P-value |
|---|
| Sex |
|
|
| 0.266 |
|
Male | 28 | 12 | 16 |
|
|
Female | 24 | 14 | 10 |
|
| Age |
|
|
| 0.578 |
| <60
years | 28 | 15 | 13 |
|
| ≥60
years | 24 | 11 | 13 |
|
| Tumor size |
|
|
| 0.779 |
| <5
cm | 22 | 10 | 12 |
|
| ≥5
cm | 30 | 16 | 14 |
|
| Smoking
history |
|
|
| 0.760 |
| <10
years | 15 | 7 | 8 |
|
| ≥10
years | 37 | 19 | 18 |
|
| Tumor
differentiation |
|
|
| 0.780 |
|
I–II | 23 | 11 | 12 |
|
|
III–IV | 29 | 15 | 14 |
|
|
Tumor-Node-Metastasis stage |
|
|
| 0.026 |
|
I–II | 24 | 8 | 16 |
|
|
III–IV | 28 | 18 | 10 |
|
| Lymph node
metastasis |
|
|
| 0.012 |
|
Negative | 27 | 9 | 18 |
|
|
Positive | 25 | 17 | 8 |
|
miR-208a inhibition attenuates the
proliferation and invasion of NSCLC cells
To clarify the biological function of miR-208a in
NSCLC progression, a miR-208a inhibitor was introduced into H460
and A549 cells, which expressed a relatively high level of miR-208
among the five NSCLC cell lines, to knock down endogenous miR-208a
expression. miR-208a revealed a significant decrease in H460 and
A549 cells following miR-208a knockdown compared with the NC
inhibitor group (P<0.05; Fig.
2A). A CCK-8 assay was performed to determine whether NSCLC
cell proliferation may be influenced by miR-208a. As expected, cell
proliferation in H460 and A549 cells was significantly decreased
with miR-208a inhibitor transfection compared with the NC inhibitor
group (P<0.05; Fig. 2B). The
effect of silencing miR-208a expression on cell invasion capacity
in NSCLC was evaluated using a Matrigel invasion assay. The results
revealed that the numbers of invaded cells were significantly
reduced in miR-208a inhibitor-transfected H460 and A549 cells
compared with the NC inhibitor groups (P<0.05; Fig. 2C). These results suggest that
miR-208a may suppress the growth of a tumor in NSCLC
progression.
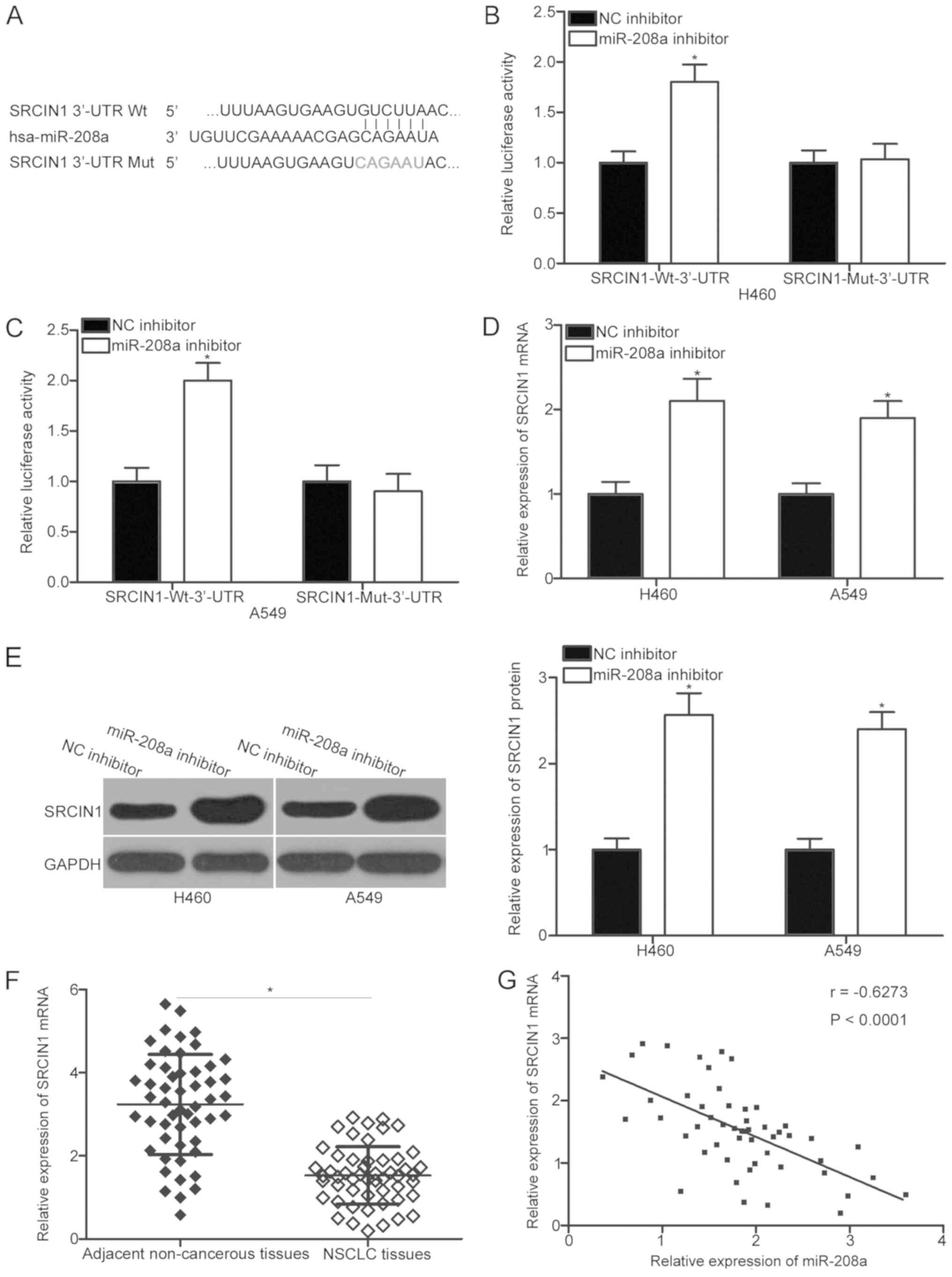
miR-208a directly targets SRCIN1 in
NSCLC
To investigate the mechanism by which miR-208a
regulates the malignant phenotypes in NSCLC, bioinformatics
analysis was performed to predict the putative target genes with
complementary sites of miR-208a in their 3′-UTR. SRCIN1, which has
been reported to be implicated in NSCLC occurrence and development
(26–28), was predicted as a major candidate
of miR-208a (Fig. 3A). To verify
whether miR-208a may directly interact with the 3′-UTR of SRCIN1, a
luciferase reporter assay was performed in H460 and A549 cells that
were co-transfected with miR-208a inhibitor or NC inhibitor and
pMIR-SRCIN1-Wt-3′-UTR or pMIR-SRCIN1-Mut-3′-UTR. miR-208a
inhibition induced a significant increase in luciferase activity
for pMIR-SRCIN1-Wt-3′-UTR in H460 and A549 cells compared with the
NC inhibitor control group (P<0.05). This effect, however, was
abrogated when harboring mutations were present at the predicted
SRCIN1 3′-UTR-binding sequences for miR-208a (Fig. 3B and C). RT-qPCR analysis and
western blot analysis were also performed to determine whether
miR-208a regulates SRCIN1 expression in NSCLC cells. The expression
levels of SRCIN1 mRNA (P<0.05) and protein (P<0.05) in
miR-208a inhibitor-transfected H460 and A549 cells were
significantly upregulated in comparison with those in NC
inhibitor-transfected cells (Fig. 3D
and E).
SRCIN1 mRNA expression was then detected in 52
primary NSCLC tissues to further investigate the association
between miR-208a and SRCIN1 in NSCLC. The results revealed that
NSCLC tissues exhibited significantly lower SRCIN1 mRNA expression
levels in comparison with those of the adjacent non-cancerous
tissues (P<0.05; Fig. 3F).
Furthermore, an significant inverse expression correlation between
miR-208a and SRCIN1 mRNA levels in NSCLC tissues was identified
through Spearman's correlation analysis (r=−0.6273, P<0.001;
Fig. 3G). Collectively, SRCIN1 was
generally revealed to be a direct target of miR-208a in NSCLC.
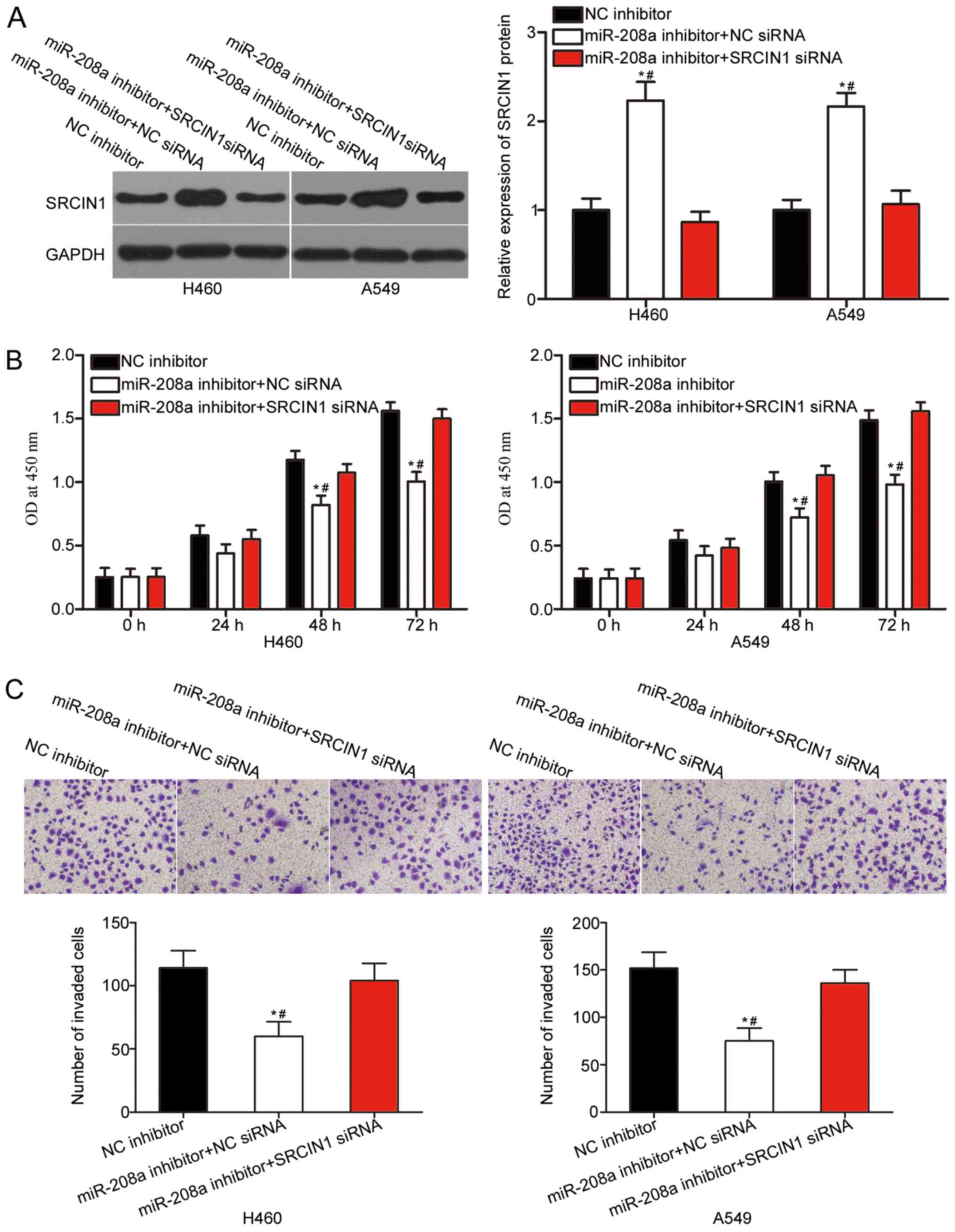
Knockdown of SRCIN1 expression
partially reverses the influence of miR-208a inhibitor on NSCLC
cells
A number of rescue experiments were applied to
determine whether the oncogenic function of miR-208a on NSCLC cell
progression is mediated by SRCIN1 upregulation. SRCIN1 siRNA or NC
siRNA along with miR-208a inhibitor were transfected into H460 and
A549 cells. The co-transfection of SRCIN1 siRNA significantly
abrogated the miR-208a inhibitor-mediated upregulation of SRCIN1 in
H460 and A549 cells, as demonstrated by western blot analysis
(P<0.05; Fig. 4A). In addition,
the knockdown of SRCIN1 expression in H460 and A549 cells
transfected with miR-208a inhibitor significantly rescued the
inhibition of cell proliferation (P<0.05; Fig. 4B) and invasion (P<0.05; Fig. 4C) caused by miR-208a
underexpression. On the basis of the aforementioned results, it was
concluded that the tumor-promoting functions of miR-208a in NSCLC
cells are, at least in part, attributable to SRCIN1 regulation.
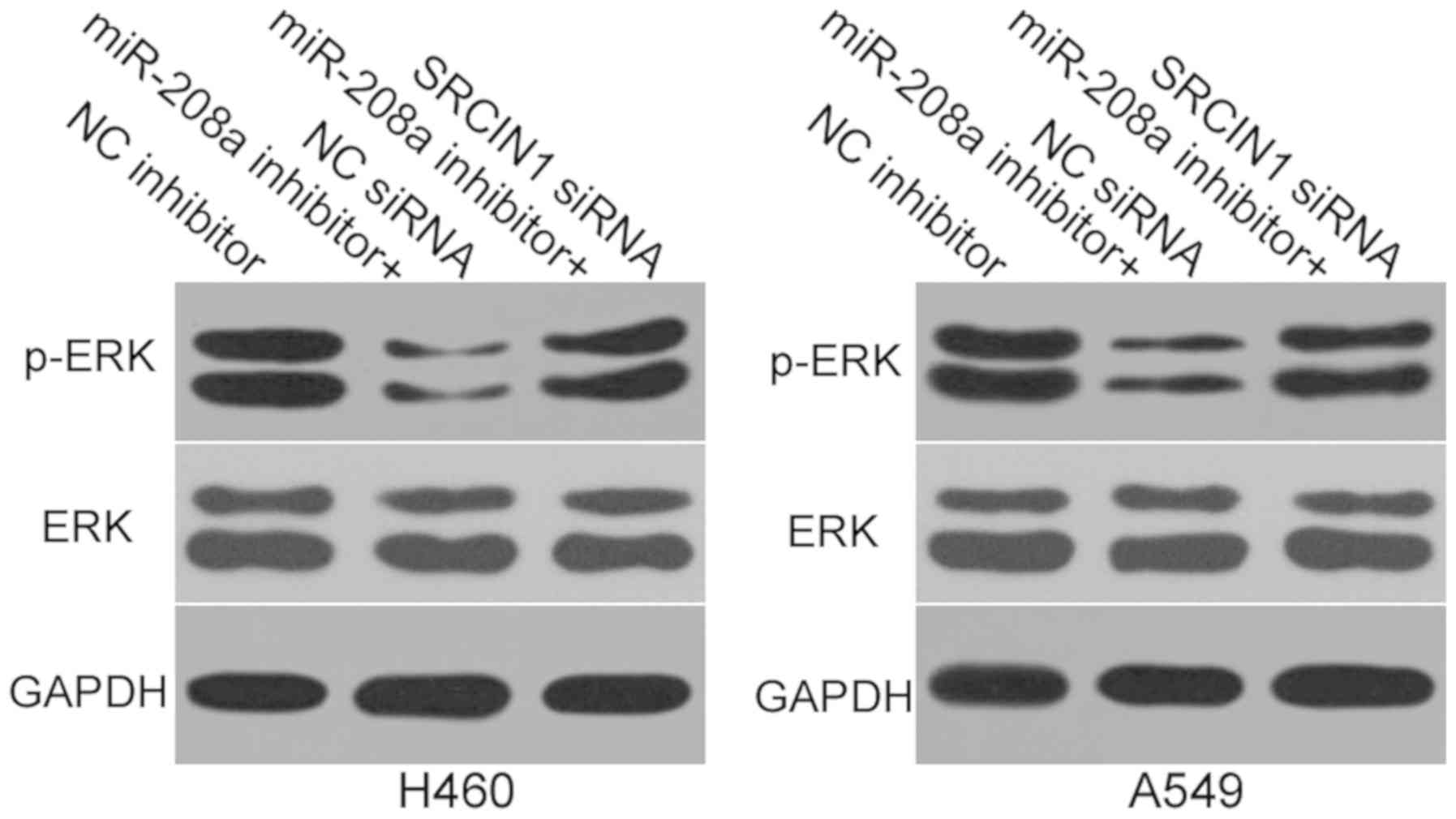
miR-208a inhibition reduces the
activity of the ERK signaling pathway in NSCLC
One previous study has demonstrated that SRCIN1
regulates the ERK pathway in lung cancer (26). Thus, the present study aimed to
determine whether miR-208a may regulate the ERK signaling pathway
in NSCLC. Western blot analysis was utilized to measure the
expression of ERK and p-ERK proteins in H460 and A549 cells
subsequent to co-transfection with miR-208a inhibitor and SRCIN1
siRNA or NC siRNA. As expected, miR-208a inhibition was revealed to
reduce p-ERK expression in H460 and A549 cells (Fig. 5), whereas the total ERK protein
expression was unaffected. In addition, the protein expression of
p-ERK in H460 and A549 cells were recovered following
co-transfection with SRCIN1 siRNA. These results suggest that
miR-208a downregulation deactivates the ERK signaling pathway via
the regulation of SRCIN1.
Discussion
Abnormal miRNA expression has been demonstrated to
serve critical functions in NSCLC formation and progression
(29,30). Therefore, understanding the
association between NSCLC and abnormally expressed miRNAs may allow
for the identification of numerous novel diagnostic and therapeutic
biomarkers for patients with NSCLC. The present study is the first
to present data, to the best of our knowledge, characterizing the
expression profile, specific functions and associated mechanisms
underlying miR-208a in NSCLC. miR-208a was notably upregulated in
NSCLC tissues and cell lines compared with the non-cancerous
controls. High expression levels of miR-208a were significantly
associated with TNM stage and lymph node metastasis in NSCLC.
Meanwhile, miR-208a downregulation attenuated the proliferation and
invasion of NSCLC cells. SRCIN1 was validated as a direct target of
miR-208a in NSCLC. Silencing SRCIN1 expression partially reversed
the oncogenic effects of miR-208a on NSCLC cell proliferation and
invasion. Inhibition of miR-208a reduced the activity of the ERK
signaling pathway in NSCLC through the regulation of SRCIN1. These
results suggest that miR-208a may exert tumor-promoting functions
in NSCLC and may be further developed as a novel target in treating
patients with NSCLC.
miR-208a has been associated with multiple types of
human cancer. For instance, miR-208a has been revealed to be
upregulated in hepatocellular carcinoma. Inhibition of miR-208a
inhibited hepatocellular carcinoma cell proliferation and invasion
in vitro and decreased tumorigenesis in vivo
(20). Li et al (21) reported that miR-208a was highly
expressed in oesophageal squamous cell carcinoma tissues and cell
lines. miR-208a upregulation facilitated the cell proliferation,
tumorigenicity and cell cycle progression of oesophageal squamous
cell carcinoma. Yin et al (22) also revealed that miR-208a was
overexpressed in gastric cancer. miR-208a overexpression attenuated
gastric cancer cell apoptosis and induced tumor growth in
vivo. Liu et al (31)
revealed that the ectopic expression of miR-208a promoted the cell
migration, invasion and epithelial-mesenchymal transition of
pancreatic cancer. Accordingly, miR-208a serves an oncogenic
function in tumorigenesis and tumor development and may be
developed as a potential target in the therapy of these specific
tumor types.
A number of target miR-208a's have been identified,
including AT-rich interactive domain-containing protein 1 in
hepatocellular carcinoma (20),
SRY-Box 6 in oesophageal squamous cell carcinoma (21) and programmed cell death 4 in
gastric cancer (22). SRCIN1, also
known as p140 cas-associated protein, has been demonstrated to be a
direct target gene of miR-208a in NSCLC. The gene contains two
coiled-coil domains, two proline-rich regions and two regions of
highly charged amino acids (32).
SRCIN was previously reported to be decreased in multiple human
malignancy types, including liver cancer (33), cutaneous squamous cell carcinoma
(34), breast cancer (35) and osteosarcoma (36). SRCIN1 was revealed to serve an
inhibitory function in tumorigenesis and tumor development. For
instance, SRCIN1 restoration repressed cell proliferation, colony
formation, invasion and epithelial-mesenchymal transition in
osteosarcoma (36). Resumption
expression of SRCIN1 prohibited the proliferation and
epithelial-mesenchymal transition in hepatocellular carcinoma
(33). Ectopic expression of
SRCIN1 in cutaneous squamous cell carcinoma suppressed the
proliferative and migratory abilities of the cells (34). In the present study, it was
demonstrated that miR-208a silencing deactivated the ERK signaling
pathway via the regulation of SRCIN1. The ERK signaling pathway
serves crucial functions in the occurrence and development of
NSCLC, and is implicated in the regulation of aggressive phenotypes
of NSCLC cells (37–39). These results suggest that restoring
SRCIN1 expression may be adopted as a novel therapeutic strategy
for anti-tumor therapy.
SRCIN1 has been demonstrated to be regulated by
multiple miRNAs in NSCLC. For example, Cao et al (26) revealed that miR-150 targeted SRCIN1
to promote the proliferation and migration of NSCLC cells. Ye et
al (27) reported that miR-211
induced cell growth in NSCLC through the negative regulation of
SRCIN1. Gao et al (28)
also identified that miR-873 increased the cell proliferation and
migration of NSCLC cells via a SRCIN1 blockade. Zhang et al
(40) indicated that miR-150
enhanced cell growth in vitro and in vivo by directly
targeting SRCIN1. The present study demonstrated that the
downregulation of miR-208a reduced NSCLC cell proliferation and
invasion through SRCIN1 upregulation. These results suggest that
the miRNA/SRCIN1 pathway may have certain clinical applications in
the management of patients with NSCLC.
In summary, miR-208a was frequently overexpressed in
NSCLC, and increased miR-208a expression was associated with TNM
stage and lymph node metastasis. miR-208a may function as an
oncogene by directly targeting SRCIN1 in NSCLC. The miR-208a/SRCIN1
axis may be used in miRNA-based therapy for the treatment of
patients with NSCLC. However, the association between miR-208 and
the overall survival or disease-free survival of patients with
NSCLC was unexplored in the present study. It was a limitation of
the present study, and survival information will be collected in
order to resolve this limitation in the near future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW designed the present study and analyzed the data.
LL performed RT-qPCR and the CCK-8 assay. Matrigel invasion assay
and luciferase reporter assay were performed by WW. SG conducted
western blotting. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The Ethics Committee of the Qilu Hospital of
Shandong University ethically approved the present study. All
patients provided written informed consent prior to the
operation.
Patient consent for publication
All patients provided written informed consent prior
to the operation. All patients provided consent for
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Latimer KM: Lung cancer: Clinical
presentation and diagnosis. FP Essent. 464:23–26. 2018.PubMed/NCBI
|
|
4
|
Gadgeel SM: New targets in non-small cell
lung cancer. Curr Oncol Rep. 15:411–423. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mott TF: Lung cancer: Management. FP
Essent. 464:27–30. 2018.PubMed/NCBI
|
|
6
|
Yu JL, Simmons C, Victor JC, Han D,
Hogeveen S, Leighl N and Verma S: Impact of new chemotherapeutic
and targeted agents on survival in stage IV non-small cell lung
cancer. Oncologist. 16:1307–1315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Parums DV: Current status of targeted
therapy in non-small cell lung cancer. Drugs Today (Barc).
50:503–525. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Simon J: Technology radiation technology
targets tumors. Surgical precision without the incision. S D Med.
67:3622014.PubMed/NCBI
|
|
9
|
Miller YE: Pathogenesis of lung cancer:
100 year report. Am J Respir Cell Mol Biol. 33:216–223. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Diniz GP and Wang DZ: Regulation of
skeletal muscle by microRNAs. Compr Physiol. 6:1279–1294. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bao W, Greenwold MJ and Sawyer RH:
Expressed miRNAs target feather related mRNAs involved in cell
signaling, cell adhesion and structure during chicken epidermal
development. Gene. 591:393–402. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hata A and Lieberman J: Dysregulation of
microRNA biogenesis and gene silencing in cancer. Sci Signal.
8:re32015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang A, Lakshmanan J, Motameni A and
Harbrecht BG: MicroRNA-203 suppresses proliferation in liver cancer
associated with PIK3CA, p38 MAPK, c-Jun, and GSK3 signaling. Mol
Cell Biochem. 441:89–98. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Z, Yang Y and Zhang X: MiR-770
inhibits tumorigenesis and EMT by targeting JMJD6 and regulating
WNT/β-catenin pathway in non-small cell lung cancer. Life Sci.
188:163–171. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang X, Ma X, An H, Xu C, Cao W, Yuan W
and Ma J: Upregulation of microRNA-125b by G-CSF promotes
metastasis in colorectal cancer. Oncotarget. 8:50642–50654.
2017.PubMed/NCBI
|
|
19
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu P, Wu D, You Y, Sun J, Lu L, Tan J and
Bie P: miR-208-3p promotes hepatocellular carcinoma cell
proliferation and invasion through regulating ARID2 expression. Exp
Cell Res. 336:232–241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li H, Zheng D, Zhang B, Liu L, Ou J, Chen
W, Xiong S, Gu Y and Yang J: Mir-208 promotes cell proliferation by
repressing SOX6 expression in human esophageal squamous cell
carcinoma. J Transl Med. 12:1962014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yin K, Liu M, Zhang M, Wang F, Fen M, Liu
Z, Yuan Y, Gao S, Yang L, Zhang W, et al: miR-208a-3p suppresses
cell apoptosis by targeting PDCD4 in gastric cancer. Oncotarget.
7:67321–67332. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chassagnon G, Bennani S and Revel MP: New
TNM classification of non-small cell lung cancer. Rev Pneumol Clin.
73:34–39. 2017.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao M, Hou D, Liang H, Gong F, Wang Y, Yan
X, Jiang X, Wang C, Zhang J, Zen K, et al: miR-150 promotes the
proliferation and migration of lung cancer cells by targeting SRC
kinase signalling inhibitor 1. Eur J Cancer. 50:1013–1024. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ye L, Wang H and Liu B: miR-211 promotes
non-small cell lung cancer proliferation by targeting SRCIN1.
Tumour Biol. 37:1151–1157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao Y, Xue Q, Wang D, Du M, Zhang Y and
Gao S: miR-873 induces lung adenocarcinoma cell proliferation and
migration by targeting SRCIN1. Am J Transl Res. 7:2519–2526.
2015.PubMed/NCBI
|
|
29
|
Cao Y, Zhao D, Li P, Wang L, Qiao B, Qin
X, Li L and Wang Y: MicroRNA-181a-5p impedes IL-17-induced nonsmall
cell lung cancer proliferation and migration through targeting
VCAM-1. Cell Physiol Biochem. 42:346–356. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang H, Zhan Y, Jin J, Zhang C and Li W:
MicroRNA-15b promotes proliferation and invasion of non-small cell
lung carcinoma cells by directly targeting TIMP2. Oncol Rep.
37:3305–3312. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu A, Shao C, Jin G, Liu R, Hao J, Song
B, Ouyang L and Hu X: miR-208-induced epithelial to mesenchymal
transition of pancreatic cancer cells promotes cell metastasis and
invasion. Cell Biochem Biophys. 69:341–346. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Di Stefano P, Cabodi S, Boeri Erba E,
Margaria V, Bergatto E, Giuffrida MG, Silengo L, Tarone G, Turco E
and Defilippi P: P130Cas-associated protein (p140Cap) as a new
tyrosine-phosphorylated protein involved in cell spreading. Mol
Biol Cell. 15:787–800. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen R, Liao JY, Huang J, Chen WL, Ma XJ
and Luo XD: Downregulation of SRC kinase signaling inhibitor 1
(SRCIN1) expression by MicroRNA-32 promotes proliferation and
epithelial-mesenchymal transition in human liver cancer cells.
Oncol Res. 26:573–579. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen B, Pan W, Lin X, Hu Z, Jin Y, Chen H,
Ma G, Qiu Y, Chang L, Hua C, et al: MicroRNA-346 functions as an
oncogene in cutaneous squamous cell carcinoma. Tumour Biol.
37:2765–2771. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang F, Luo LJ, Zhang L, Wang DD, Yang SJ,
Ding L, Li J, Chen D, Ma R, Wu JZ and Tang JH: MiR-346 promotes the
biological function of breast cancer cells by targeting SRCIN1 and
reduces chemosensitivity to docetaxel. Gene. 600:21–28. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang P, Wang H, Li X, Liu Y, Zhao C and
Zhu D: SRCIN1 suppressed osteosarcoma cell proliferation and
invasion. PLoS One. 11:e01555182016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang H, Wu C, Wan S, Zhang H, Zhou S and
Liu G: Shikonin attenuates lung cancer cell adhesion to
extracellular matrix and metastasis by inhibiting integrin β1
expression and the ERK1/2 signaling pathway. Toxicology.
308:104–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qi M, Tian Y, Li W, Li D, Zhao T, Yang Y,
Li Q, Chen S, Yang Y, Zhang Z, et al: ERK inhibition represses
gefitinib resistance in non-small cell lung cancer cells.
Oncotarget. 9:12020–12034. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cao Q, Mao ZD, Shi YJ, Chen Y, Sun Y,
Zhang Q, Song L and Peng LP: MicroRNA-7 inhibits cell
proliferation, migration and invasion in human non-small cell lung
cancer cells by targeting FAK through ERK/MAPK signaling pathway.
Oncotarget. 7:77468–77481. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang L, Lin J, Ye Y, Oba T, Gentile E,
Lian J, Wang J, Zhao Y, Gu J, Wistuba II, et al: Serum MicroRNA-150
predicts prognosis for early-stage non-small cell lung cancer and
promotes tumor cell proliferation by targeting tumor suppressor
gene SRCIN1. Clin Pharmacol Ther. 103:1061–1073. 2018. View Article : Google Scholar : PubMed/NCBI
|