Introduction
According to the latest statistical data from the
International Agency for Research on Cancer, there were 528,000 new
cases of cervical cancer and 266,000 associated mortalities
worldwide in 2012 (1). Cervical
cancer is the most common gynecological malignant tumor that
seriously threatens the health and life of women. The majority of
cervical cancer cases are reported in developing countries,
accounting for approximately 85% of the total number of patients
worldwide (2). Despite advances in
screening for cervical cancer, surgical techniques, radiotherapy
and chemotherapy, the efficacy of clinical treatment of advanced
and recurrent cervical cancer remains unsatisfactory.
With the development of modern molecular biology and
genomics, targeted therapy has become one of the research hotspots
for the treatment of advanced or recurrent cervical cancer
(3). During the targeted therapy
process, an effective therapeutic target enables the drug to
specifically bind to the oncogenic site, thereby promoting the
specific death of tumor cells. Thus, the discovery of therapeutic
targets has become a research focus. Currently, in addition to
coding genes, certain non-coding genes have also been identified as
effective targets for the treatment of tumors, including cervical
cancer. Accumulated studies have suggested that long non-coding
RNAs (lncRNAs) exhibit an important role in the occurrence and
progression of tumors (4,5). It has been reported that lncRNAs
regulate gene expression by affecting epigenetic, transcriptional
and post-transcriptional levels (4,5).
Several lncRNAs have also been demonstrated to be effective
therapeutic targets for cervical cancer. For instance, abnormal
upregulation of the lncRNA RP11-396F22.1 was closely associated
with poor prognosis of cervical cancer patients (6). In addition, Liu et al
(7) revealed that the lncRNA SNHG1
was upregulated in cervical cancer, resulting in enhanced tumor
cell proliferation, migration and invasion. The lncRNA HOTAIR was
also found to enhance cervical cancer cell proliferation and
migration via inhibition of microRNA (miR)-326 (8). Furthermore, reduced lncRNA GAS5 level
may be associated with poor prognosis of patients with cervical
cancer (9,10).
TP73 antisense RNA 1 (TP73-AS1) is a member of the
lncRNA family, and its expression and effects in several tumors
have been studied (11,12). However, whether TP73-AS1 regulates
cervical cancer has not been reported to date. The present study
aimed to explore the expression and influence of TP73-AS1 in
cervical cancer, and further investigated the underlying molecular
mechanisms to determine whether this lncRNA is an effective
therapeutic target and provide a theoretical basis for the
treatment of cervical cancer.
Materials and methods
Ethics statement
All patients voluntarily participated in the present
study and signed informed consent. The study was performed with the
approval of the Ethics Committee of People's Hospital of Chongqing
Hechuan.
Patients and cervical cancer
tissues
A total of 56 patients were involved in the present
study, who were first diagnosed with cervical cancer between April
2011 and August 2015, and underwent surgery at People's Hospital of
Chongqing Hechuan. During surgery, 56 tumor tissues and 56
corresponding adjacent normal tissues were obtained and stored in
liquid nitrogen. Following surgery, all patients were followed up
for 5 years, and the follow-up was completed in the case of
mortality during this 5-year period. Based on the follow-up
records, the 5-year overall survival of patients was analyzed by
Kaplan-Meier survival analysis.
Cell culture
A human cervical epithelial cell line (HCvEpC) and
two cervical cancer cell lines (HeLa and CaSki cells) were all
provided by the Type Culture Collection of the Chinese Academy of
Sciences. All cells were maintained in Dulbecco's Modified Eagle's
medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) in an
incubator with 5% CO2 at 37°C. The medium was
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.), as well as penicillin (100 U/ml) and
streptomycin (100 U/ml).
Cell transfection
Small interfering RNA (siRNA) targeting TP73-AS1,
miR-607 mimics, miR-607 inhibitors and the corresponding negative
controls were synthetized by Thermo Fisher Scientific, Inc.
(Invitrogen; Thermo Fisher Scientific, Inc.). For CCND2
overexpression, the coding sequence of the cyclin D2 (CCND2) gene
was constructed into a pcDNA3 vector (Invitrogen; Thermo Fisher
Scientific, Inc.). All transfections were performed using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol, at a
concentration of 50 nM for siRNAs or miRNAs, and 1 µg/well for the
pcDNA3 vector. After 48 h at 37°C, the transfection efficiency was
validated using reverse transcription-quantitative polymerase chain
reaction (RT-qPCR), as described later in the methods.
Cell proliferation detection by Cell
Counting Kit-8 (CCK-8) assay
The transfected cells of each group were seeded into
96-well plates for routine culture at 37°C and 5% CO2,
with each well containing 1×105 cells. After 24, 48 and
72 h of culture, 10 µl CCK-8 solution (Dojindo Molecular
Technologies, Inc.) was added to each well. After 4 h of
incubation, a microplate reader was used to detect the optical
density (OD) value of each well at a wavelength of 450 nm.
Cell migration and invasion Transwell
assays
A Transwell chamber was inserted into a 24-well
plate containing 600 µl DMEM (10% FBS) in each well. Serum-free
cell suspensions with a volume of 150 µl were added to the upper
chamber of the Transwell chamber. For the cell invasion assay, the
upper layer of the chamber was pre-coated with a thin layer of
Matrigel. For the cell migration assay, a similar experiment was
conducted, with the exception that Matrigel was not used. After 2
days, all chambers were removed and the residual liquid was
discarded. Matrigel and any remaining cells in the upper layer of
the chamber were gently wiped off with a wet cotton swab. Cells in
the lower layer of the chamber were washed twice with PBS and fixed
with 90% ethanol for 30 min. Crystal violet (0.1%) was subsequently
used to stain cells for 30 min, and the invading or migrating cells
were counted under a microscope in five random, nonoverlapping
fields of view.
Luciferase reporter gene assay
The interaction between TP71-AS1 and miR-607 was
predicted using the miRDB online database (http://mirdb.org/miRDB/index.html). The interaction
between miR-607 and CCND2 was predicted using the TargetScan
version 7.1 tool (http://www.targetscan.org/vert_71/). For the
luciferase reporter assay, mutant (Mut) and wild-type (WT)
sequences of TP73-AS1 or CCND2 were constructed into the pGL3-basic
luciferase reporter vector (Promega Corporation). Subsequently,
miR-607 mimics and reporters were transfected into HeLa cells.
After 48 h, the relative luciferase activity was measured using a
dual-luciferase reporter assay system (Promega Corportation) and
normalized to the Renilla luciferase activity.
RT-qPCR assay
TRIzol reagent (Thermo Fisher Scientific, Inc.) was
used to extract total RNA from the tissues and cells, strictly
following the manufacturer's protocol. The RNA concentration was
determined using a Nanodrop' 2000 spectrophotometer (Nanodrop
Technologies; Thermo Fisher Scientific, Inc.) A total of 1 µg RNA
samples were collected and used to perform an RT reaction with the
High Capacity cDNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). qPCR was then conducted with Fast
Start Universal SYBR Green Master (Roche Diagnostics). The qPCR
program was set as follows: 50°C for 2 min initially, followed by
95°C for 10 min, and 38 cycles of 95°C for 15 sec and 60°C for 1
min. The 2−ΔΔCq method (13) was subsequently used as the common
method for analyzing the relative gene expression. U6 and GAPDH
served as the internal reference for miRNA and mRNA detection,
respectively. Primer sequences were as follows: TP73-AS1, forward
5′-AGAAGGTGAGAGGAATGGCTACC-3′, reverse 5′-TGGACCCAGGCGAGAAGGAT-3′;
U6, forward 5′-AACGAGACGACGACAGAC-3′, reverse
5′-GCAAATTCGTGAAGCGTTCCATA-3′; miR-607, forward
5′-AACGAGACGACGACAGAC-3′, reverse 5′-GTTCAAATCCAGATCTATAAC-3′;
CCND2, forward 5′-ACCTTCCGCAGTGCTCCTA-3′, reverse
5′-CCCAGCCAAGAAACGGTCC-3′; and GAPDH, forward
5′-ATGTTGCAACCGGGAAGGAA-3′, reverse 5′-AGGAAAAGCATCACCCGGAG-3′.
Western blot analysis
Using RIPA buffer (Thermo Fisher Scientific, Inc.),
total proteins were extracted from HeLa and CaSki cells, followed
by determination of the protein concentrations using a BCA Protein
Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Next, 30 µg of
each protein sample was used to conduct sodium dodecyl
sulfate-polyacrylamide gel electrophoresis, and then the separated
proteins were transferred to a PVDF membrane. Subsequent to
blocking with 5% skimmed milk for 1 h at room temperature, the PVDF
membrane was probed with anti-CCND2 primary antibody (1:1,000; cat.
no. ab207604; Abcam) overnight at 4°C. TBS-Tween-20 was used to
wash the membrane prior to incubation with horseradish
peroxidase-conjugated goat anti-rabbit IgG secondary antibody
(1:5,000; cat. no. ab7090; Abcam). An enhanced chemiluminescence
reagent (Thermo Fisher Scientific, Inc.) was used to detect the
signals, followed by analysis using Image Lab 3.0 software (Bio-Rad
Laboratories, Inc.). With GAPDH (1:1,000; cat. no. ab9485; Abcam)
serving as the internal reference, the relative CCND2 protein
expression was determined.
Statistical analysis
All data in this study were processed using SPSS
software (version 19.0; IBM Corp.), and are expressed in the form
of the mean ± standard deviation. Student's t-test was used to
analyze differences between two groups, while one-way analysis of
variance followed by Tukey's post hoc test was used to analyze
differences among multiple groups for statistical significance.
Pearson's correlation analysis was used to analyze the expression
correlation between two genes. For survival rate analysis, the
samples were divided into the low or high expression groups for
TP73-AS1, miR-607 or CCND2 based on their median expression value.
The Kaplan-Meier method was used to draw survival curves, and the
log-rank test was used to determine statistical significance. All
experiments were repeated three times independently. P<0.05 was
set as the threshold for statistically significant differences.
Results
High expression of TP73-AS1 in
cervical cancer tissues predicts poor 5-year overall survival
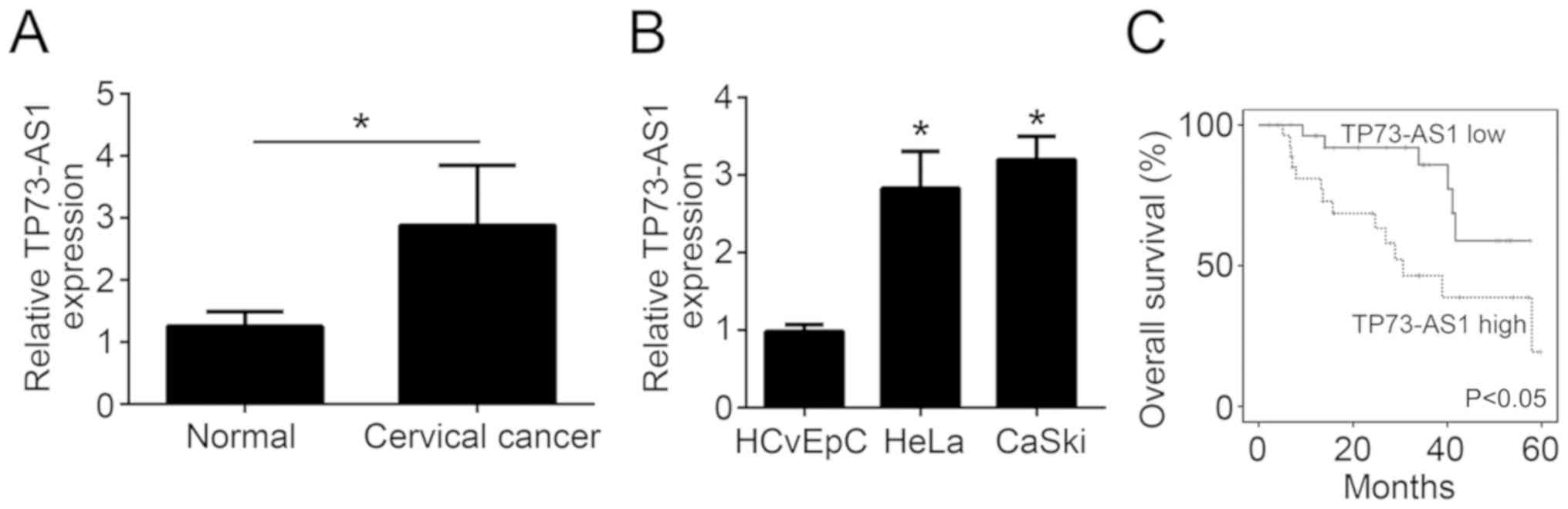
According to the results of RT-qPCR, TP73-AS1
expression was significantly increased in cervical cancer tissues
as compared with the adjacent normal tissues (P<0.05; Fig. 1A). Subsequently, the current study
further researched TP73-AS1 expression in cervical cancer cells
through in vitro experiments. Compared with the normal
HCvEpC cells, HeLa and CaSki cervical cancer cells exhibited a
significantly higher relative TP73-AS1 expression (P<0.05;
Fig. 1B). Thus, these findings
indicated that TP73-AS1 was upregulated in cervical cancer tissues
and cells. The 5-year overall survival of patients was then
analyzed by Kaplan-Meier survival analysis. As shown in Fig. 1C, patients with high TP73-AS1
expression exhibited a markedly reduced 5-year overall survival
rate in comparison with those exhibiting low TP73-AS1 expression
(P<0.05).
TP73-AS1 knockout hinders the
proliferation, migration and invasion of cervical cancer cells
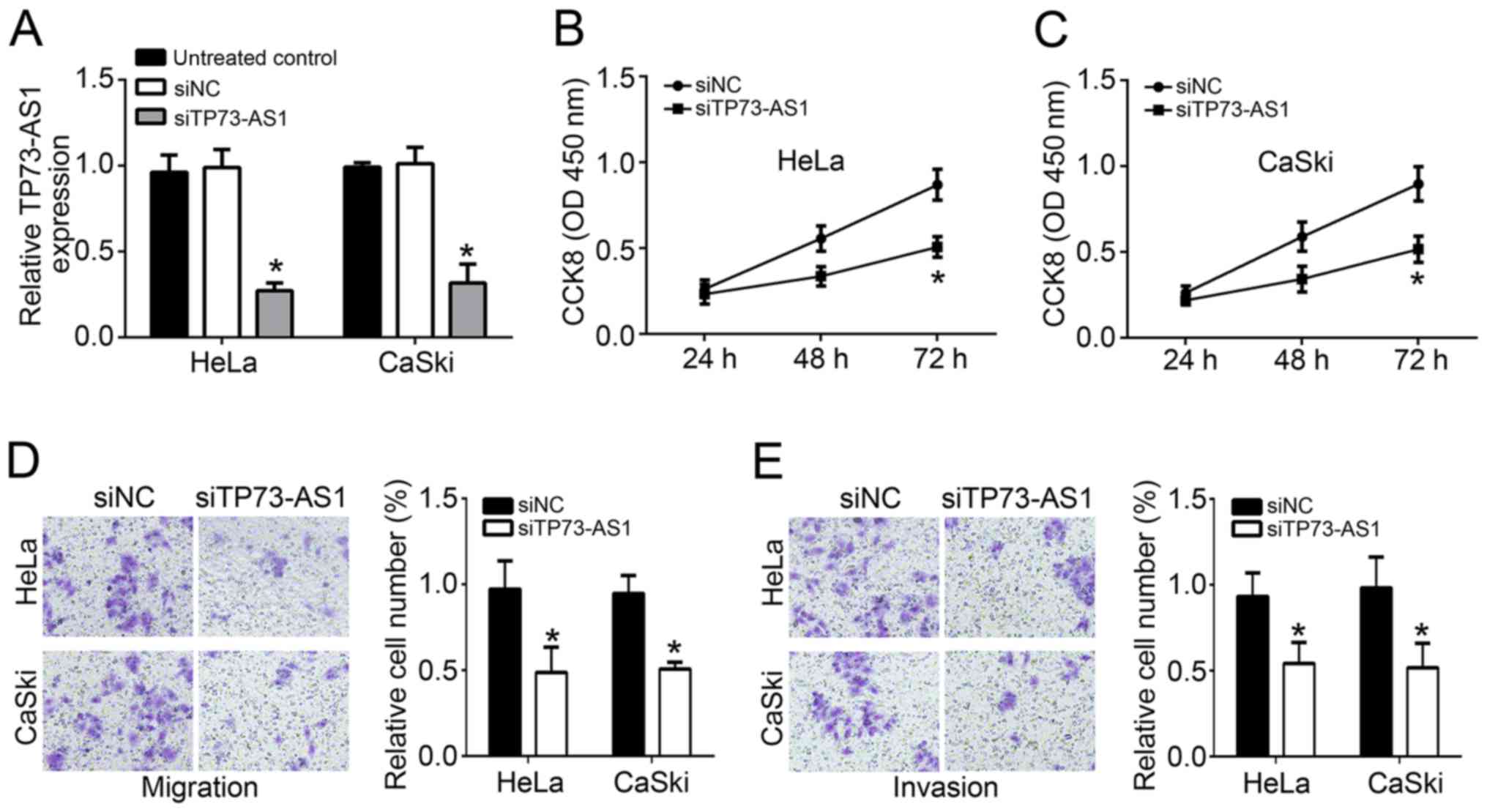
In subsequent experiments, HeLa and CaSki cells were
transfected with TP73-AS1 siRNA (siTP73-AS1 group) or TP73-AS1
siRNA negative control (siNC group). As displayed in Fig. 2A, TP73-AS1 relative expression in
cells of the siTP73-AS1 group was significantly lower compared with
that in untreated or siNC-treated cells (P<0.05), revealing that
TP73-AS1 expression in HeLa and CaSki cells was successfully
downregulated by transfection with TP73-AS1 siRNA. Following the
transfection, the proliferation ability of the two cell lines was
measured by a CCK-8 assay. Compared with the siNC group, cells in
the siTP73-AS1 group exhibited a markedly lower OD450
value at 72 h, indicating significant reduction in proliferation
(P<0.05; Fig. 2B and C).
Furthermore, according to Transwell experiments, the relative
number of migrating and invading cells in the siTP73-AS1 group also
significantly declined when compared with that in the untreated and
siNC groups (P<0.05; Fig. 2D and
E). Taken together, TP73-AS1 knockout hindered the
proliferation, migration and invasion of cervical cancer cells.
TP73-AS1 functions as a sponge for
miR-607
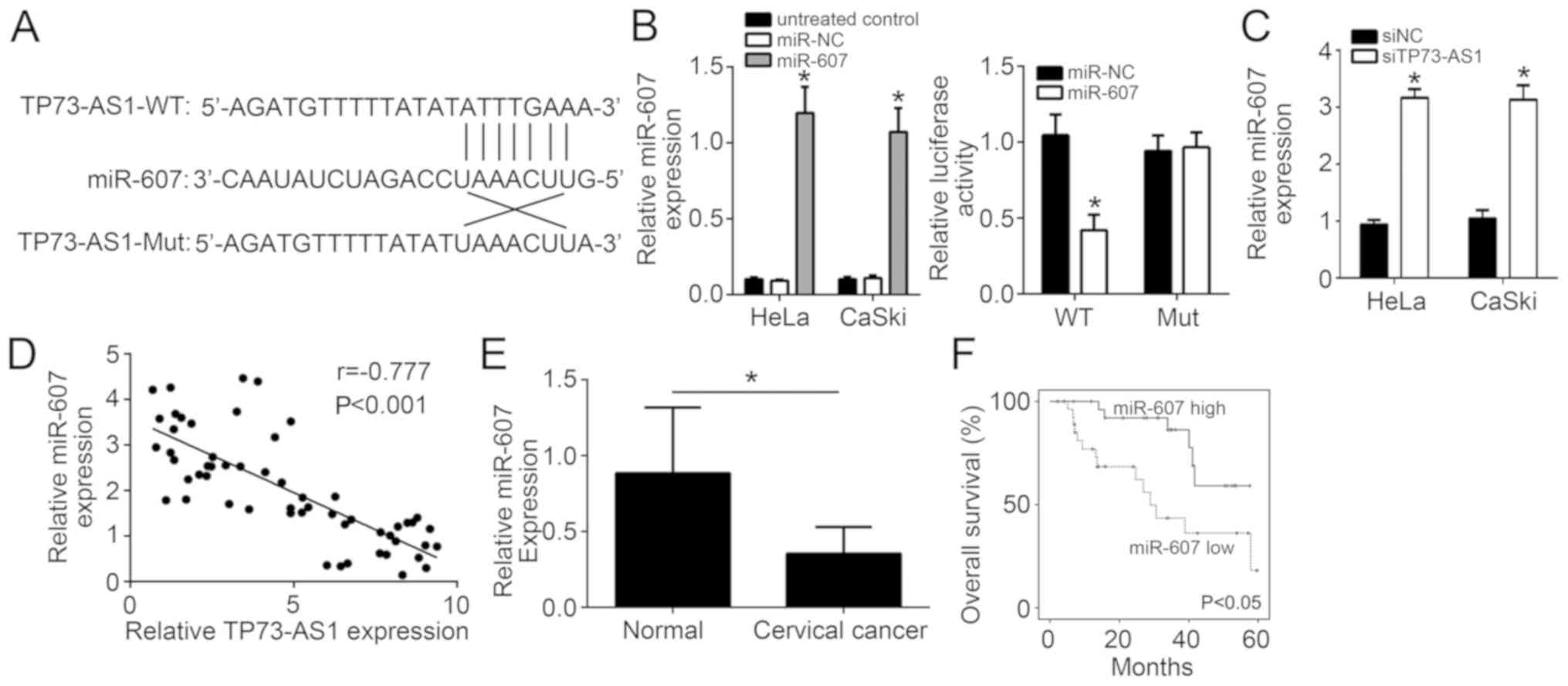
Through bioinformatics analysis, it was observed
that TP-73-AS1 may function as a sponge for miR-607, since miR-607
achieved the highest score among all candidates. In addition, among
all potential candidates, only miR-607 has previously been reported
to inhibit tumor progression (14). Thus, only miR-607 was selected in
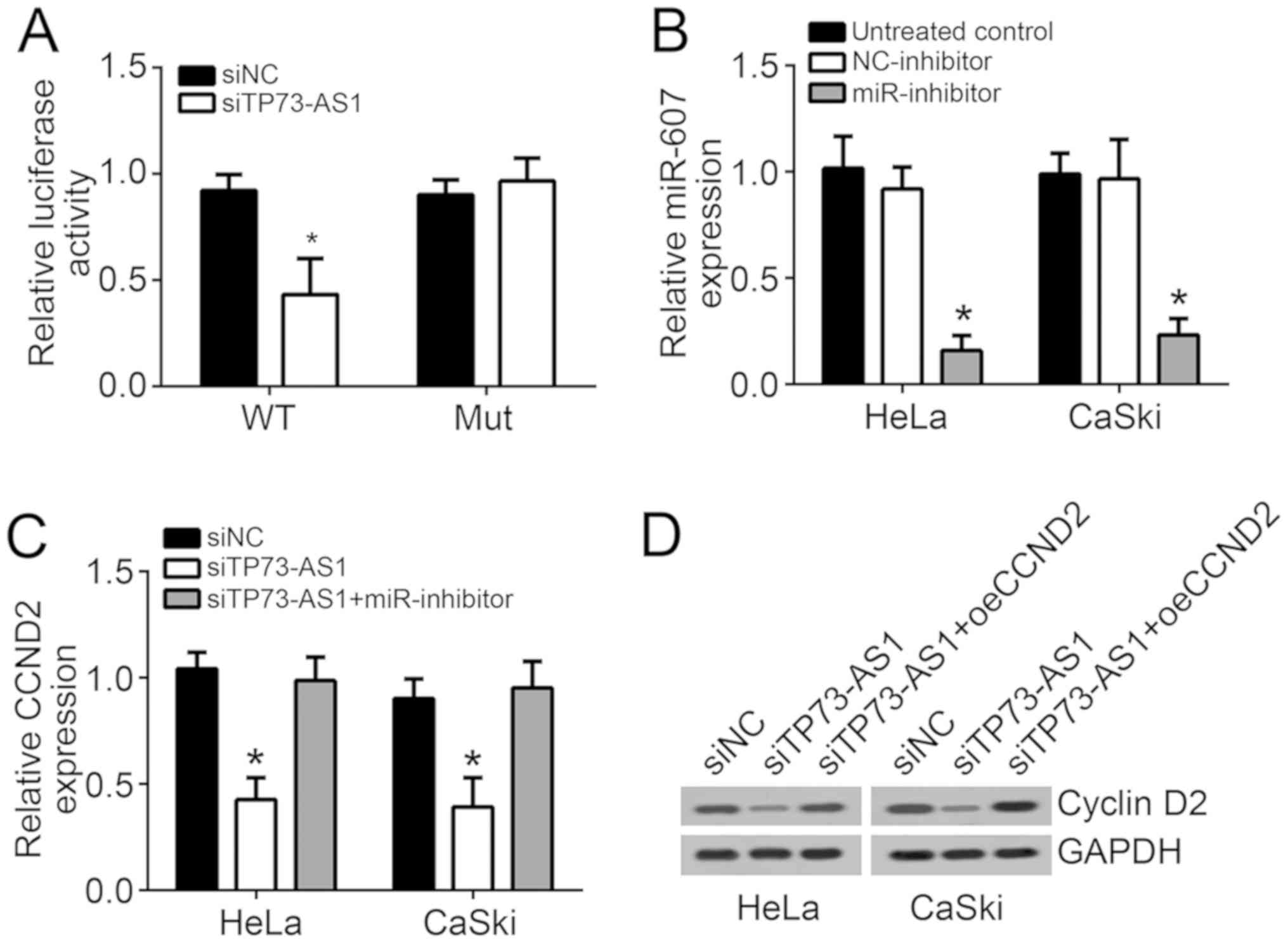
the present study for further investigation. TP73-AS1 wild-type and
mutant sequences were designed and synthesized, and the binding
sites to miR-607 are shown in Fig.
3A. Based on the results of luciferase reporter gene assay, the
insertion of the mutant TP73-AS1 sequence had little effect on the
relative luciferase activity of HeLa cells in the miR-NC and
miR-607 groups. However, the insertion of the wild-type TP73-AS1
sequence significantly decreased the relative luciferase activity
of HeLa cells in the miR-607-transfected group when compared with
that in the miR-NC group (P<0.05; Fig. 3B). At the same time, miR-607
relative expression in HeLa and CaSki cells of the siTP73-AS1 group
was significantly higher in comparison with that of the siNC group
(P<0.05; Fig. 3C). Furthermore,
there was a negative correlation between TP73-AS1 and miR-607
levels (r=−0.777; Fig. 3D).
Besides, it was observed that miR-607 expression was significantly
lower in normal tissues (Fig. 3E),
while high expression of miR-607 predicted a high survival rate for
cervical cancer patients (Fig.
3F). These data indicated that TP73-AS1 functions as a sponge
for miR-607.
miR-607 directly inhibits CCND2
expression
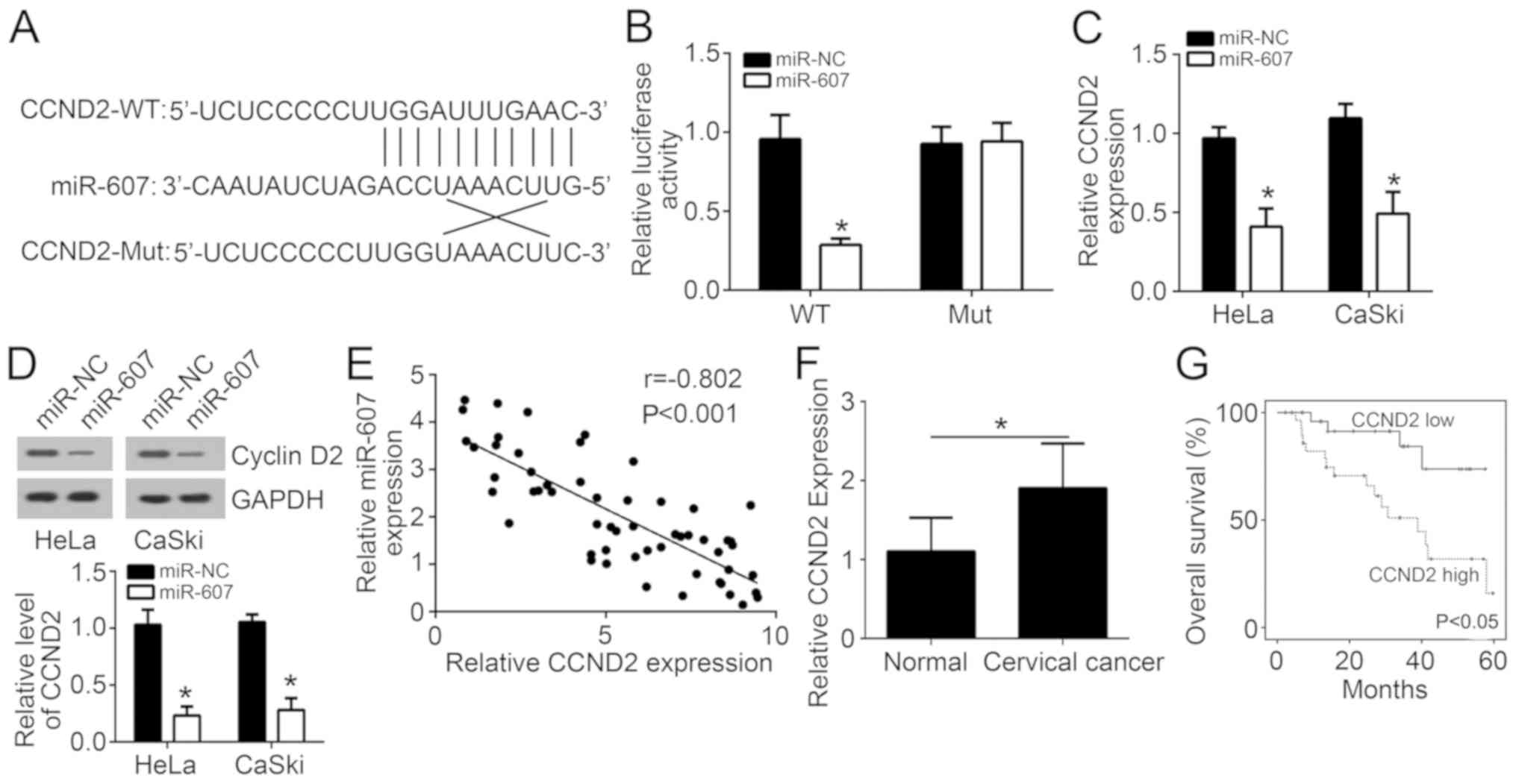
The binding sites of CCND2 and miR-607 are listed in
Fig. 4A. For HeLa cells in the
miR-NC and miR-607 groups, there was no significant difference in
relative luciferase activity following insertion of a CCND2 mutant
sequence into the cells. However, CCND2 wild-type sequence
insertion markedly reduced the relative luciferase activity of HeLa
cells in the miR-607 group as compared with that in the miR-NC
group (P<0.05; Fig. 4B),
suggesting that CCND2 expression was directly inhibited by miR-607.
Evidently lower CCND2 mRNA and protein expression levels were also
observed in HeLa and CaSki cells of the miR-607 group when compared
with the miR-NC group (P<0.05; Fig.
4C and D). Pearson's correlation analysis revealed a negative
correlation between miR-607 and CCND2 mRNA expression (r=−0.802;
Fig. 4E). Furthermore, CCND2
expression was found to be significantly higher in tumor tissues
compared with the adjacent normal tissues (Fig. 4F), while high expression of CCND2
was associated with a lower survival rate (Fig. 4G).
Downregulation of TP73-AS1 suppresses
CCND2 expression via promoting miR-607
Bioinformatics analysis was further used to search
for the potential targets of miR-607, and the data identified that
CCND2 achieved a very high score and was a classical oncogene
(15). Thus, this gene was
selected for subsequent investigation. As displayed in Fig. 5A, the insertion of the CCND2 mutant
sequence did not affect the relative luciferase activity of HeLa
cells in the siNC and siTP73-AS1 groups. However, it was noted that
CCND2 wild-type insertion significantly reduced the relative
luciferase activity of HeLa cells in the siTP73-AS1 group when
compared with that in the siNC group (P<0.05), indicating that
TP73-AS1 knockout inhibited CCND2 expression. Subsequently, miR-607
inhibitor was used, and its transfection efficiency was confirmed,
as shown in Fig. 5B. HeLa and
CaSki cells were subjected to co-transfection with TP73-AS1 siRNA
and miR-607 inhibitor (siTP73-AS1 + miR-inhibitor group). According
to Fig. 5C and D, CCND2 mRNA and
protein expression in HeLa and CaSki cells of the siTP73-AS1 group
was markedly lower compared with that in the siNC and siTP73-AS1 +
miR-inhibitor groups (P<0.05). However, no significant changes
were observed in CCND2 mRNA and protein expression levels between
the siNC and siTP73-AS1 + miR-inhibitor groups (Fig. 5C and D). These results illustrated
that downregulation of TP73-AS1 suppressed CCND2 expression via
promoting miR-607.
TP73-AS1 knockout hinders the
proliferation, migration and invasion of cervical cancer cells via
suppressing CCND2
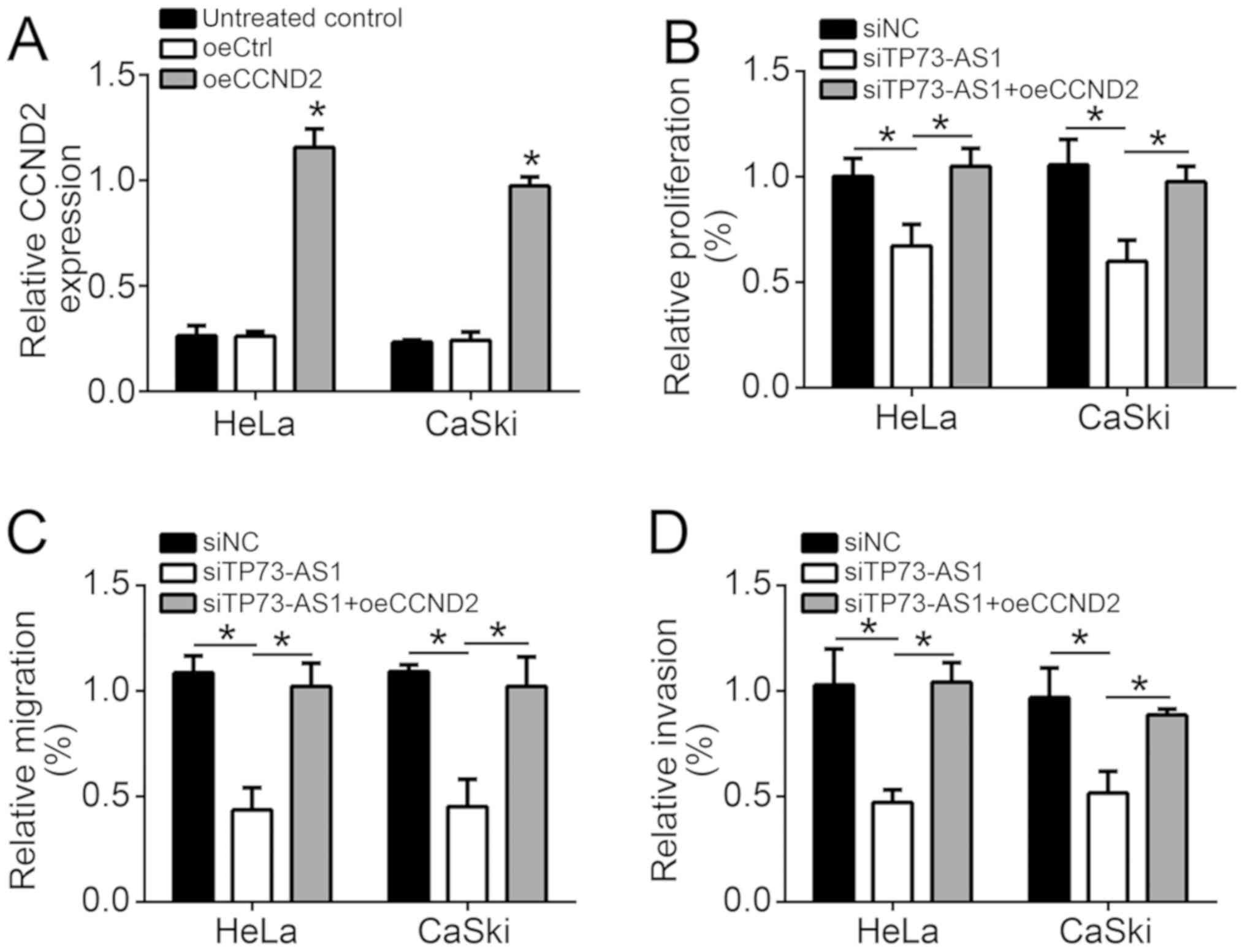
Next, CCND2 control and overexpression vectors were
used to transfect HeLa and CaSki cells, referred to as the oeCtrl
and oeCCND2 groups, respectively. Compared with the oeCtrl group,
significantly elevated CCND2 mRNA expression was observed in HeLa
and CaSki cells of the oeCCND2 group (P<0.05), revealing that
HeLa and CaSki cells were successfully transfected (Fig. 6A). Co-transfection with TP73-AS1
siRNA and CCND2 overexpression vectors was also performed in HeLa
and CaSki cells (siTP73-AS1 + oeCCND2 group). HeLa and CaSki cells
in the siTP73-AS1 group exhibited significantly lower relative
proliferation, migration and invasion in comparison with those in
the siNC and siTP73-AS1 + oeCCND2 groups (P<0.05; Fig. 6B-D). However, no evident difference
was identified in the relative proliferation, migration and
invasion of cells in the siTP73-AS1 + oeCCND2 group as compared
with the siNC group (Fig.
6B-D).
Discussion
In the present study, it was observed that TP73-AS1
was aberrantly overexpressed in cervical cancer tissues and cells,
which promoted cervical cancer progression by promoting CCND2
through the suppression of miR-607 expression. Researchers have
recently reported that lncRNAs are closely associated with
tumorigenesis (16). In patients
with brain glioma, high TP73-AS1 expression was correlated with
poor prognosis, and TP73-AS1 functioned as a carcinogenic lncRNA in
this tumor, contributing to brain glioma cell invasion and
proliferation (11). It was also
demonstrated that downregulated TP73-AS1 inhibited breast cancer
cell proliferation (12). A study
by Li et al (17) revealed
that ovarian cancer patients with higher TP73-AS1 expression
exhibited lower survival when compared with those with lower
TP73-AS1 expression. To the best of our knowledge, no study in the
literature has thus far defined the role of TP73-AS1 in cervical
cancer. Therefore, the present study is the first to discover that
TP73-AS1 was upregulated in cervical cancer and promoted tumor
progression by upregulating CCND2 through the inhibition of
miR-607.
miRNAs are a class of non-coding small RNAs with a
length of approximately 19–23 nucleotides, which are important
post-transcriptional regulators in vivo and widely involved
in various biological behaviors of cells (18). Studies have identified that miRNAs
are closely associated with the biological processes of cervical
cancer cells, such as proliferation, apoptosis, invasion and
angiogenesis, which are considered to be new targets for the
diagnosis, treatment and prognosis of this tumor (19). Several miRNAs exert a carcinogenic
role in cervical cancer, such as miR-92, miR-150, miR-21, miR-494
and miR-155, among others (20–24).
In addition, certain miRNAs serve as tumor suppressor in cervical
cancer, including miR-138, miR-195, miR-362, miR-218, miR-744 and
so on (25–29). The main factors associated with
cervical cancer prognosis include age, pathological type, FIGO
stage and lymph node metastasis (24,30,31).
Recent studies have identified that miRNA expression levels are
closely correlated with the FIGO stage, tumor differentiation,
human papillomavirus (HPV) infection and lymph node metastasis, and
may thus be used as prognostic indicators and independent
prognostic factors for cervical cancer (24,30,31).
CCND2 is a cell cycle-associated gene that encodes
the CCND2 protein. Abnormal cell cycle is an important early event
leading to cervical cancer. In a previous study, researchers
reported that CCND2, an important factor in regulating the
transition from G1 to S phase in the cell cycle, was abnormally
overexpressed in several tumors, including in cervical cancer
(15). CCND2 protein can bind to
cyclin-dependent kinase 4 (CDK4) and CDK6 to form a Cyclin/CDK
complex. This complex then facilitates a series of processes that
ultimately lead to tumor cells entering the DNA synthesis phase
(32). Carcinogenic factors may
cause abnormal expression of CCND2 protein, which leads to
uncontrolled cell cycle, infinite proliferation of cells and loss
of apoptotic ability, and ultimately promotes cell malignant
transformation and tumor formation (33). The current study results indicated
that CCND2 transcription and translation in cervical cancer was
aberrantly activated by upregulated TP73-AS1, which enhanced the
progression of this tumor.
However, the current study also has several
limitations. For example, the correlation of TP73-AS1 expression
with the clinical stage or HPV status of patients could not be
examined. Furthermore, the effect of TP73-AS1 on cell cycle
progression was not analyzed. Additionally, whether overexpression
of TP73-AS1 in the normal cell line HCvEpC was able to induce a
cancer-like phenotype was not investigated in the present
manuscript.
In conclusion, the present article first
demonstrated that TP73-AS1 was abnormally overexpressed in cervical
cancer, which promoted cervical cancer progression by promoting
CCND2 via the suppression of miR-607 expression. Thus, TP73-AS1 may
be considered as a novel target for cervical cancer treatment.
Acknowledgements
Not applicable.
Funding
This study was supported by the Youth Fund of Jining
Medical College (grant no. JYQ2011KM055).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HZ and TF initiated and designed the study, analyzed
and interpreted the results, and wrote the manuscript. HZ and BX
participated in all the experiments. SW and XL performed western
blotting. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Jining No. 1 People's Hospital, and all
enrolled patients signed a written informed consent document.
Patient consent for publication
All patients included in this study provided consent
for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Freitas AC, Gomes Leitão Mda C and
Coimbra EC: Prospects of molecularly-targeted therapies for
cervical cancer treatment. Curr Drug Targets. 16:77–91. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang Q, Huang H, Gong Z, Xiong W, Zeng Z
and Li G: Advances in regulation of gene expression mediated by
lncRNAs. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 39:91–95. 2014.(In
Chinese). PubMed/NCBI
|
|
5
|
Liu W, Ma R and Yuan Y:
Post-transcriptional regulation of genes related to biological
behaviors of gastric cancer by long noncoding RNAs and MicroRNAs. J
Cancer. 8:4141–4154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao Y, Huang J, Liu T, He S, Shang C, Guo
L, Du Q and Yao S: Overexpression of long non-coding RNA
RP11-396F22.1 correlates poor prognosis of patients with
early-stage cervical cancer. Am J Transl Res. 10:684–695.
2018.PubMed/NCBI
|
|
7
|
Liu Y, Yang Y, Li L, Liu Y, Geng P, Li G
and Song H: lncRNA SNHG1 enhances cell proliferation, migration,
and invasion in cervical cancer. Biochem Cell Biol. 96:38–43. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu X, Cao X and Chen F: WITHDRAWN:
lncRNA-HOTAIR activates tumor cell proliferation and migration by
suppressing MiR-326 in cervical cancer. Oncol Res. Aug
31–2017.(Epub ahead of print). View Article : Google Scholar :
|
|
9
|
Cao S, Liu W, Li F, Zhao W and Qin C:
Decreased expression of lncRNA GAS5 predicts a poor prognosis in
cervical cancer. Int J Clin Exp Pathol. 7:6776–6783.
2014.PubMed/NCBI
|
|
10
|
Zhu FY, Chen MX, Ye NH, Shi L, Ma KL, Yang
JF, Cao YY, Zhang Y, Yoshida T, Fernie AR, et al: Proteogenomic
analysis reveals alternative splicing and translation as part of
the abscisic acid response in Arabidopsis seedlings. Plant J.
91:518–533. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang R, Jin H and Lou F: The long
non-coding RNA TP73-AS1 interacted with miR-142 to modulate brain
glioma growth through HMGB1/RAGE pathway. J Cell Biochem.
119:3007–3016. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zou Q, Zhou E, Xu F, Zhang D, Yi W and Yao
J: A TP73-AS1/miR-200a/ZEB1 regulating loop promotes breast cancer
cell invasion and migration. J Cell Biochem. 119:2189–2199. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xia L, Wu L, Bao J, Li Q, Chen X, Xia H
and Xia R: Circular RNA circ-CBFB promotes proliferation and
inhibits apoptosis in chronic lymphocytic leukemia through
regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem Biophys Res
Commun. 503:385–390. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du X, Lin LI, Zhang L and Jiang J:
microRNA-195 inhibits the proliferation, migration and invasion of
cervical cancer cells via the inhibition of CCND2 and MYB
expression. Oncol Lett. 10:2639–2643. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao XL, Zhao ZH, Xu WC, Hou JQ and Du XY:
Increased expression of SPRY4-IT1 predicts poor prognosis and
promotes tumor growth and metastasis in bladder cancer. Int J Clin
Exp Pathol. 8:1954–1960. 2015.PubMed/NCBI
|
|
17
|
Li X, Wang X, Mao L, Zhao S and Wei H:
lncRNA TP73-AS1 predicts poor prognosis and promotes cell
proliferation in ovarian cancer via cell cycle and apoptosis
regulation. Mol Med Rep. 18:516–522. 2018.PubMed/NCBI
|
|
18
|
Fan X, Chen W, Fu Z, Zeng L, Yin Y and
Yuan H: MicroRNAs, a subpopulation of regulators, are involved in
breast cancer progression through regulating breast cancer stem
cells. Oncol Lett. 14:5069–5076. 2017.PubMed/NCBI
|
|
19
|
González-Quintana V, Palma-Berré L,
Campos-Parra AD, López-Urrutia E, Peralta-Zaragoza O, Vazquez-Romo
R and Pérez-Plasencia C: MicroRNAs are involved in cervical cancer
development, progression, clinical outcome and improvement
treatment response (Review). Oncol Rep. 35:3–12. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Su Z, Yang H, Zhao M, Wang Y, Deng G and
Chen R: MicroRNA-92a promotes cell proliferation in cervical cancer
via inhibiting p21 expression and promoting cell cycle progression.
Oncol Res. 25:137–145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Z, Wang J, Li J, Wang X and Song W:
MicroRNA-150 promotes cell proliferation, migration, and invasion
of cervical cancer through targeting PDCD4. Biomed Pharmacother.
97:511–517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Z, Wang J, Wang X, Song W, Shi Y and
Zhang L: MicroRNA-21 promotes proliferation, migration, and
invasion of cervical cancer through targeting TIMP3. Arch Gynecol
Obstet. 297:433–442. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang YK, Xi WY, Xi RX, Li JY, Li Q and Gao
YE: MicroRNA-494 promotes cervical cancer proliferation through the
regulation of PTEN. Oncol Rep. 33:2393–2401. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang H, Shuang D, Yi Z, Sheng H and Liu Y:
Up-regulated microRNA-155 expression is associated with poor
prognosis in cervical cancer patients. Biomed Pharmacother.
83:64–69. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin YF, Li LH, Lin CH, Tsou MH, Chuang MT,
Wu KM, Liao TL, Li JC, Wang WJ, Tomita A, et al: Selective
retention of an inactive allele of the DKK2 tumor suppressor gene
in hepatocellular carcinoma. PLoS Genet. 12:e10060512016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang N, Wei H, Yin D, Lu Y, Zhang Y, Zhang
Q, Ma X and Zhang S: MicroRNA-195 inhibits proliferation of
cervical cancer cells by targeting cyclin D1a. Tumour Biol.
37:4711–4720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shi C and Zhang Z: MicroRNA-362 is
downregulated in cervical cancer and inhibits cell proliferation,
migration and invasion by directly targeting SIX1. Oncol Rep.
37:501–509. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang Z, Song Q, Zeng R, Li J, Li J, Lin
X, Chen X, Zhang J and Zheng Y: MicroRNA-218 inhibits EMT,
migration and invasion by targeting SFMBT1 and DCUN1D1 in cervical
cancer. Oncotarget. 7:45622–45636. 2016.PubMed/NCBI
|
|
29
|
Chen CL, Wang Y, Pan QZ, Tang Y, Wang QJ,
Pan K, Huang LX, He J, Zhao JJ, Jiang SS, et al:
Bromodomain-containing protein 7 (BRD7) as a potential tumor
suppressor in hepatocellular carcinoma. Oncotarget. 7:16248–16261.
2016.PubMed/NCBI
|
|
30
|
Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y
and Mu N: MicoRNA-425-5p is a potential prognostic biomarker for
cervical cancer. Ann Clin Biochem. 54:127–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang Y, Song KL, Chang H and Chen L:
Decreased expression of microRNA-126 is associated with poor
prognosis in patients with cervical cancer. Diagn Pathol.
9:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Deshpande A, Sicinski P and Hinds PW:
Cyclins and cdks in development and cancer: A perspective.
Oncogene. 24:2909–2915. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ely S, Di Liberto M, Niesvizky R, Baughn
LB, Cho HJ, Hatada EN, Knowles DM, Lane J and Chenkiang S: Mutually
exclusive cyclin-dependent kinase 4/cyclin D1 and cyclin-dependent
kinase 6/cyclin D2 pairing inactivates retinoblastoma protein and
promotes cell cycle dysregulation in multiple myeloma. Cancer Res.
65:11345–11353. 2005. View Article : Google Scholar : PubMed/NCBI
|