Introduction
Those with a history of chronic alcohol abuse are at
a higher risk of developing alcoholic fatty liver disease (AFLD),
cirrhosis and, ultimately, liver failure (1). AFLD is represented by abnormal
hepatic lipid accumulation in the early stages and steatohepatitis
in the latter stages (1,2). Lipids storage in lipid droplets is
influenced by synthesis and degradation in the liver (3). Therefore, an increase in lipid
degradation in lipid droplets via autophagy or lipolysis could
ameliorate AFLD (4,5).
Autophagy is a lysosome-dependent degradation
process of intracellular organelles in mammalian cells. It is
activated during stressful periods, such as energy deficiency
(6), and is also influenced by
cellular cholesterol levels and alcohol intake (6–8). The
relationship between alcoholic beverages and autophagy is complex
(9). Autophagy ameliorates liver
steatosis in vivo (5,10,11).
Alterations in cholesterol levels in cells can
regulate autophagy (8,12). By extension, the expression level
of the cholesterol transporter Niemann-Pick C1-like 1 (NPC1L1) in
the liver may also influence autophagy. NPC1L1 is a transmembrane
cholesterol absorption transporter expressed in the liver and
intestines in humans (13). The
function of hepatic NPC1L1 is to prevent excessive cholesterol loss
in humans (14,15). By contrast, mouse NPC1L1 is
expressed only in the intestines (16). The expression of human NPC1L1 in
the mouse liver results in an elevation of the total plasma
cholesterol level, and hepatic triglyceride secretion (17,18).
The hepatic cholesterol level is increased in the livers of
apolipoprotein E-knockout mice with NPC1L1 overexpression (19). Furthermore, inhibition of NPC1L1 by
ezetimibe can significantly activate autophagy in human hepatocytes
(20). It is indicated that NPC1L1
may inhibit hepatic autophagy. In humans, NPC1L1 may be a potential
factor contributing to alcoholic steatosis by reducing
autophagy.
The present study assessed the function of NPC1L1 in
the activation of hepatic autophagy in a mouse model expressing
human NPC1L1 in the liver under conditions of alcohol consumption.
It was concluded that NPC1L1 expression reduced hepatic
autophagy.
Materials and methods
Generation of recombinant
adenoviruses
cDNA of human NPC1L1 was cloned and NPC1L1
adenoviral vectors (Ad-L1) were constructed using the AdMax system
(Hanbio Biotechnology Co., Ltd.) and purified through sucrose
gradient ultracentrifugation (21). A control adenovirus (Ad-null)
without encoded cDNA was also constructed. Adenoviral vectors were
administered at a dose of 1×1011 particles
(10.5×109 plaque-forming units) via the retro-orbital
vein into the mice. The sequences of the primers used to amplify
human (h)NPC1L1 from the mouse liver for reverse
transcription-quantitative PCR (RT-qPCR) are listed in Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Name | Forward
(5′-3′) | Reverse
(3′-5′) | Primer melting
temperature (°C) |
|---|
| hNPC1L1 |
AGAGTGAGCCTTACACAACCA |
GCAGGACACGTTGGAGAGT | 60 |
| mAtg5 |
TGTGCTTCGAGATGTGTGGTT |
ACCAACGTCAAATAGCTGACTC | 60 |
| mAtg7 |
TCTGGGAAGCCATAAAGTCAGG |
GCGAAGGTCAGGAGCAGAA | 60 |
| mAtg12 |
TGGCCTCGGAACAGTTGTTTA |
GGGCAAAGGACTGATTCACAT | 60 |
| GAPDH |
GAGCCAAACGGGTCATCATC |
CATCACGCCACAGCTTTCCA | 60 |
Animals and feeding
A total of 24 male mice (C57BL/6J, weighting 20–22
g) at the age of 8 weeks (Jackson Laboratory; stock no. 002207)
were housed in standard cages at 4 mice were per cage. The
conditions were 23°C, 50% humidity with a 12 h light/dark cycle and
free access to water and a chow diet. A 10 day feeding plan was
used to induce alcoholic liver steatosis during NPC1L1 expression
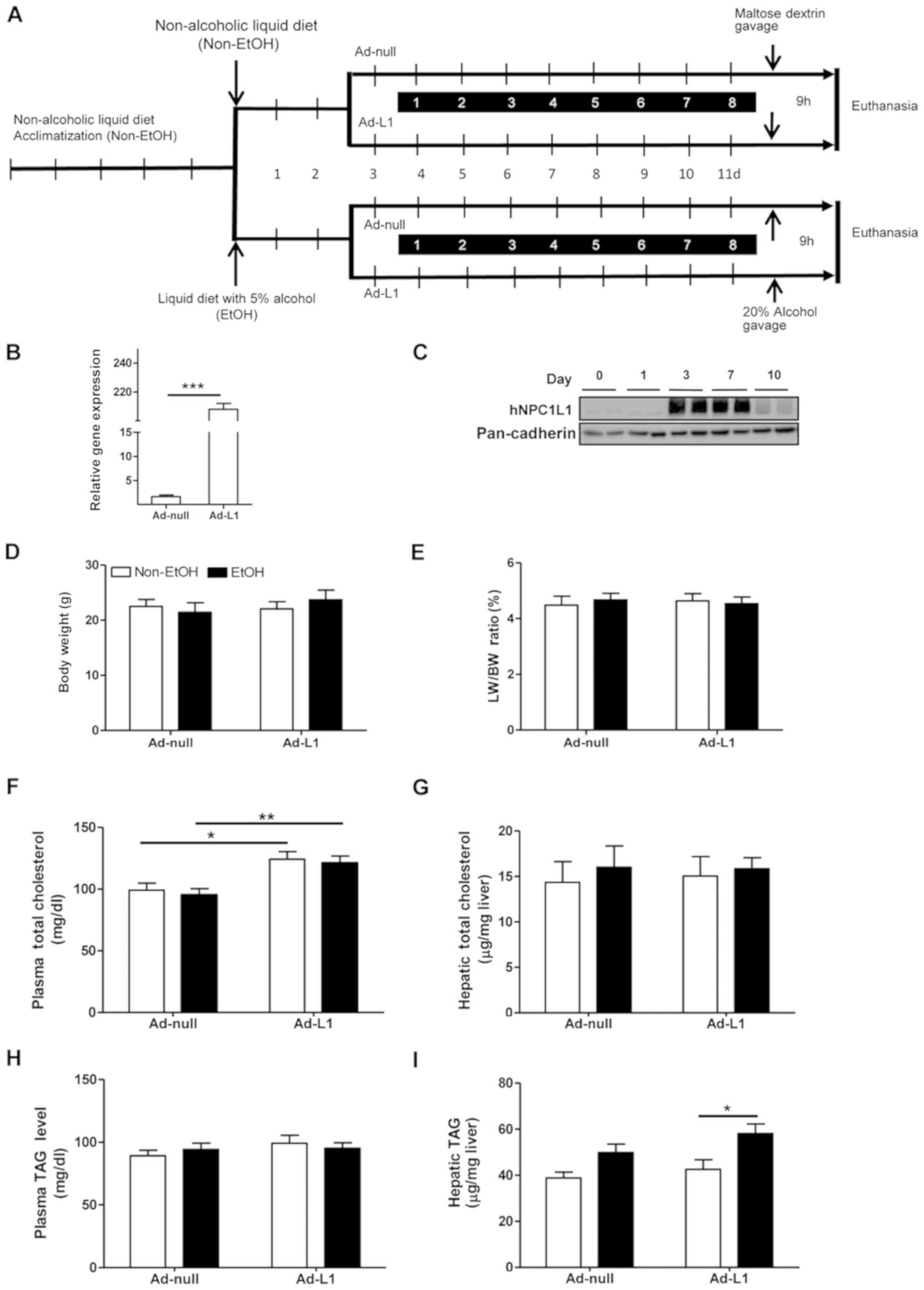
(Fig. 1A). At the beginning, the
mice were fed on a non-alcoholic liquid diet ad libitum
(Bio-Serv; product no. F1259SP) for acclimatization. A total of 5
days later, 12 mice were changed to the Lieber-DeCarli alcoholic
liquid diet (Bio-Serv; product no. F1258SP; 5 g/kg body weight).
The other 12 mice continued to be administered the non-alcoholic
diet. After 3 days, Ad-L1 and Ad-null adenoviruses were injected
retro-orbitally into 6 mice on the alcohol diet and 6 mice on the
non-alcoholic diet. At the end of feeding, mice on the alcoholic
diet were euthanized at 9 h after a single binge gavage (20%
alcohol). Instead of alcohol, the non-alcohol-fed mice were also
euthanized at the same time after receiving a single dose of
maltodextrin for energy balance. All mice were euthanized by 5%
isoflurane (Hairui Chemical; cat. no. HR135327).
SDS-PAGE and western blot
analysis
Liver tissues or primary hepatocytes were
homogenized in the RIPA buffer (cat. no. 89900; Thermo Fisher
Scientific, Inc.) followed by centrifugation at 12,000 × g for 10
min at 4°C to isolate the proteins for western blotting. Then, the
protein concentrations were quantified by BCA protein assay kit
(cat. no. 23227; Thermo Fisher Scientific, Inc) via absorbance
measurement using the LAS-4000 lumino-image analyzer (Fujifilm).
Then, 30 µg protein was loaded to each lane of 10% SDS-PAGE gel.
Western blotting was performed following protocols described
previously (17). Data were
normalized against β-actin. The antibodies used in the present
study were: Anti-microtubule-associated proteins 1A/1B light chain
3 (LC3) A/B antibody (cat. no. ab128025; Abcam); anti-β-actin
antibody (cat. no. ab8227; Abcam); anti-p62/sequestosome-1 (SQSTM1)
antibody (cat. no. ab91526; Abcam), anti-hNPC1L1 antibody (cat. no.
ab124801; Abcam); anti-pan-cadherin antibody (cat. no. ab6528;
Abcam); goat anti-mouse IgG (cat. no. ab6788; Abcam); and goat
anti-rabbit IgG (cat. no. ab6720; Abcam). Primary antibodies were
diluted as 1:3,000 in 5% BSA for overnight incubation at 4°C.
Secondary antibodies were diluted as 1:10,000 in 5% BSA for 1 h
incubation at room temperature.
Duration of NPC1L1 expression
To determine how long the hNPC1L1 expression lasted
in the liver, a total of 10 C57BL/6J male mice received Ad-L1
injection, 2 mice were sacrificed at each time point (0, 1, 3, 7
and 10 days) after injection and liver samples were harvested.
Western blotting was performed using the aforementioned protocol
and anti-hNPC1L1 antibody. The pan-cadherin antibody was used as a
loading control.
Evaluation of liver injury by
histopathology and biochemistry assay
Hematoxylin and eosin (H&E) and Sirius red
staining were performed to evaluate pathological changes in the
liver. All specimens were fixed overnight at room temperature by
10% formalin and were embedded in paraffin wax. Paraffin were
cleared from 5 µm thick sections in three changes of xylene for 2
min per change at room temperature. The samples were hydrated at
room temperature through three changes of 100% ethanol for 2 min
per change followed by 95% ethanol and 70% ethanol for 2 min,
respectively. The slides were rinsed in running water at room
temperature for 5 min. The sections were stained in hematoxylin
solution (cat. no. 143350, Shanghai Seebio Biotech, Inc.) for 3 min
at room temperature, then rinsed under running water at room
temperature for 10 min. The sections were stained working eosin Y
solution (cat. no. C0850110535, Nanjing reagent) for 2 min before
dehydration by transfer from 95% ethanol (20 min) to 100% ethanol
(2 min each change), then three changes in xylene for 2 min per
change. Slides were covered by glass coverslip with mounting medium
(22). Picro-sirius red solution
(cat. no. ab150681; Abcam) was using for 1 h staining at room
temperature after de-waxing and hydration as described above
Histological analyses were evaluated at ×200 magnification using a
Leica DM 2700P microscope (Leica Microsystems GmbH). Aspartate
transaminase (AST) and alanine transaminase (ALT) were measured
using an AST activity colorimetric assay kit (cat. no. K753;
Bio-Vision, Inc.) and an ALT activity colorimetric assay kit (cat.
no. K752; Bio-Vision, Inc.), respectively.
Primary hepatocyte isolation and
fluorescence microscopy
Primary hepatocytes were isolated from
non-alcohol-fed and alcohol-fed mice, as described previously
(23). The hepatocytes were seeded
onto collagen-coated coverslips in six-well plates. Cells were
collected and fixed with 4% paraformaldehyde for 20 min at room
temperature. LC3 protein was detected by immunofluorescence
antibody (cat. no. ab128025; Abcam) as previously described.
Briefly, slides were incubated for 10 min with PBS containing 0.1%
Triton X-100 then rinsed three times in PBS. The cells were
incubated with 10% goat serum (cat. no. ab7481; Abcam) for 30 min
at room temperature, then incubated cells in diluted primary
antibody (1:100) in 1% BSA in PBST overnight at 4°C. Then, cells
were incubated with goat anti-rabbit IgG (1:1,000, cat. no.
ab150079; Abcam) at room temperature for 1 h. The cell nucleus was
stained with DAPI in Prolong Diamond Antifade Mountant (cat. no.
P36962; Thermo Fisher Scientific, Inc.) while the slides were
sealed. LC3 protein (red dot) in the cells was observed under a
Leica DM2500 florescence microscope (magnification ×1,000; Leica
Microsystems GmbH). Quantification was determined in 20 cells from
captured images using Image J software (version 1.8.0, National
Institutes of Health).
Ezetimibe treatment in cultured
hepatocytes
To determine whether inhibition of NPC1L1 expression
may restore the autophagy in alcohol-fed mice, primary hepatocytes
were incubated in 5% CO2 at 37°C) in different
concentrations of ezetimibe (0, 10, 20 and 40 µM; HR138776, Hairui
Chemical, Co., Ltd.). After 1 h, hepatocytes were collected for the
protein expression of LC3 by western blotting.
RT-qPCR
RNA extraction from hepatocytes of Ad-L1 mice and
Ad-null mice (n=6) were performed as described using a RNeasy Mini
kit 250 (cat. no. 74106; Qiagen, Inc.). Purified RNA was
transcribed at 42°C by QuantiTectRev. Transcription kit (cat. no.
205313; Qiagen GmbH), and qPCR was performed using a
SYBR® Green PCR Master Mix kit (cat. no. 4309155; Thermo
Fisher Scientific, Inc.). Thermocycling conditions were 95°C for 2
min, then 40 cycles of 95°C for 15 sec, annealing for 15 sec, 72°C
for 45 sec. After amplification, a melting curve was used to assess
product purity. Annealing temperatures were optimized for each
primer pair (Table I). GAPDH was
used as an internal control and mRNA expression levels were
calculated based on the 2−ΔΔCq method (24). mRNA levels for each gene represent
the amount relative to that in control mice, which was arbitrarily
standardized to 1. Sequences of the primers used for qPCR are given
in Table I.
Statistical analysis
Data are presented as the mean ± SEM of at least
four independent experiments. Densitometric analysis was performed
on the immunoblots using ImageJ 1.8.0 (National Institutes of
Health). Statistical differences were determined using the
Student's t-test and one-way ANOVA (post hoc test: Tukey's multiple
comparison test) or two-way ANOVA (post hoc test: Bonferroni). All
statistics were performed using GraphPad Prism 5 (GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Ethics statement
Experimental procedures and animal use and care
protocols were in accordance with the guidelines of the Northwest
A&F University Institutional Committee for the Care and Use of
Laboratory Animals, and were approved by the Committee on Ethical
Use of Animals of Northwest A&F University.
Results
Expression of human NPC1L1 (Ad-L1) in
the mouse liver via adenovirus transfection
According to the experimental workflow (Fig. 1A), adenovirus-packaged human NPC1L1
cDNA was transfected retro-orbitally at day 3 after the
commencement of the alcohol diet. NPC1L1 mRNA expression levels
were evaluated in the mouse liver 3 days post-injection. The
results revealed that mRNA expression in the Ad-L1 mice was
~200-fold higher than that in the Ad-null mice (Fig. 1B; P<0.001). Western blot
analysis showed that NPC1L1 protein was markedly expressed in the
mouse liver starting from day 3 until day 7 after adenoviral
injection (Fig. 1C), and was
decreased at about 10 days post-injection.
The Ad-L1 mice behaved normally under the
experimental conditions. The alcohol consumption and adenoviral
treatment did not result in significant changes in body weight,
liver weight or the liver/body weight ratio in Ad-null and Ad-L1
mice (Fig. 1D and E).
Lipids in the blood and liver were measured. Plasma
total cholesterol was significantly increased in Ad-L1 mice
regardless of alcohol feeding (Fig.
1F; P<0.05). However, total hepatic cholesterol levels were
similar under both non-alcoholic and alcohol feeding conditions
(Fig. 1G). In contrast with the
total cholesterol, total plasma triglyceride levels were similar
between Ad-null and Ad-L1 mice (Fig.
1H) under both feeding states. Hepatic triglyceride levels were
significantly higher in alcohol-fed mice in the Ad-L1 group
(Fig. 1I; P<0.05).
Chronic-binge alcohol uptake causes
mild liver injury and steatosis in mice
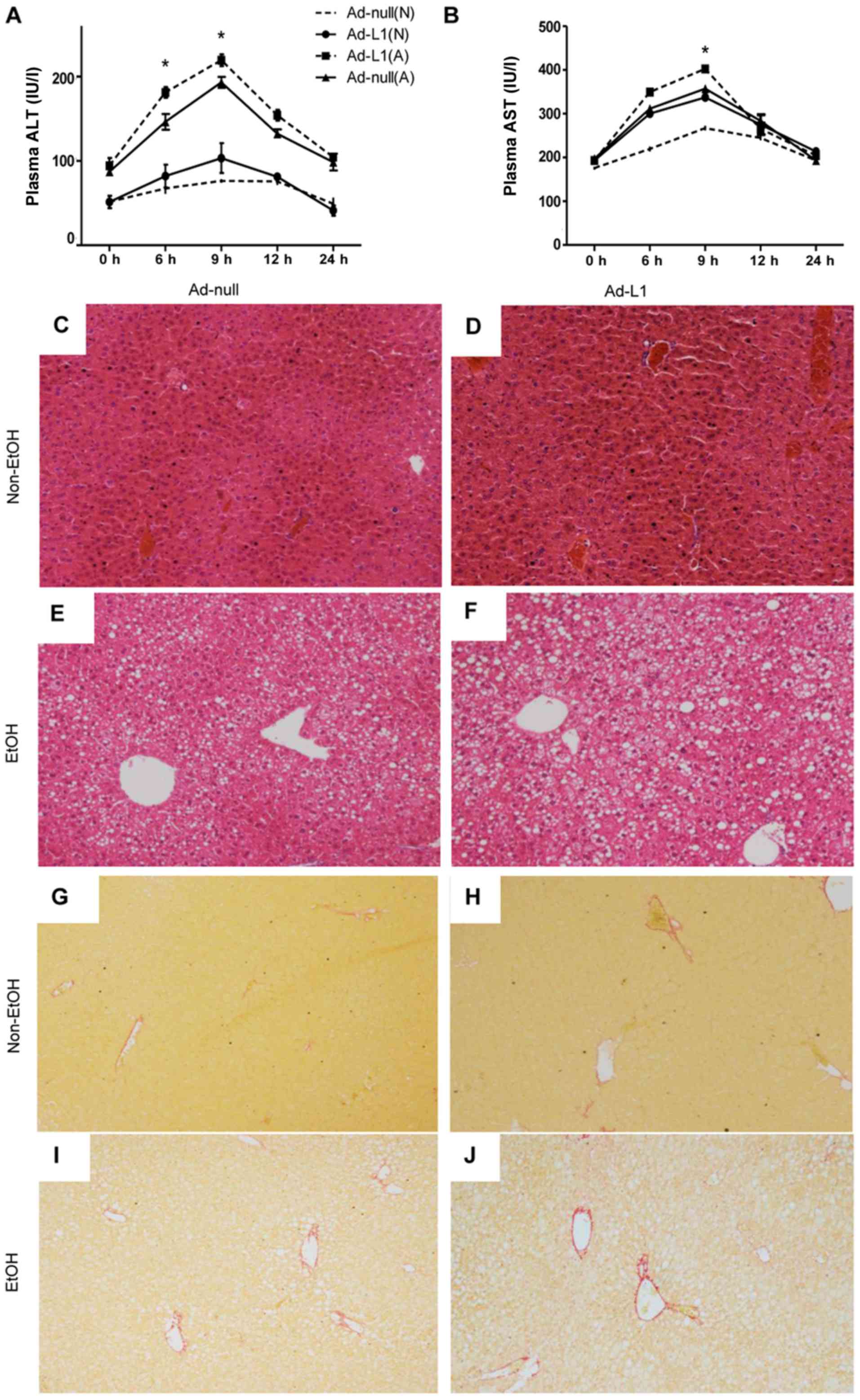
A chronic-binge alcohol feeding plan was followed in
the present study to induce alcoholic liver injury. Plasma AST and
ALT activity were measured to evaluate liver injury. The results
revealed that alcohol feeding caused increases in AST and ALT
activity in the blood of Ad-L1 and Ad-null mice. Compared with the
Ad-null mice after alcohol feeding, the Ad-L1 mice had
significantly higher ALT activity (Fig. 2A; P<0.05). A small but
significant difference was detected in plasma AST activity between
Ad-L1 and Ad-null mice at 9 h after gavage (Fig. 2B; P<0.05). This suggested that 9
h after the gavage was the appropriate time to evaluate liver
injury.
Histologically, H&E staining showed mild lipid
accumulation in the livers of alcohol-fed mice (Fig. 2C-F). The lipid accumulation was
more pronounced in the livers of AD-L1 mice than that in Ad-null
mice. This demonstrated that alcohol uptake induced mild liver
steatosis. Sirius red staining showed no detectable fibrosis in the
livers of alcohol-fed mice (Fig.
2G-J).
Autophagy is reduced in the
hepatocytes of Ad-null mice after alcohol feeding
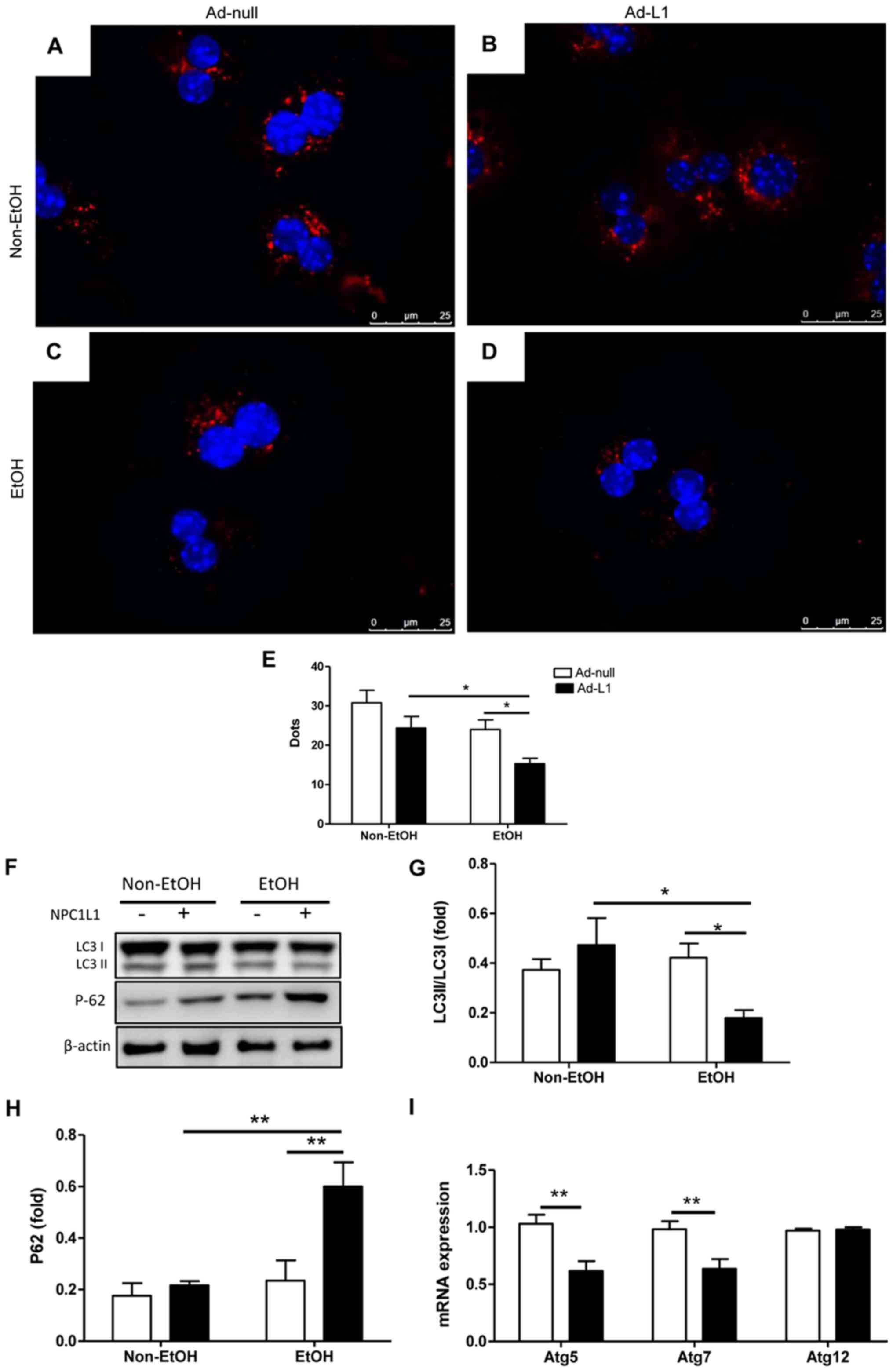
Fluorescence microscopy was used to monitor the
formation of autophagic particles in isolated primary hepatocytes
(Fig. 3A-D). Red dots indicate the
intracellular autophagy marker LC3I/II, and blue dots stained with
DAPI indicate the nuclei of the hepatocytes. Quantification of 20
hepatocytes from captured images (Fig.
3A-D) showed fewer red dots in the alcohol-fed mice (Fig. 3D and E; P<0.05). The result was
confirmed through analysis of the protein expression of the
autophagy marker p62/SQSTM1 (Fig. 3F
and H; P<0.01). The ratio of LC3II/LCI in the liver was
unchanged in the alcohol-fed mice compared with the non-alcohol-fed
mice.
Autophagy is attenuated in the
hepatocytes of Ad-L1 mice
According to the immunofluorescence images, Ad-L1
mice had significantly less LC3 expression (Fig. 3D and E) compared with Ad-null mice
under both alcoholic and non-alcoholic feeding conditions (Fig. 3B and E). These results were
confirmed by western blot analysis (Fig. 3F). The LC3II expression was
reduced. By contrast, p62/SQSTM1 protein expression was increased
in Ad-L1 mice compared with Ad-null mice. This indicated that
autophagy was inhibited. Quantification by densitometry showed that
the LC3II/LC3I expression ratio (Fig.
3G) was significantly decreased and p62/SQSTM1 protein content
(Fig. 3H) was significantly
increased in Ad-L1 mice compared with Ad-null mice after alcohol
feeding. The mRNA expression of autophagy-related markers atg5,
atg7 and atg12 was evaluated after alcohol feeding in both Ad-L1
and Ad-null mice. The expression of atg5 and atg7, but not that of
atg12, was significantly reduced in Ad-L1 mice compared with
Ad-null mice after alcohol feeding (Fig. 3I; P<0.01). Together, these
results indicated that NPC1L1 expression caused the inhibition of
autophagy in the livers of mice after alcohol feeding.
Ezetimibe activates autophagy in
alcohol-fed mouse hepatocytes
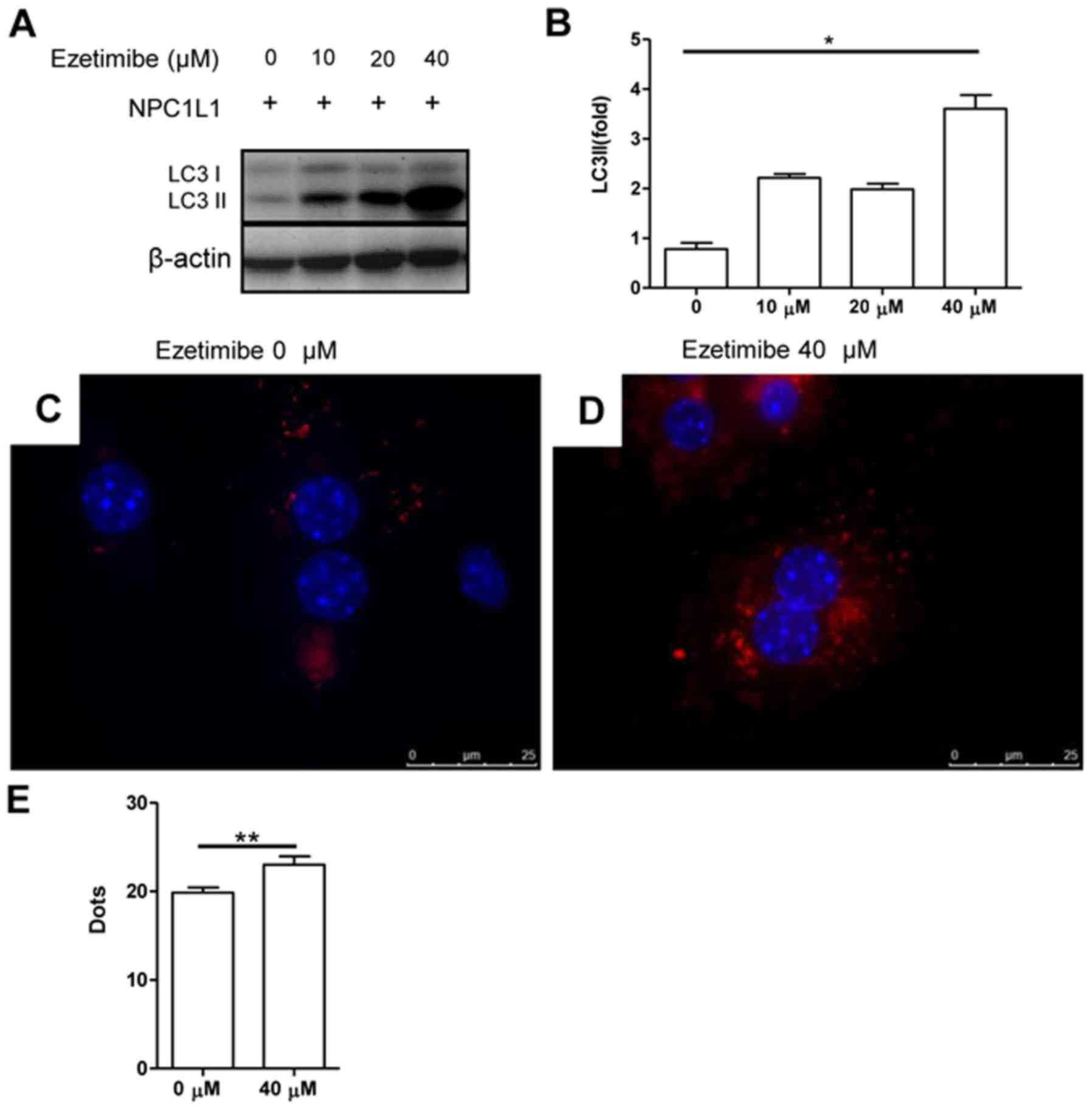
To further demonstrate that the reduction in
autophagy was due to the NPC1L1 expression, primary hepatocytes
from Ad-L1 mice were treated with different doses of ezetimibe, an
inhibitor of NPC1L1 (Fig. 4A and
B). The results indicated that LC3II protein expression was
restored in Ad-L1 mice after ezetimibe treatment. The increase in
the LC3II expression level was dose-dependent and reached its
highest level at 40 µM (Fig. 4A and
B; P<0.05). Quantification of the LC3 particles in
fluorescence images indicated an increase at 40 µM of ezetimibe
(Fig. 4C-E; P<0.01).
Discussion
Autophagy is the process of eliminating unnecessary
or toxic intracellular components, including extra lipids, through
activation of the lysosomal degradation process. Increased
understanding of autophagy in hepatocytes after alcohol abuse could
contribute to the treatment of AFLD (6). The effects of alcohol on hepatic
autophagy have been studied in mouse models (25,26).
However, the findings from these studies are not entirely
applicable to humans, because the expression of NPC1L1 is
restricted to the intestines in rodents (27). The present study investigated
hepatic autophagy in a more appropriate mouse model in which NPC1L1
was expressed in the liver, as it is in humans. It was also
demonstrated that 10 day alcohol feeding plus a single binge gavage
is an effective alcohol treatment procedure, because liver injury
and steatosis were detected via elevation of blood AST and ALT
activity, and by H&E staining. This result was consistent with
a previously reported binge model for AFLD (28,29).
Alcohol treatment resulted in a reduction of hepatic
autophagy in Ad-null mice, as supported by the results of the
fluorescence microscopy in hepatocytes and liver p62 western
blotting. The fluorescence microscopy images showed that there was
a statistically significant reduction in LC3 in the hepatocytes of
alcohol-fed mice compared with non-alcohol-fed mice. Moreover, p62
protein expression was also significantly increased in alcohol-fed
mice. These results were in accordance with the consensus on
alcoholic autophagy: That chronic alcohol treatment impairs
autophagy (30), while acute
alcohol treatment increases autophagy (5,11).
In comparison to that in Ad-null mice, the
expression of NPC1L1 in the liver led to a more pronounced
reduction in autophagy in both the non-alcohol and alcohol feeding
states. These results were confirmed by the expression of LC3 using
fluorescence microscopy and western blotting. Other autophagic
markers, p62/SQSTM1 protein expression and the mRNA level of atg5
and atg7, also indicated that hepatic autophagy was inhibited in
the livers of Ad-L1 mice. The reduction in autophagy in primary
hepatocytes of Ad-L1 mice could be restored by ezetimibe treatment.
It was concluded that the expression of NPC1L1 impaired hepatic
autophagy in Ad-L1 mice treated with chronic alcohol feeding.
A reasonable explanation for the autophagy
inhibition mediated by NPC1L1 may involve changes in cellular
cholesterol homeostasis. It is known that autophagy is highly
sensitive to variations in cellular cholesterol homeostasis, and
cholesterol-lowering drugs such as statins, cholestyramine and
ezetimibe increase autophagy in hepatocytes (9). Hepatic NPC1L1 prevents cholesterol
loss from bile in humans (15,18).
Overexpression of NPC1L1 in the liver results in free cholesterol
absorption (30) and further
impairs autolysosome clearance (9).
The experiments in human hepatocytes demonstrated
that NPC1L1 was associated with the inhibition of autophagy, and
ezetimibe treatment activated autophagy (15). To the best of our knowledge, this
was the first in vivo study to show that overexpression of
NPC1L1 in the liver impairs hepatic autophagy under alcohol feeding
conditions. The findings of the present study suggested that
activation of autophagy may be a potential therapy for AFLD via
inhibition of NPC1L1. However, long-term investigation in other
animal models, and clinical studies, are necessary before the
treatment of AFLD patients with NPC1L1 inhibition.
Acknowledgements
Not applicable.
Funding
This study was funded by the National Natural
Science Foundation of China (grant no. 31372280).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YW, PY and QL made substantial contributions to the
conception and design of the study. YW, PY, BZ, YD and SL were
involved in data analysis and interpretation. YH and XG performed
histological examination. YW, PY and QL drafted the manuscript and
revised it critically for important intellectual content. All
authors approved the final version of the manuscript and agreed to
be accountable for all aspects of the work in ensuring that
questions related to the accuracy or integrity of any part of the
work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Experimental procedures and animal use and care
protocols were in accordance with the guidelines of the Northwest
A&F University Institutional Committee for the Care and Use of
Laboratory Animals, and were approved by the Committee on Ethical
Use of Animals of Northwest A&F University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ishak KG, Zimmerman HJ and Ray MB:
Alcoholic liver disease: Pathologic, pathogenetic and clinical
aspects. Alcohol Clin Exp Res. 15:45–66. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramaiah S, Rivera C and Arteel G:
Early-phase alcoholic liver disease: An update on animal models,
pathology, and pathogenesis. Int J Toxicol. 23:217–231. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Walther TC and Farese RV Jr: Lipid
droplets and cellular lipid metabolism. Ann Rev Biochem.
81:687–714. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lass A, Zimmermann R, Haemmerle G,
Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG,
Gorkiewicz G and Zechner R: Adipose triglyceride lipase-mediated
lipolysis of cellular fat stores is activated by CGI-58 and
defective in chanarin-dorfman syndrome. Cell Metab. 3:309–319.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao
W, Lu B, Stolz DB, Clemens DL and Yin XM: Autophagy reduces acute
ethanol-induced hepatotoxicity and steatosis in mice.
Gastroenterology. 139:1740–1752. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Noh BK, Lee JK, Jun HJ, Lee JH, Jia Y,
Hoang MH, Kim JW, Park KH and Lee SJ: Restoration of autophagy by
puerarin in ethanol-treated hepatocytes via the activation of
AMP-activated protein kinase. Biochem Biophys Res Commun.
414:361–366. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thomes PG, Ehlers RA, Trambly CS, Clemens
DL, Fox HS, Tuma DJ and Donohue TM: Multilevel regulation of
autophagosome content by ethanol oxidation in HepG2 cells.
Autophagy. 9:63–73. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh R, Kaushik S, Wang Y, Xiang Y, Novak
I, Komatsu M, Tanaka K, Cuervo AM and Czaja MJ: Autophagy regulates
lipid metabolism. Nature. 458:1131–1135. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Y, Wang S, Ni HM, Huang H and Ding WX:
Autophagy in alcohol-induced multiorgan injury: Mechanisms and
potential therapeutic targets. Biomed Res Int. 2014:4984912014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rasineni K, Donohue TM Jr, Thomes PG, Yang
L, Tuma DJ, McNiven MA and Casey CA: Ethanol-induced steatosis
involves impairment of lipophagy, associated with reduced Dynamin2
activity. Hepatol Commun. 1:501–512. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ni HM, Du K, You M and Ding WX: Critical
role of FoxO3a in alcohol-induced autophagy and hepatotoxicity. Am
J Pathol. 183:1815–1825. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng J, Ohsaki Y, Tauchi-Sato K, Fujita A
and Fujimoto T: Cholesterol depletion induces autophagy. Biochem
Biophys Res Commun. 351:246–252. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Altmann SW, Davis HR, Zhu LJ, Yao X, Hoos
LM, Tetzloff G, Iyer SP, Maguire M, Golovko A, Zeng M, et al:
Niemann-Pick C1 Like 1 protein is critical for intestinal
cholesterol absorption. Science. 303:1201–1204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Davis HR Jr, Zhu LJ, Hoos LM, Tetzloff G,
Maguire M, Liu J, Yao X, Iyer SP, Lam MH, Lund EG, et al:
Niemann-Pick C1 Like 1 (NPC1L1) is the intestinal phytosterol and
cholesterol transporter and a key modulator of whole-body
cholesterol homeostasis. J Biol Chem. 279:33586–33592. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Temel RE, Tang W, Ma Y, Rudel LL,
Willingham MC, Ioannou YA, Davies JP, Nilsson LM and Yu L: Hepatic
Niemann-Pick C1-like 1 regulates biliary cholesterol concentration
and is a target of ezetimibe. J Clin Invest. 117:1968–1978. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Davis HR Jr and Altmann SW: Niemann-Pick
C1 like 1 (NPC1L1) an intestinal sterol transporter. Biochim
Biophys Acta. 1791:679–683. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Tang W, Yang P, Shin H and Li Q:
Hepatic NPC1L1 promotes hyperlipidemia in LDL receptor deficient
mice. Biochem Biophys Res Commun. 499:626–633. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang W, Jia L, Ma Y, Xie P, Haywood J,
Dawson PA, Li J and Yu L: Ezetimibe restores biliary cholesterol
excretion in mice expressing Niemann-Pick C1-like 1 only in liver.
Biochim Biophys Acta. 1811:549–555. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kurano M, Hara M, Tsuneyama K, Okamoto K,
Iso-O N, Matsushima T, Koike K and Tsukamoto K: Modulation of lipid
metabolism with the overexpression of NPC1L1 in mouse liver. J
Lipid Res. 53:2275–2285. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamamura T, Ohsaki Y, Suzuki M, Shinohara
Y, Tatematsu T, Cheng J, Okada M, Ohmiya N, Hirooka Y, Goto H and
Fujimoto T: Inhibition of niemann-pick-type C1-like1 by ezetimibe
activates autophagy in human hepatocytes and reduces mutant
α1-antitrypsin Z deposition. Hepatology. 59:1591–1599. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Simpson IA and Sonne O: A simple, rapid,
and sensitive method for measuring protein concentration in
subcellular membrane fractions prepared by sucrose density
ultracentrifugation. Anal Biochem. 119:424–427. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fischer AH, Jacobson KA, Rose J and Zeller
R: Hematoxylin and eosin staining of tissue and cell sections. CSH
Protoc 2008: pdb.prot4986. 2008.
|
|
23
|
Li WC, Ralphs KL and Tosh D: Isolation and
culture of adult mouse hepatocytes. Methods Mol Biol. 633:185–196.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ding W X, Manley S and Ni HM: The emerging
role of autophagy in alcoholic liver disease. Exp Biol Med
(Maywood). 236:546–556. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dolganiuc A, Thomes PG, Ding WX, Lemasters
JJ and Donohue TM Jr: Autophagy in alcohol-induced liver diseases.
Alcohol Clin Exp Res. 36:1301–1308. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang W, Ma Y and Yu L: Plasma cholesterol
is hyperresponsive to statin in ABCG5/ABCG8 transgenic mice.
Hepatology. 44:1259–1266. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ki SH, Park O, Zheng M, Morales-Ibanez O,
Kolls JK, Bataller R and Gao B: Interleukin-22 treatment
ameliorates alcoholic liver injury in a murine model of
chronic-binge ethanol feeding: Role of signal transducer and
activator of transcription 3. Hepatology. 52:1291–1300. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bertola A, Mathews S, Ki SH, Wang H and
Gao B: Mouse model of chronic and binge ethanol feeding (the NIAAA
model). Nat Protoc. 8:627–637. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Betters JL and Yu L: NPC1L1 and
cholesterol transport. FEBS Lett. 584:2740–2747. 2010. View Article : Google Scholar : PubMed/NCBI
|