Introduction
Liver cancer is a one of the most common types of
cancer with high morbidity and mortality rates, particularly in
China (1,2). Therapeutic options for liver cancer
are stage-dependent (3). Liver
cancer treatment is challenging since more than two-thirds of
patients are diagnosed at advanced stages with local or distant
metastasis and poor liver function caused by underlying cirrhosis
(4,5). Considerable progress in understanding
the molecular biology of liver cancer has been made in recent years
(6). However, few effective
treatments are available for advanced liver cancer. Therefore,
efficient biomarkers are essential for improving the diagnosis and
treatment of liver cancer.
MicroRNAs (miRNAs) are small non-coding RNAs
approximately 19–23 nucleotides in length and are associated with
cell proliferation, development and apoptosis (7). Generally, miRNAs can serve oncogene
or tumor suppressor roles by repressing translation or inducing
degradation of target messenger RNAs. For example, miR-744
expression is positively associated with poor survival of
pancreatic cancer patients (8).
miR-126 can inhibit non-small-cell lung carcinoma progression in
vitro and in vivo by targeting the epidermal growth
factor-like domain 7 (9). miR-29b
and miR-195 suppress liver cancer angiogenesis, metastasis and
invasion (10,11), whereas miR-10b initiates cancer
cell invasion and metastasis in breast cancer (12). These data suggest that miRNAs are
potential markers for targeted therapies in cancer.
miR-373 is transcribed from the location on
chromosome 19q13.4 (13). Previous
studies have indicated the potential prognostic role of miR-373 in
cancers, including gliomas (14),
bladder (15), and breast cancer
(16). However, the association
between miR-373 expression and liver cancer progression remains to
be elucidated. The present study focused on the role of miR-373 in
liver cancer development. The prognostic value of miR-373 in liver
cancer and its influence in cancer cell growth and metastasis were
investigated. The results indicated that miR-373 may serve as a
novel anti-cancer target for liver cancer treatment.
Materials and methods
Patients and tissue samples
A total of 96 human liver cancer specimens and the
matched adjacent normal tissue samples were obtained from the
Seventh People's Hospital (Shanghai, China) (Table I). The diagnosis of all patients
(enrolled between January 2002 and January 2012) was based on
histological examination. All patients underwent surgical
resections. Additionally, follow-up data were available for all
patients. Tumor stages were determined according to 2007 World
Health Organization (WHO) classification (17). All patients included in the present
study signed written informed consents, and collection of patients'
samples was authorized by the Ethical Committee for Clinical
Research of the Seventh People's Hospital (Shanghai, China).
 | Table I.Correlations between miR-373 and
clinicopathological factors. |
Table I.
Correlations between miR-373 and
clinicopathological factors.
|
|
|
| miR-373 |
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
factor | Number | Percentage | Low | High |
P-valuea |
|---|
| Age (years) |
|
|
|
|
|
|
<50 | 58 | 60.4 | 33 | 25 | 0.384 |
|
≥50 | 38 | 39.6 | 25 | 13 |
|
| Sex |
|
|
|
|
|
|
Female | 26 | 27.1 | 18 | 8 | 0.282 |
|
Male | 70 | 72.9 | 40 | 30 |
|
| AFP (U/ml) |
|
|
|
|
|
|
<400 | 34 | 35.4 | 19 | 15 | 0.501 |
|
≥400 | 62 | 64.6 | 39 | 23 |
|
| Tumor number |
|
|
|
|
|
|
Single | 67 | 69.8 | 33 | 34 | 0.001b |
|
Multiple | 29 | 30.2 | 25 | 4 |
|
| Tumor size
(cm) |
|
|
|
|
|
| ≤5 | 46 | 47.9 | 19 | 27 |
<0.001b |
|
>5 | 50 | 52.1 | 39 | 11 |
|
| Tumor location |
|
|
|
|
|
|
Unilateral | 85 | 88.5 | 51 | 34 | 0.816 |
|
Bilateral | 11 | 11.5 | 7 | 4 |
|
| Histopathological
grade |
|
|
|
|
|
|
Well/moderate | 46 | 47.9 | 23 | 23 | 0.045b |
|
Poor/undifferentiated | 50 | 52.1 | 35 | 15 |
|
| Surgical margin
(mm) |
|
|
|
|
|
|
≤10 | 50 | 52.1 | 27 | 23 | 0.180 |
|
>10 | 46 | 47.9 | 31 | 15 |
|
| TNM stage |
|
|
|
|
|
|
I/II | 47 | 49.0 | 20 | 27 |
<0.001b |
|
III/IV | 49 | 51.0 | 38 | 11 |
|
Cell culture and transfection
Human liver cancer cells (Hep3B and HepG2) were
obtained from the Cell Bank of the Chinese Academy of Sciences
(Beijing, China) and maintained with RPMI-1640 containing 10% fetal
bovine serum (BSA; both from HyClone; GE Healthcare Life Sciences,
Logan, UT, USA), 1% streptomycin and penicillin (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and incubated in a humidified
atmosphere of 5% CO2 at 37°C. The miR-373 mimics and
negative control (NC) used in the present study were produced by
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The sequence of
miR-373 mimics was 5′-GAAGUGCUUCGAUUUUGGGGUGU-3′. The NC sequence
was 5′-UGGGCGUAUAGACGUGUUACAC-3′. Both miRNAs were used at a final
concentration of 50 nM. The transfections were carried out using
Lipofectamine® 2000 in accordance with the
manufacturer's protocols (Invitrogen; Thermo Fisher Scientific,
Inc.). Cells were harvested after 48 h for further experiments.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from tissues (cell density,
106 cells/ml) using TRIzol reagent (Life Technologies;
Thermo Fisher Scientific, Inc.). The absorbance at 260 and 280 nm
was used to validate RNA purity and concentration using a
spectrophotometer (Nanodrop 2000; Thermo Fisher Scientific, Inc.,
Wilmington, DE, USA). Total miRNA was synthesized using miRNA First
Strand cDNA Synthesis (Tailing Reaction) kit supplied by Sangon
Biotech Co., Ltd. (Shanghai, China) following the manufacturer's
protocols (18). RT-qPCR was
performed with SYBR Premix Ex Taq II (Takara Biotechnology Co.,
Ltd., Dalian, China) to detect the expression of mature miRNAs. The
qPCR reactions were performed with an initial denaturation at 95°C
for 30 sec, followed by 40 cycles of 95°C for 5 sec, and 60°C for
35 sec using the ABI 7500 real-time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The 2−∆∆Cq method was
applied to calculate the transcription level of miR-373 with U6
small nuclear RNA as the normalization reference gene (19). Primers used were: miR-373 forward,
5′-GTCGTATCCAGTGCAGGGTCCGAGGT-3′; U6 forward,
5′-CGAATTTGCGTGTCATCCT-3′. The universal PCR reverse primer was
provided by the cDNA Synthesis kit. The experiments were repeated
three times.
Cell proliferation assay
Hep3B and HepG2 cells were reseeded on 96-well
plates at 3×103 cells/well following transfection with
miRNA mimics for 24 h. The effect of miR-373 on the proliferation
of liver cancer cells was evaluated by MTT assay at a daily
interval for 4 days; 20 µl of 5 mg/ml MTT was added to each well
and incubated for 4 h at each time point. The medium was discarded,
and the precipitated formazan was dissolved in 150 µl DMSO.
Absorbance was measured at 450 nm using a 96-well plate reader
(Thermo Fisher Scientific, Inc.).
Transwell invasion assay
Invasion assays were performed using Transwell
chambers (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) (20). Cells (1×105) were
re-suspended and added in the upper chamber in serum-free RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.). Then, 20% fetal
calf serum (Gibco; Thermo Fisher Scientific, Inc.) was added to the
lower chamber. The membranes were precoated with Matrigel for 1 h
at 37°C. Following invasion across the Matrigel-coated membranes
for 48 h, invasive cells were observed by 0.5% crystal violet
staining for 10 min at room temperature.
Plasmids, transfection, and luciferase
activity assays
The Ras-related protein Rab22a plasmid was purchased
from Addgene (Cambridge, MA, USA) and subcloned into pGL3
luciferase vector, the mutant of Rab22a was constructed by
site-specific mutagenesis strategy as described by others (21).
Hep3B cells were seeded into a 24-well plate at the
density of 1×104 cells/well. Cells were transfected with
miR-373 mimics and Rab22a plasmids using Lipofectamine®
2000 in accordance with the manufacturer's protocols (Invitrogen;
Thermo Fisher Scientific, Inc.). Briefly, a blank vector was
introduced as the control of miR-373, which was also co-transfected
with either Rab22a-WT (wild-type) or Rab22a-mutant plasmids
containing firefly luciferase. The value of relative luciferase
activity was evaluated using a dual luciferase assay kit (Promega
Corporation, Madison, WI, USA) (22). For each well, luciferase activity
was normalized to Renilla luciferase activity.
Western blot analysis
Immunoblotting was conducted to test protein levels
as described elsewhere (23).
Briefly, cultured cells were lysed using RIPA buffer supplemented
with protease inhibitor cocktails (Roche Diagnostics, Indianapolis,
IN, USA). Following the determination of the extracted protein
concentration by a BCA kit, 20 µg of total proteins was subjected
to 12% SDS-PAGE and transferred onto nitrocellulose membranes.
Following blocking with 5% BSA (Sigma-Aldrich; Merck KGaA) in
tris-buffered saline/Tween (0.05%) buffer for 1 h at room
temperature, the membranes were incubated with primary antibodies
(dilution, 1:1,000) at 4°C overnight. The antibodies were purchased
from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA), including
Rab22a (cat. no. sc-390726), E-cadherin (cat. no. sc-71009), and
β-actin (cat. no. sc-517582). Corresponding horseradish peroxidase
(HRP)-conjugated goat anti-mouse secondary antibody (cat. no.
sc-2039; Santa Cruz Biotechnology, Inc.; 1:10,000 dilution) were
added and incubated for another 1 h at room temperature. The
immunoreactivity was detected using the Pierce enhanced
electrochemiluminescence western blotting substrate (Thermo Fisher
Scientific, Inc.) and X-ray film. Data were analyzed using ImageJ
Software (National Institutes of Health, Bethesda, MD, USA).
β-actin was used as the loading control. Each experiment was
performed three times.
Target prediction
TargetScan tool (version 7.1; www.targetscan.org) was used to investigate the
potential targets of miR-373. Rab22a gene was one of the predicted
targets and was selected for further analysis. Subsequently, the
predicted miRNAs targeting Rab22a were investigated using
TargetScan (version 7.1) and various miRNAs were identified,
including miR-373-3p.
Statistical analysis
Statistical analyses were performed by SPSS software
version 18.0 (SPSS, Inc., Chicago, IL, USA). The data were
presented as mean ± standard deviation from ≥3 separate
experiments. Difference among two groups was compared by Student's
t-test (two-tailed). Comparison between multiple groups was
performed by one-way analysis of variance followed by least
significant difference post hoc test. The prognosis of patients was
evaluated by the Kaplan-Meier method and log-rank test, and
multivariate analysis was conducted to test potential clinical
variables using Cox regression test. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-373 transcription level is
downregulated in liver cancer tumors
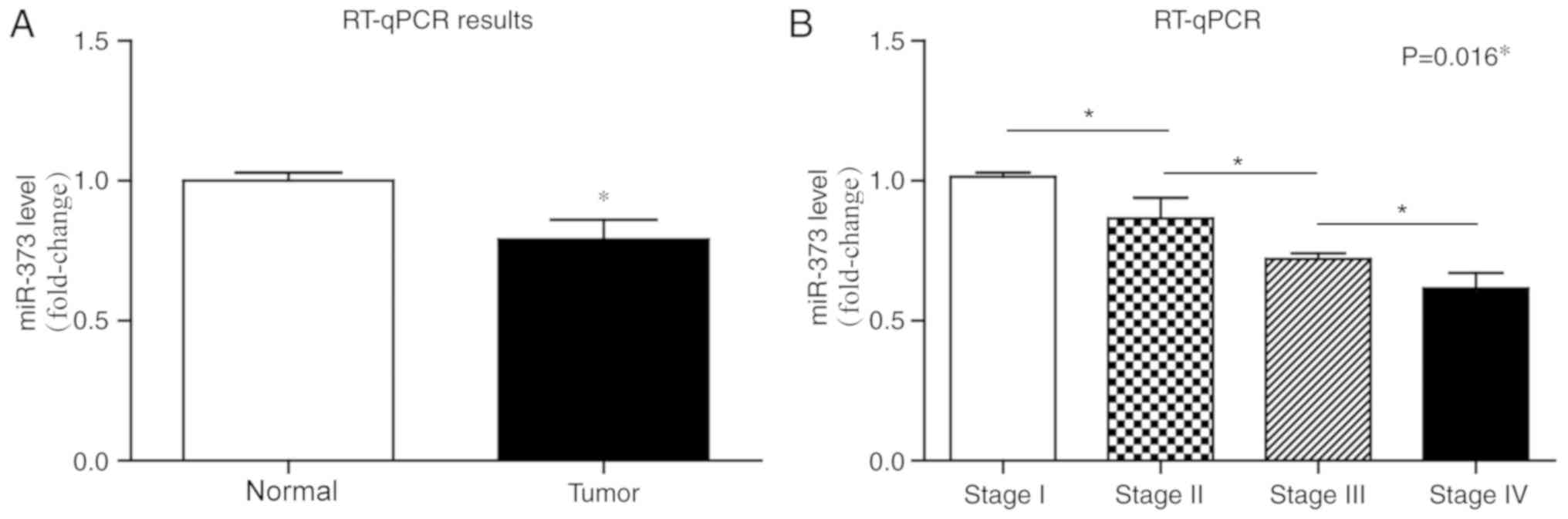
RT-qPCR was performed to determine the miR-373
expression in liver cancer and adjacent normal tissues. miR-373
transcription level was significantly downregulated in 96 liver
cancer tissues compared with the adjacent tissues (Fig. 1A; P<0.05). In addition, the
expression of miR-373 was negatively associated with cancer stages
(Fig. 1B).
Correlation between miR-373 expression
and clinicopathological factors in liver cancer patients
As shown in Table
I, all 96 liver cancer patients were grouped in high- or
low-miR-373 groups based on the median value of miR-373 level in
tumor tissues. miR-373 level was significantly associated with
tumor number (P=0.001), tumor size (P<0.001), histopathological
grade (P=0.045) and TNM stage (P<0.001), but was not correlated
to age (P=0.384), sex (P=0.282), α-fetoprotein (AFP; P=0.501),
tumor location (P=0.816) or surgical margin (P=0.180).
Prognostic value of miR-373 for liver
cancer patients
Survival analysis was performed to test whether
clinicopathological factors and miR-373 expression level were
associated with liver cancer prognosis. As shown in Fig. 2 and Table II, according to a univariate
analysis, tumor number (P=0.014), surgical margin (P<0.001), TNM
stage (P<0.001) and miR-373 level (P<0.001) were all
significant prognostic factors. Additionally, Cox multivariate
analysis indicated that surgical margin [hazard risk (HR)=3.1, 95%
confidence interval (CI)=1.52–6.43, P=0.002], TNM stage (HR=3.2,
95% CI=1.24–8.22, P=0.016) and miR-373 level (HR=0.43, 95%
CI=0.17–0.94, P=0.048) were independent prognostic indicators for
liver cancer (Table III).
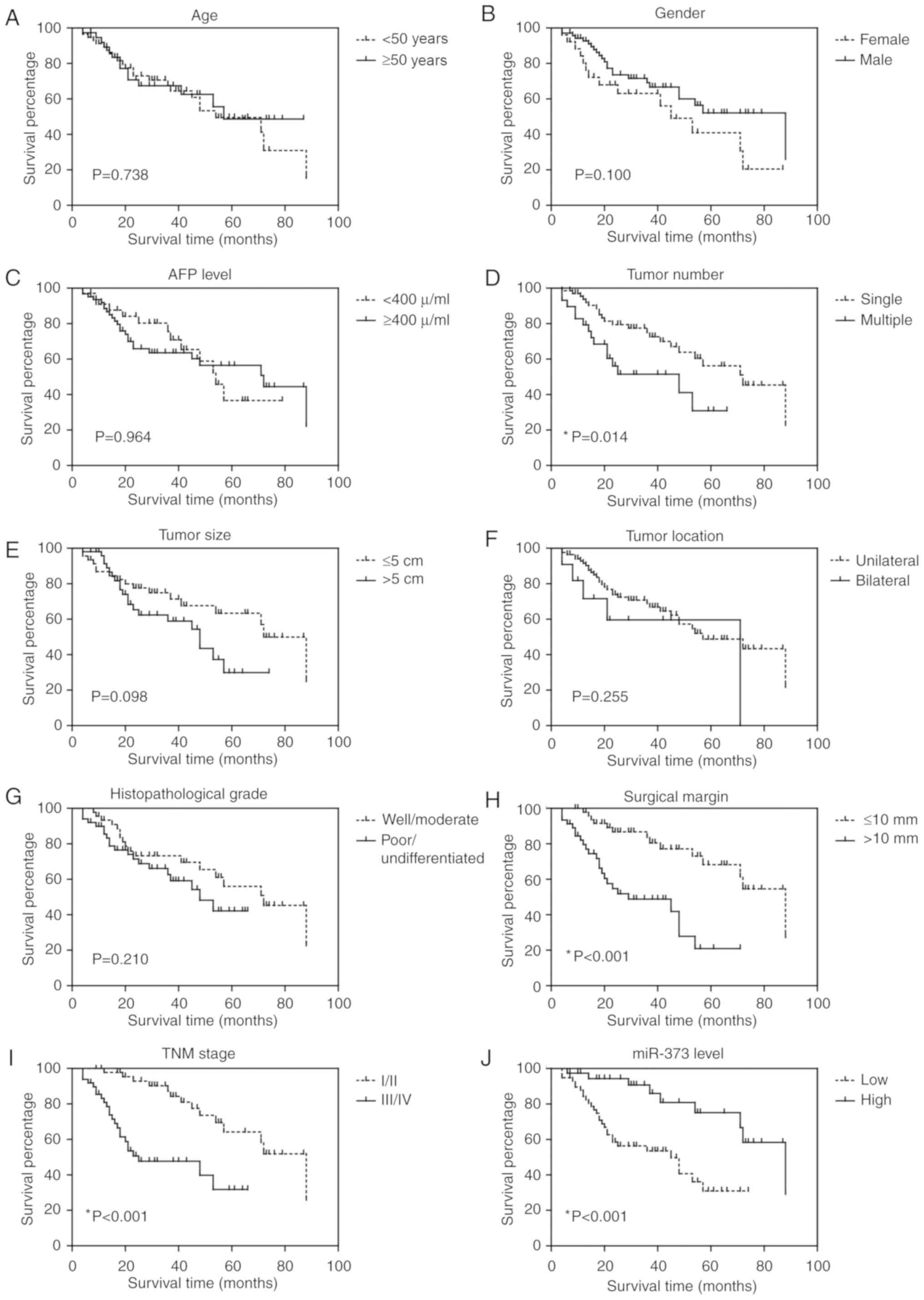 | Figure 2.Overall survival curve of liver
cancer patients. Kaplan-Meier analysis of the overall survival in
96 liver cancer patients in relation to (A) age, (B) sex, (C) AFP
level, (D) tumor number, (E) tumor size, (F) tumor location, (G)
histopathological grade, (H) surgical margin, (I) TNM stage and (J)
miR-373 level. AFP, α-fetoprotein; miR, microRNA. |
 | Table II.Correlation between clinical factors
and survival rates. |
Table II.
Correlation between clinical factors
and survival rates.
| Clinical
factor | 5-year survival
rate (%) |
P-valuea |
|---|
| Age (years) |
|
|
|
<50 | 49.5 | 0.738 |
|
≥50 | 48.7 |
|
| Sex |
|
|
|
Female | 40.9 | 0.100 |
|
Male | 52.2 |
|
| AFP (U/ml) |
|
|
|
<400 | 36.6 | 0.964 |
|
≥400 | 56.5 |
|
| Tumor number |
|
|
|
Single | 56.2 | 0.014b |
|
Multiple | 30.9 |
|
| Tumor size
(cm) |
|
|
| ≤5 | 63.4 | 0.098 |
|
>5 | 29.8 |
|
| Tumor location |
|
|
|
Unilateral | 48.8 | 0.255 |
|
Bilateral | 59.7 |
|
| Histopathological
grade |
|
|
|
Well/moderate | 56.0 | 0.210 |
|
Poor/undifferentiated | 42.2 |
|
| Surgical margin
(mm) |
|
|
|
≤10 | 68.3 |
<0.001b |
|
>10 | 20.9 |
|
| TNM stage |
|
|
|
I/II | 64.2 |
<0.001b |
|
III/IV | 31.8 |
|
| miR-373 level |
|
|
|
Low | 31.0 |
<0.001b |
|
High | 75.1 |
|
 | Table III.Cox multivariate analysis. |
Table III.
Cox multivariate analysis.
| Clinical
factor | HR | 95% CI |
P-valuea |
|---|
| Tumor number |
|
|
|
|
Single | 1 |
|
|
|
Multiple | 0.63 | 0.26–1.51 | 0.301 |
| Surgical margin
(mm) |
|
|
|
|
≤10 | 1 |
|
|
|
>10 | 3.1 | 1.52–6.43 | 0.002b |
| TNM stage |
|
|
|
|
I/II | 1 |
|
|
|
III/IV | 3.2 | 1.24–8.22 | 0.016b |
| miR-373 level |
|
|
|
|
Low | 1 |
|
|
|
High | 0.43 | 0.17–0.94 | 0.048b |
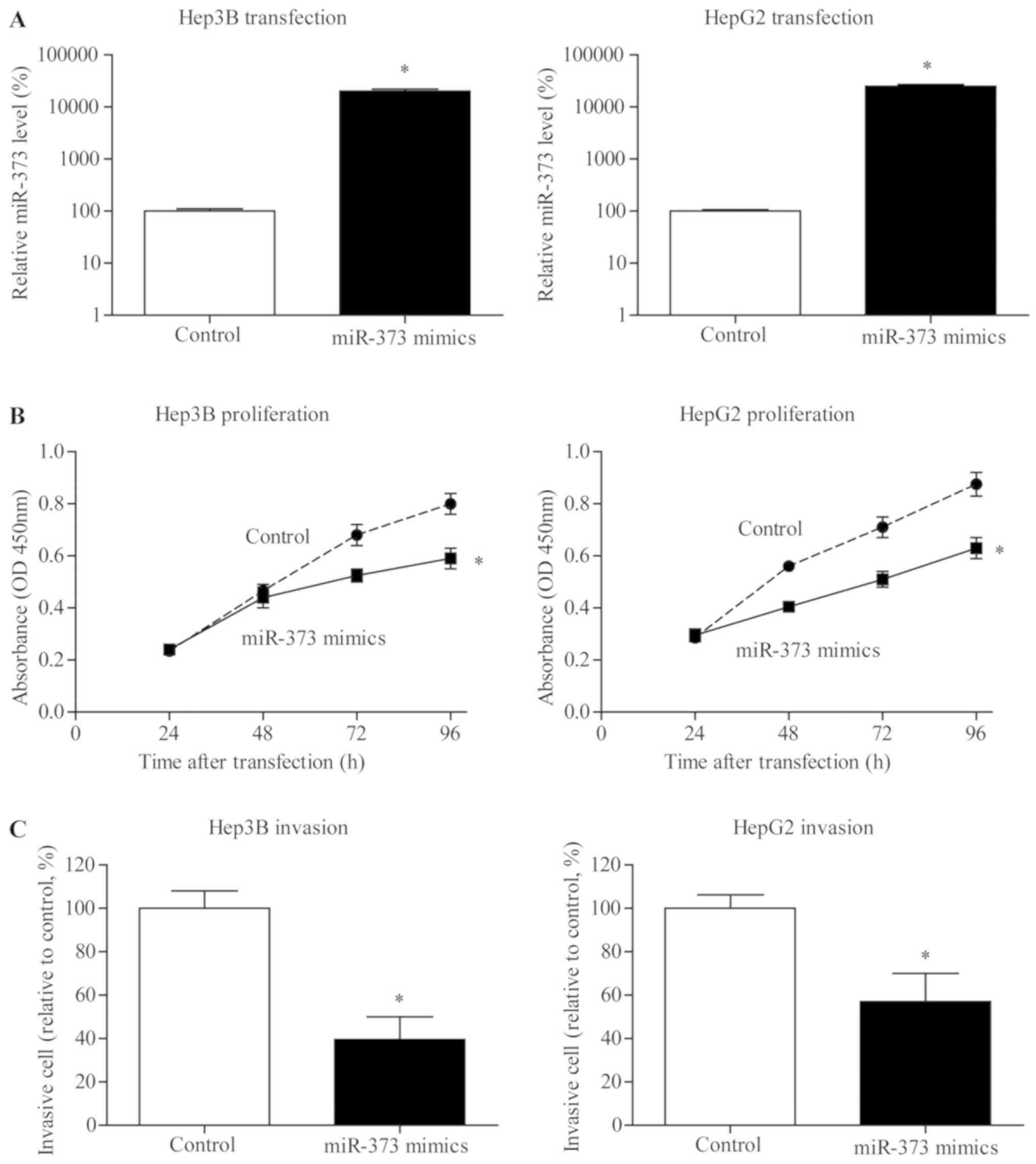
miR-373 inhibits the viability of
liver cancer cells
To further explore possible the effect of miR-373 on
the proliferation of liver cancer, miR-373 transiently transfected
cells were established (Fig. 3A).
MTT results indicated that proliferation of Hep3B and HepG2 cells
in the miR-373 transfection group was notably inhibited compared
with NC groups (Fig. 3B).
miR-373 inhibits the invasive capacity
of liver cancer cells
To investigate whether miR-373 upregulation served
critical roles in the metastasis of liver cancer cells, miR-373
mimic was transfected in Hep3B and HepG2 cell lines. Fig. 3C demonstrates a substantial
reduction in the invasion of liver cancer cells following the
upregulation of miR-373 expression level.
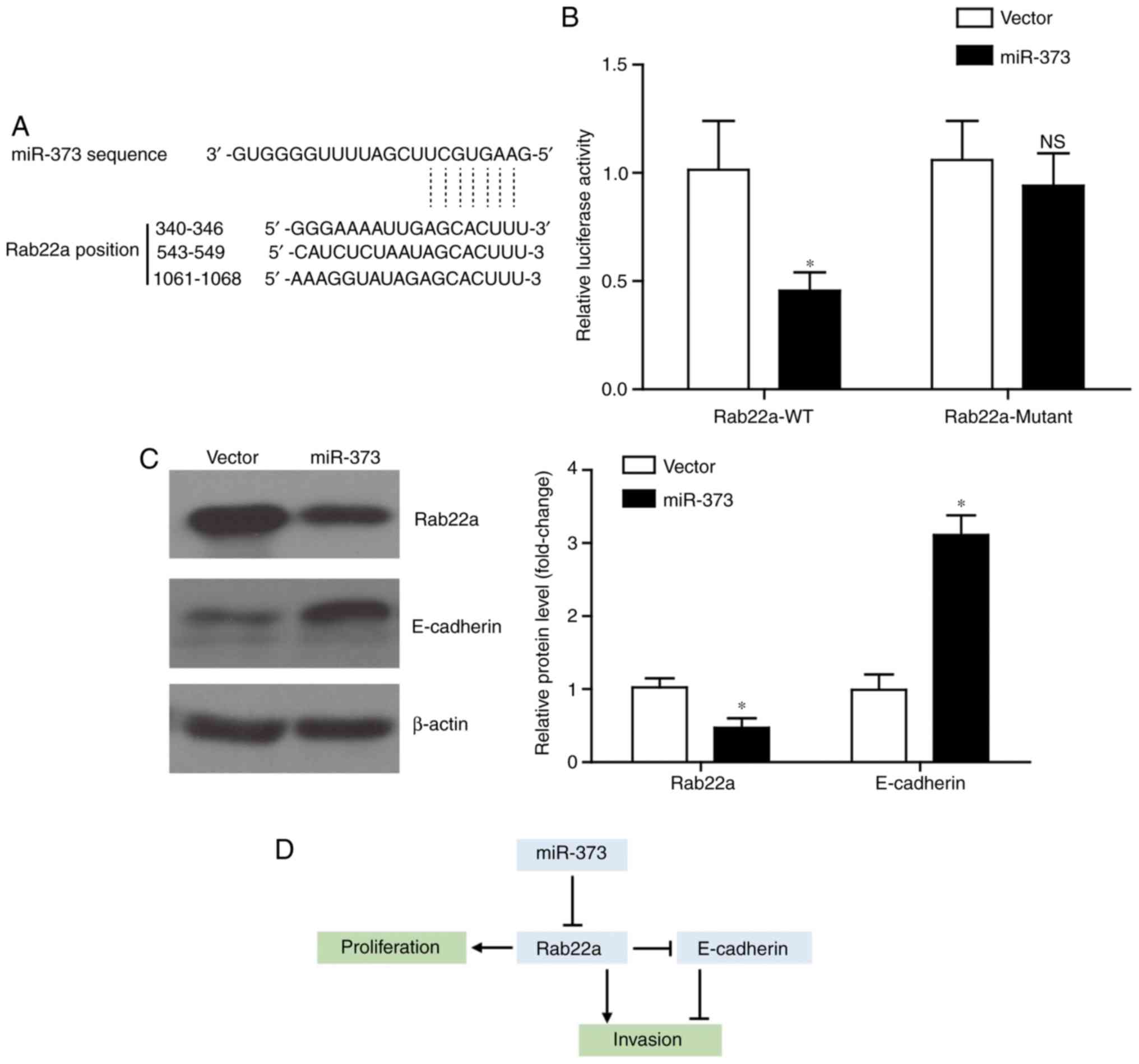
miR-373 targets Rab22a and elevates
E-cadherin level
The present study next explored the underlying
mechanism of miR-373 in inhibiting tumor progression in liver
cancer. By using the TargetScan tool (www.targetscan.org), it was identified that Rab22a
possessed three potential binding sites with miR-373 (Fig. 4A), which is consistent with a
recent study (24). Furthermore,
it was verified by using luciferase assays that miR373 can directly
regulate the transcription of Rab22a (Fig. 4B). By contrast, following the
mutation of the binding sites on Rab22a, miR-373 co-transfection
exhibited little effect on its luciferase activity. Western blot
analysis results demonstrated that miR-373 overexpression can
inhibit the expression of the oncoprotein Rab22a, while increasing
the protein level of E-cadherin (Fig.
4C). Considering the regulatory role of Rab22a on E-cadherin
expression (25), an
Rab22a-dependent role for miR-373 on inhibiting liver cancer
proliferation and invasion was hypothesized (Fig. 4D).
Discussion
A previous study demonstrated that miRNAs serve
non-negligible roles in the progression of various tumors,
including liver cancer (26).
Previous studies have reported that miR-373 serves as either an
oncogene or antioncogene in several cancer types with variable
expression. For instance, an upregulated level of miR-373
attenuates TGF-β-induced metastasis of breast cancer cells in
vivo, indicating the tumor suppressor activity of miR-373
(27). Another study demonstrated
that a low level of miR-373 is associated with poorer survival
rates in hilar cholangiocarcinoma (28). Conversely, it has been noted that
miR-373 is upregulated in human cervical cancer tissues and its
overexpression promotes the tumorigenicity of cervical cancer cells
by targeting the YOD1 gene (29). Consistently, miR-373 can enhance
tumor metastasis of breast cancer by directly suppressing
thioredoxin-interacting protein (30).
The current study established the
clinicopathological role of miR-373 in liver cancer patients.
RT-qPCR was performed to evaluate the endogenous transcription of
miR-373 and indicated that it was significantly decreased in liver
cancer tissues compared with the adjacent normal tissues. Further
investigation demonstrated that low miR-373 levels were associated
with multiple tumor number, larger tumor size, poorly
differentiated histopathological and advanced TNM stages.
Kaplan-Meier survival analysis demonstrated that high expression
levels of miR-373 in the tissues of patients was a predictor of
improved prognosis while low miR-373 expression levels suggested a
worse prognosis. Cox multivariate analysis also supported the
hypothesis that miR-373 expression level was an independent
prognostic factor for the survival rates of liver cancer patients.
In addition, transient high expression of miR-373 in Hep3B and
HepG2 cells attenuated the proliferation of liver cancer. miR-373
exerted an inhibitory effect in the invasion of liver cancer cells,
confirming the antioncogenic role of miR-373 in liver cancer.
Previous studies have revealed specific targets of
miR-373 in other tumor types. For example, miR-373 targets the
transforming growth factor-β type II receptor and reduces its
protein expression, therefore suppressing breast cancer migration
and invasion (8,27). miR-373 also demonstrates an
inhibitory effect on the expression of estrogen receptor in breast
cancer, which subsequently regulates the downstream matrix
metalloproteinases signaling and suppresses tumor progression
(31). Therefore, the targets and
mechanisms could be distinctive in liver cancer due to the
histospecificity of miRNAs. According to the data of the present
study, the tumor inhibiting effect of miR-373 in liver cancer was
exerted, at least partly, by suppressing the Rab22a signaling
pathway, which is consistent with its functions in ovarian cancer
(24). However, Wu et al
(32) reported that miR-373 is
upregulated in human liver cancer tissues and promotes tumor
progression by targeting the protein phosphatase 6 catalytic
subunit. It is reasonable to hypothesize that specific miRNAs may
serve multiple roles even in the same tumor type. For example,
miR-708-5p has been shown to exert different roles in lung cancer
by targeting distinct downstream targets (33,34).
Another possible explanation for the differences
between Wu et al (32) and
the present study may be due to the hepatitis effect. The majority
of the Chinese liver cancer patients were the result of hepatitis,
and the expression of miR-373 has been reported to exhibit
significant crosstalk with hepatitis B and C (35–37).
However, neither the present study nor that of Wu et al
retrieved the hepatitis information of enrolled patients.
Similarly, whether the patients were treated with anti-hepatitis
drugs or anti-tumor drugs prior to specimen collection may also
affect the results. Additionally, the results from the present
study and those of Wu et al were drawn from different
medical centers, therefore it may also reflect bias due to limited
patient resources in specific regions. Besides, Wu et al
used pri-miR-373-expressing pcDNA3 vector to overexpress miR-373 in
HepG2 cells, while the present study directly transfected cells
with miR-373 mimics, which may also be a reason for significant
differences. Although a previous study reported that co-expression
of miR371, miR372 and miR373 clusters can promote liver cancer
progression (38), the authors did
not test the individual roles of each miRNA. Therefore, further
in vivo studies are necessary to verify the exact and
multifaceted roles of miR-373 in liver cancer. In summary, the
present study revealed that downregulated miR-373 predicted poor
prognosis in liver cancer patients. In addition, miR-373 inhibited
the proliferation and invasion of liver cancer cells in
vitro. The outcomes represent a potential drug target for liver
cancer therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from The
National Natural Science Foundation of China (grant no. 81873178),
Shanghai Municipal Commission of Health and Family Planning (grant
no. 201740084), Key Specialty Construction Project of Pudong Health
and Family Planning Commission of Shanghai (grant no.
PWZxk2017-06), Science and Technology Development Fund of Shanghai
Pudong New Area (grant no. PKJ2017-Y14) and Talents Training
Program of Seventh People's Hospital of Shanghai University of TCM
(grant no. XX2017-01).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WX designed the present study. YY, LZ, YS and JZ
performed the in vitro experiments and interpreted the data.
GW, JN and SZ analyzed the clinical datasets and performed western
blotting. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All patients included in the present study signed
written informed consents and collection of patients' samples was
authorized by the Ethical Committee for Clinical Research of the
Seventh People's Hospital (approval no. v1.0-2015-04-23).
Patient consent for publication
All patients included in this study signed written
informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM: Global cancer statistics in the
year 2000. Lancet Oncol. 2:533–543. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruix J and Sherman M; Practice Guidelines
Committee, : American Association for the Study of Liver Diseases:
Management of hepatocellular carcinoma. Hepatology. 42:1208–1236.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park KW, Park JW, Choi JI, Kim TH, Kim SH,
Park HS, Park HS, Lee WJ, Park SJ, Hong EK and Kim CM: Survival
analysis of 904 patients with hepatocellular carcinoma in a
hepatitis B virus-endemic area. J Gastroenterol Hepatol.
23:467–473. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Llovet JM, Fuster J and Bruix J;
Barcelona-Clínic Liver Cancer Group, : The Barcelona approach:
Diagnosis, staging, and treatment of hepatocellular carcinoma.
Liver Transpl. 10 (Suppl 1):S115–S120. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou W, Li Y, Gou S, Xiong J, Wu H, Wang
C, Yan H and Liu T: miR-744 increases tumorigenicity of pancreatic
cancer by activating Wnt/β-catenin pathway. Oncotarget.
6:37557–37569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun Y, Bai Y, Zhang F, Wang Y, Guo Y and
Guo L: miR-126 inhibits non-small cell lung cancer cells
proliferation by targeting EGFL7. Biochem Biophys Res Commun.
391:1483–1489. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fang JH, Zhou HC, Zeng C, Yang J, Liu Y,
Huang X, Zhang JP, Guan XY and Zhuang SM: MicroRNA-29b suppresses
tumor angiogenesis, invasion, and metastasis by regulating matrix
metalloproteinase 2 expression. Hepatology. 54:1729–1740. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang R, Zhao N, Li S, Fang JH, Chen MX,
Yang J, Jia WH, Yuan Y and Zhuang SM: MicroRNA-195 suppresses
angiogenesis and metastasis of hepatocellular carcinoma by
inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology.
58:642–653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Voorhoeve PM, le Sage C, Schrier M, Gillis
AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A,
et al: A genetic screen implicates miRNA-372 and miRNA-373 as
oncogenes in testicular germ cell tumors. Adv Exp Med Biol.
604:17–46. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jing SY, Jing SQ, Liu LL, Xu LF, Zhang F
and Gao JL: Down-expression of miR-373 predicts poor prognosis of
glioma and could be a potential therapeutic target. Eur Rev Med
Pharmacol Sci. 21:2421–2425. 2017.PubMed/NCBI
|
|
15
|
Zhang Q, Wang C, Miao S, Li C, Chen Z and
Li F: Enhancing E-cadherin expression via promoter-targeted miR-373
suppresses bladder cancer cells growth and metastasis. Oncotarget.
8:93969–93983. 2017.PubMed/NCBI
|
|
16
|
Ding W, Fan XL, Xu X, Huang JZ, Xu SH,
Geng Q, Li R, Chen D and Yan GR: Epigenetic silencing of ITGA2 by
miR-373 promotes cell migration in breast cancer. PLoS One.
10:e01351282015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martins-Filho SN, Paiva C, Azevedo RS and
Alves VAF: Histological grading of hepatocellular carcinoma - a
systematic review of literature. Front Med (Lausanne). 4:1932017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou D, Li Z and Bai X: BRAFV600E and
RET/PTC promote proliferation and migration of papillary thyroid
carcinoma cells in vitro by regulating nuclear factor-κB. Med Sci
Monit. 23:5321–5329. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu H, Zhang Q, Li K, Gong Z, Liu Z, Xu Y,
Swaney MH, Xiao K and Chen Y: Prognostic significance of USP33 in
advanced colorectal cancer patients: New insights into
β-arrestin-dependent ERK signaling. Oncotarget. 7:81223–81240.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carter P: Site-directed mutagenesis.
Biochem J. 237:1–7. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu H, Xu Y, Zhang Q, Li K, Wang D, Li S,
Ning S, Yang H, Shi W, Liu Z and Chen Y: Correlations between
TBL1XR1 and recurrence of colorectal cancer. Sci Rep. 7:442752017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu H, Liu Z, Li K, Li S, Song L, Gong Z,
Shi W, Yang H, Xu Y, Ning S, et al: TBL1XR1 predicts isolated tumor
cells and micrometastasis in patients with TNM stage I/II
colorectal cancer. J Gastroenterol Hepatol. 32:1570–1580. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Zhao FJ, Chen LL, Wang LQ, Nephew
KP, Wu YL and Zhang S: miR-373 targeting of the Rab22a oncogene
suppresses tumor invasion and metastasis in ovarian cancer.
Oncotarget. 5:12291–12301. 2014.PubMed/NCBI
|
|
25
|
Wei F, Cao C, Xu X and Wang J: Diverse
functions of miR-373 in cancer. J Transl Med. 13:1622015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Baranwal S and Alahari SK: miRNA control
of tumor cell invasion and metastasis. Int J Cancer. 126:1283–1290.
2010.PubMed/NCBI
|
|
27
|
Keklikoglou I, Koerner C, Schmidt C, Zhang
JD, Heckmann D, Shavinskaya A, Allgayer H, Gückel B, Fehm T,
Schneeweiss A, et al: MicroRNA-520/373 family functions as a tumor
suppressor in estrogen receptor negative breast cancer by targeting
NF-κB and TGF-β signaling pathways. Oncogene. 31:4150–4163. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Y, Luo J, Tian R, Sun H and Zou S:
miR-373 negatively regulates methyl-CpG-binding domain protein 2
(MBD2) in hilar cholangiocarcinoma. Dig Dis Sci. 56:1693–1701.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang LQ, Zhang Y, Yan H, Liu KJ and Zhang
S: MicroRNA-373 functions as an oncogene and targets YOD1 gene in
cervical cancer. Biochem Biophys Res Commun. 459:515–520. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen D, Dang BL, Huang JZ, Chen M, Wu D,
Xu ML, Li R and Yan GR: miR-373 drives the
epithelial-to-mesenchymal transition and metastasis via the
miR-373-TXNIP-HIF1alpha-TWIST signaling axis in breast cancer.
Oncotarget. 6:32701–32712. 2015.PubMed/NCBI
|
|
31
|
Lu S, Zhu Q, Zhang Y, Song W, Wilson MJ
and Liu P: Dual-functions of miR-373 and miR-520c by differently
regulating the activities of MMP2 and MMP9. J Cell Physiol.
230:1862–1870. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu N, Liu X, Xu X, Fan X, Liu M, Li X,
Zhong Q and Tang H: MicroRNA-373, a new regulator of protein
phosphatase 6, functions as an oncogene in hepatocellular
carcinoma. FEBS J. 278:2044–2054. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jang JS, Jeon HS, Sun Z, Aubry MC, Tang H,
Park CH, Rakhshan F, Schultz DA, Kolbert CP, Lupu R, et al:
Increased miR-708 expression in NSCLC and its association with poor
survival in lung adenocarcinoma from never smokers. Clin Cancer
Res. 18:3658–3667. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu X, Liu T, Fang O, Dong W, Zhang F,
Leach L, Hu X and Luo Z: MicroRNA-708-5p acts as a therapeutic
agent against metastatic lung cancer. Oncotarget. 7:2417–2432.
2016.PubMed/NCBI
|
|
35
|
Varnholt H, Drebber U, Schulze F,
Wedemeyer I, Schirmacher P, Dienes HP and Odenthal M: MicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinoma. Hepatology. 47:1223–1232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo H, Liu H, Mitchelson K, Rao H, Luo M,
Xie L, Sun Y, Zhang L, Lu Y, Liu R, et al: MicroRNAs-372/373
promote the expression of hepatitis B virus through the targeting
of nuclear factor I/B. Hepatology. 54:808–819. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mukherjee A, Di Bisceglie AM and Ray RB:
Hepatitis C virus-mediated enhancement of microRNA miR-373 impairs
the JAK/STAT signaling pathway. J Virol. 89:3356–3365. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cairo S, Wang Y, de Reyniès A, Duroure K,
Dahan J, Redon MJ, Fabre M, McClelland M, Wang XW, Croce CM and
Buendia MA: Stem cell-like micro-RNA signature driven by Myc in
aggressive liver cancer. Proc Natl Acad Sci USA. 107:20471–20476.
2010. View Article : Google Scholar : PubMed/NCBI
|