Introduction
Hepatocellular carcinoma (HCC) is a common malignant
tumor that is harmful to human health. HCC ranks fifth highest in
incidence and second in cancer-related mortality worldwide, with
>50% of HCC cases occurring in China (1,2).
Approximately 75% of HCC cases are caused by chronic infection with
hepatitis B virus or hepatitis C virus (3,4).
There are almost 1,400,000 cases of chronic liver
disease-associated mortality annually (5), and HCC is the predominant cause of
mortality in patients with cirrhosis (6). When diagnosed early, the prognosis of
HCC is good, with a 5-year survival rate of >70%; however, the
prognosis of HCC is poor when diagnosed late, with a 5-year
survival rate of <16% (5). As
HCC is highly resistant to chemotherapy, immune therapy is an
attractive treatment option, as the inflammatory tumor
microenvironment is associated with improved survival (7,8).
Previous studies have shown that the aggregation of activated
regulatory T (Treg) cells in the tumor microenvironment is one of
the inhibitory mechanisms leading to immune evasion of HCC
(9–11). However, inhibitory receptors on
immune cells may contribute to the suppression of antitumor
immunity in the tumor microenvironment.
A novel inhibitory receptor, T cell immunoglobulin
and ITIM domain (TIGIT), has been shown to be expressed on active T
cells, natural killer cells (NK cells) (7) and tumor cells, including gastric
cancer cells (8). Human poliovirus
receptor, CD155, is the physical ligand of TIGIT. TIGIT/CD155
engagement can inhibit T cell responses in a cell-intrinsic manner
by directly targeting the T cell receptor (TCR) signaling cascade
and inducing tolerogenic dendritic cells (DCs) (9–11).
Treg cells selectively inhibit proinflammatory T helper 1 (Th1) and
T helper 17 (Th17) cell responses, also via expression of
coinhibitory molecule TIGIT (12,13).
Treg cells are important for maintaining immune homeostasis and
exerting immunosuppressive effects by directly recognizing tumor
cells and releasing the anti-inflammatory cytokines interleukin-2
(IL-2), interleukin-10 (IL-10), transforming growth factor-β
(TGF-β), and interleukin-35 (IL-35) (14). Previous studies have shown that the
frequency of Treg cells in tumor tissue is closely associated with
the recovery of patients. In patients with HCC, an increase in the
frequency of Treg cells indicates a poor prognosis (15). Furthermore, TIGIT-expressing T
follicular helper cells provide support to B cell functions
(16). Previous studies have shown
that genetic ablation or antibody blockade of TIGIT enhances
CD4+ T cell priming and exacerbates the severity of
experimental autoimmune encephalitis and rheumatoid arthritis
(17–19). In addition, the TIGIT pathway has
been shown to serve an important role in regulation in other
disease models, including cancer and chronic viral infection
(20). However, the role of TIGIT
in the pathogenesis of HCC remains to be elucidated.
In the present study, the expression of TIGIT and
its ligand, CD155, were examined in the cancerous tissues of
patients with HCC. Correlations between TIGIT+
CD4+ T cell and TIGIT+ Treg cell frequencies
and clinical pathological features were also analyzed. The results
showed that the TIGIT/CD155 signaling pathway may be a potential
target for the diagnosis and treatment of HCC.
Materials and methods
Ethics statement
The tissues and peripheral blood of patients with
HCC were collected in accordance with a protocol approved by the
Ethics Committee for the Conduct of Human Research at Ningxia
Medical University (Ningxia, China; no. 2015-074). Investigators
obtained informed consent prior to enrolling participants in
clinical trials. Patients who met the following eligibility
criteria were included in the study: i) diagnosis of primary HCC
identified by histopathological examination; ii) treatment with
radical resection; iii) availability of complete follow-up data;
iv) no pre-operative anticancer treatment, such as chemotherapy or
radiotherapy; v) no history of familial malignancy or other
synchronous malignancy; and vi) the patient was alive 3 months
after surgery. The exclusion criteria were as follows: i)
preoperative with acute infectious disease and/or blood system
disease and associated complications; ii) preoperative with severe
hypersplenism, kidney disease, cardiovascular and/or
cerebrovascular disease, or autoimmune disease; iii) receiving
adjuvant chemotherapy, such as radiotherapy and/or chemotherapy,
prior to surgery; and iv) a history of steroid hormone
administration in the 15 days before surgery.
HCC tissue samples and peripheral
blood
A total of 77 patients with HCC (40 with liver
cirrhosis and 37 with chronic hepatitis) were enrolled in the
present study. Immunohistochemical analysis was performed on
samples from 120 patients, including the aforementioned 77
patients, which comprised 73 cancer tissue samples and 47 adjacent
tissue samples (from paracellular tissues that were >5 cm from
the tumor), who had undergone surgical resection at the General
Hospital of Ningxia Medical University between September 2014 and
January 2017. The clinical characteristics of the patients are
summarized in Table I. Venous
blood was obtained from 31 patients with HCC (age, 29–80 years;
male: female, 17:14) and 30 healthy controls (age, 30–55 years;
male: female, 18:12).
 | Table I.Clinicopathologic features of
patients with hepatocellular carcinoma. |
Table I.
Clinicopathologic features of
patients with hepatocellular carcinoma.
| Variable | n |
|---|
| Virus
infection |
|
|
Hepatitis B virus | 42 |
|
Hepatitis C virus | 4 |
|
Hepatitis B (+) and C
virus | 3 |
| Liver
histopathology |
|
| Liver
cirrhosis | 40 |
| Chronic
hepatitis | 34 |
| Pathological
staging |
|
| Highly
differentiated group | 7 |
| High
and medium differentiation group | 6 |
| Medium
differentiation group | 40 |
| Medium
and low differentiation group | 11 |
| Low
differentiation group | 9 |
| Size of cancer
tissue (mm) |
|
|
<50 | 45 |
|
>50 | 28 |
|
α-fetoprotein >25 µg/l | 44 |
Isolation of peripheral blood
mononuclear cells (PBMCs)
Peripheral blood samples were collected from healthy
volunteers. The blood was diluted 1:1 with phosphate-buffered
saline prior to the separation of PBMCs by centrifugation at 500 ×
g at room temperature by Ficoll-Paque (Pharmacia, Uppsala, Sweden)
density gradient centrifugation. The cells were cryopreserved in
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham
MA, USA), supplemented with 10% dimethyl sulfoxide, and stored in
liquid nitrogen.
Flow cytometric analysis
Isolated PBMCs were washed with PBS and centrifuged
for 5 min at 500 g. The supernatant was discarded and the cells
were resuspended in PBS to a cell density of 1×106
cells/100 µl. Cell surface staining was performed on PBMCs using
the following anti-human monoclonal antibodies: CD127-AF647 (1:100;
clone HIL-7R-M21, cat. no. 558598), CD4-v450 (1:500; clone RPA-T4,
cat. no. 560346), CD25-APC-cy7 (1:100; clone M-A251, cat. no.
561782) and TIGIT-PE (1:300; clone 1G9, cat. no. 565168). All
antibodies were purchased from BD Pharmingen™. All data were
collected on a FACS Aria III™ (BD Biosciences) and were analyzed
with Flow Jo software version 7.5 (Tree Star, Inc.).
Liquid chip technology
The levels of cytokines in the plasma and
supernatants were determined using a cytometry bead array flex set
(BD Biosciences) in accordance with the manufacturer's
instructions. The assays were conducted for
granulocyte-macrophage-colony-stimulating factor, interferon-γ,
interleukin (IL)-β, IL-4, IL-6, IL-21, IL-10, IL-23, IL-15,
IL-2IL-17A, IL-33 and tumor necrosis factor (TNF)-α using 50 µl of
sample and a 10-point standard curve (0–2,500 pg/ml) for each
cytokine. The samples were analyzed using a flow fluorescence
detector Luminex 200 (Luminex Corporation). All data were collected
analyzed using Milliplex Analyst. V5.1 (Merck KGaA).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted using TRIzol total RNA
isolation reagent (Takara Biotechnology Co., Ltd., Dalian, China),
and reverse transcription was performed with the RT-PCR synthesis
kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The cDNA was stored at −20°C. PCR
amplification for quantification was performed with specific
primers using SYBR Premix Ex Taq II (Takara Biotechnology Co.,
Ltd.) on the PikoReal 96 qPCR system (Thermo Fisher Scientific,
Inc.) using the following thermocycling conditions: 95°C for 10
min, followed by 40 cycles of 95°C for 30 sec, 57°C for 45 sec and
72°C for 30 sec. The average of three independent analyses for each
gene and sample was calculated and normalized to the endogenous
reference control gene, β-actin. All samples were analyzed using
the 2−∆∆Cq analysis method (21). The primers used for qPCR were as
follows: β-actin, forward 5′-GCCAACACAGTGCTGTCTGG-3′ and reverse
5′-CTCAGGAGGAGCAATGATCTTG-3′; CD4, forward
5′-AGGAAGTGAACCTGGTGGTG-3′ and reverse 5′-TTGCCTCCTTGTTCTCCAGT-3′;
TIGIT, forward 5′-ATGGGACGTACACTGGGAGA-3′ and reverse
5′-ACTGTCGTGCAGATGACCAC-3′; CD155, forward
5′-ACTGTCACCAGCCTCTGGAT-3′ and reverse
5′-GGTGAGGTTCACAGTCAGCA-3′.
Western blot analysis
The tissue total protein was extracted using RIPA
buffer (Beyotime Institute of Biotechnology, Shanghai, China), and
the quantity of protein was measured using an Enhanced BCA Protein
Assay kit (Beyotime Institute of Biotechnology,). A total of 50 µg
of protein was loaded and separated on 12% SDS-PAGE gels (Beyotime
Institute of Biotechnology). The total protein was then transferred
onto polyvinylidene difluoride (PVDF) membranes. The PVDF membranes
were blocked using 5% skim milk powder and were then incubated with
antibodies overnight at 4°C. Rabbit CD155 monoclonal antibody (cat.
no. ab103630; Abcam) and rabbit TIGIT monoclonal antibody (cat. no.
ab106311; Abcam) were diluted in antibody diluent (1:500/1:1,000,
respectively; Beyotime Institute of Biotechnology) and rabbit GAPDH
antibody (cat. no. bs-2188R; BIOSS, Beijing, China) was diluted in
antibody diluent (1:4,000). The PVDF membranes were washed four
times (5 min each time) with TBST and then incubated with goat
anti-mouse or rabbit IgG horseradish peroxidase (HRP)-conjugated
secondary antibodies (1:4,000; cat. nos. A21010 and A21020; Abbkine
Scientific Co., Ltd.) for 1 h at room temperature. The PVDF
membranes were washed another four times, and the protein bands
were visualized by ECL chemiluminescence and then measured using
ImageJ software 1.8.0. (National Institutes of Health, Bethesda,
MD, USA). The expression levels of proteins of interest were
normalized with GAPDH. Independent experiments were repeated three
times for each sample.
Immunohistochemistry
The tissues were dehydrated and embedded in paraffin
wax. The sections were dewaxed in xylene and dehydrated in alcohol.
Antigen retrieval was performed by incubating the slides in citric
acid buffer for 10 min. Following blocking with 3% hydrogen
peroxide for 15 min and 5% bovine serum albumin (Beijing Solarbio
Science and Technology Co., Ltd., Beijing, China), the slides were
incubated overnight at 4°C with mouse polyclonal antibodies against
CD4 (1:100; cat. no. ab133616; Abcam), CD155 (1:100; cat. no.
ab103630; Abcam) and TIGIT (1:100; cat. no. ab243903; Abcam).
Following rinsing with PBS, the sections were incubated with
secondary antibodies from the Polink-2 plus Polymer HRP Detection
System (cat. nos. ZB-2301 and ZB-2305; ZSGB-BIO) for 30 min at room
temperature. The expression of CD4, CD155 and TIGIT was visualized
by diaminobenzidine tetrahydrochloride solution (Cowin Bioscience)
staining. The sections were then counterstained with hematoxylin
for 5 min. All slides were observed and images were captured under
a fluorescence microscope.
Statistical analysis
All statistical calculations were performed with
SPSS 23.0 software (IBM Corp.). Data are expressed as the mean ±
standard deviation (SD). Statistical differences between groups
were analyzed using the Mann-Whitney U test. Spearman's rank
correlation test for nonparametric data was used to analyze the
relationship between the two factors. Statistical analysis was
performed using GraphPad Prism software version 6.0 (GraphPad
Software Inc., San Diego, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of TIGIT/CD155 in cancerous
tissue is significantly elevated in patients with HCC
To determine whether TIGIT/CD155 is involved in the
pathogenesis of HCC, the expression of TIGIT/CD155 in paracancerous
and cancerous tissues was assessed by immunohistochemistry. A
previous study showed that TIGIT was expressed on CD4+ T
cells in patients with rheumatoid arthritis (20) and on CD8+ T cells in
both healthy individuals and in patients with melanoma (22,23).
In order to further understand the aggregation of inflammation in
tumor sites, the expression of CD4+ T cells was also
examined in patients with HCC. As shown in Fig. 1A and B, the expression of CD4 was
distinctly elevated in the liver tumor tissues compared with that
in the paracancerous tissues. In the cancerous tissue, as the
degree of malignancy increased, the number of CD4+ cells
gradually increased, indicating that the interaction between the
tumor cells and effector cells determines disease prognosis. The
expression levels of TIGIT and CD155 were increased in liver cancer
tissues. Of note, the expression of TIGIT gradually increased in
liver cancer tissues as the degree of tumor cell differentiation
changed from high to low, although the expression of CD155 was
highest in the high-medium differentiation stage; generally, the
expression of CD155 gradually decreased as differentiation
increased (Fig. 1C-F). The
molecular mRNA expression levels of TIGIT and CD155 were detected
by RT-qPCR analysis. The results showed that the mRNA expression
levels of CD4, TIGIT and CD155 were higher in cancerous tissues
than those in the paracancerous tissues (Fig. 1G). This suggests that the
TIGIT/CD155 pathway may be involved in the pathogenesis of HCC.
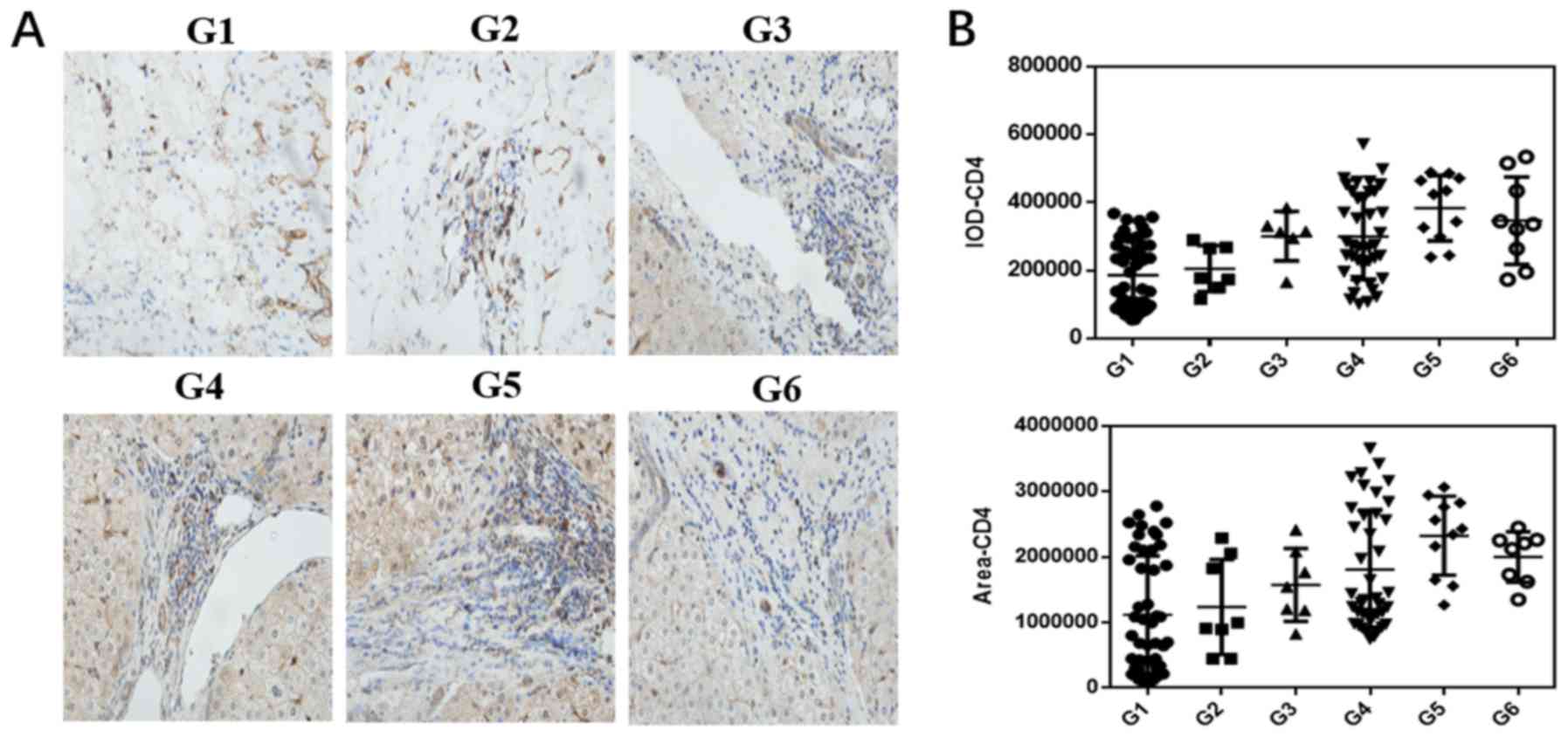 | Figure 1.Expression of CD4, TIGIT and CD155 in
HCC tissues at different pathological stages and adjacent tissues.
Paracancerous tissues (G1) and HCC tissues (G2-6) were stained by
immunohistochemistry (×400 magnification). (A) Staining of CD4 and
(B) statistical analysis of CD4-positive dots in paracancerous and
HCC tissues. (C) Staining of TIGIT and (D) statistical analysis of
TIGIT-positive dots in paracancerous and HCC tissues. (E) Staining
of CD155 and (F) statistical analysis of CD155-positive dots in
paracancerous and HCC tissues. (G) Relative mRNA expression levels
of CD4, TIGIT and CD155. All data from at least three independent
experiments were measured by the band density, which were
normalized to GAPDH. Bar graphs (mean ± SEM) and representative
images are shown. *P<0.05. G1, paracancerous tissue; G2, highly
differentiated group; G3, high and medium differentiation; G4,
medium differentiation; G5, medium and low differentiation; G6, low
differentiation. HCC, hepatocellular carcinoma; TIGIT, T cell
immunoglobulin and ITIM domain. |
Expression of TIGIT and CD155 in
different patients with HCC
In order to confirm the roles of TIGIT and CD155 in
HCC, the protein expression levels of TIGIT and CD155 were examined
via western blotting in cancerous and adjacent tissues from
different patients. As shown in Fig.
2A and B, the expression levels of TIGIT and CD155 in the
cancerous tissues were higher than those in the paracancerous
tissues.
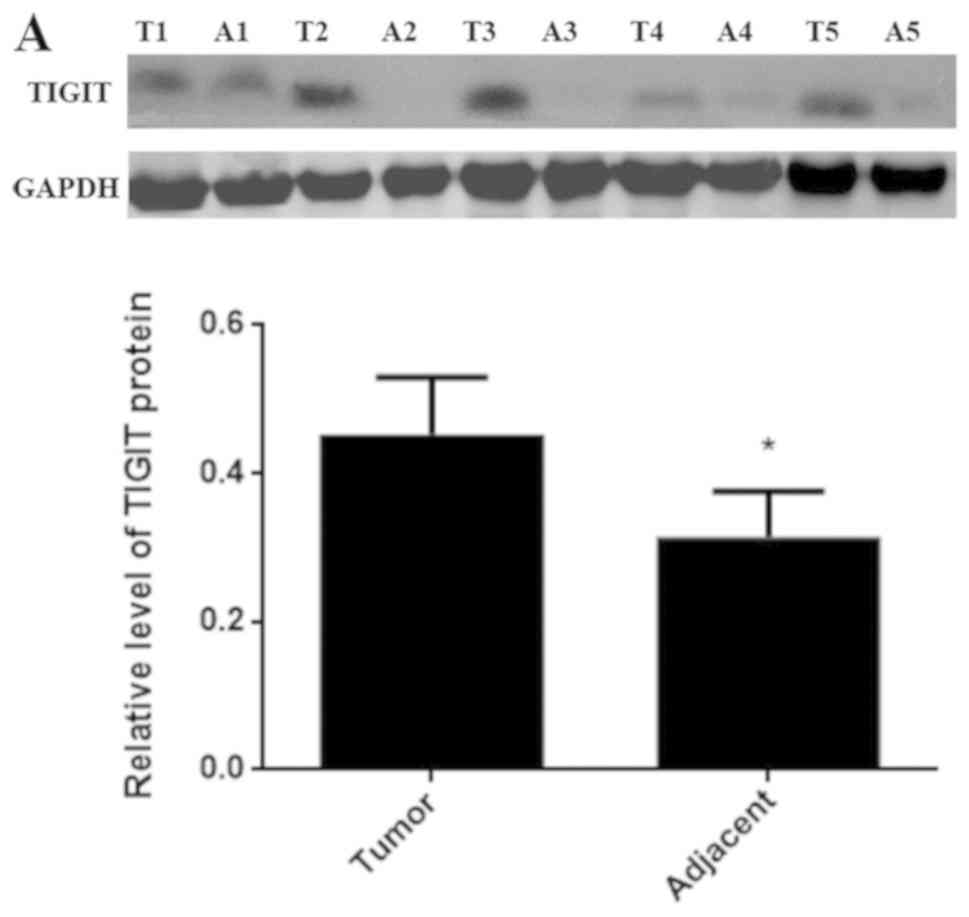 | Figure 2.Expression of TIGIT and CD155 in
different patients with HCC. (A) Protein expression of TIGIT was
detected by western blotting. (B) Protein expression of CD155 was
detected by western blotting. Data are expressed as the mean ± SD
(*P<0.05 vs. tumor). 1, 2, 3, 4 and 5 refer to patients 1, 2,
3,4 and 5, respectively. T, tumor tissue; A, adjacent tissue;
TIGIT, T cell immunoglobulin and ITIM domain. |
TIGIT+ CD4+ T
cell and TIGIT+ Treg cell frequency shift in patients
with HCC
It is well known that TIGIT is expressed on
activated T cells, including CD4+ T and Treg cells. In
the present study, the frequency of TIGIT+
CD4+ T and TIGIT+ Treg cells was altered in
patients with HCC. PBMCs were extracted from the peripheral blood 5
days before surgery and 5 days after surgery for flow cytometry.
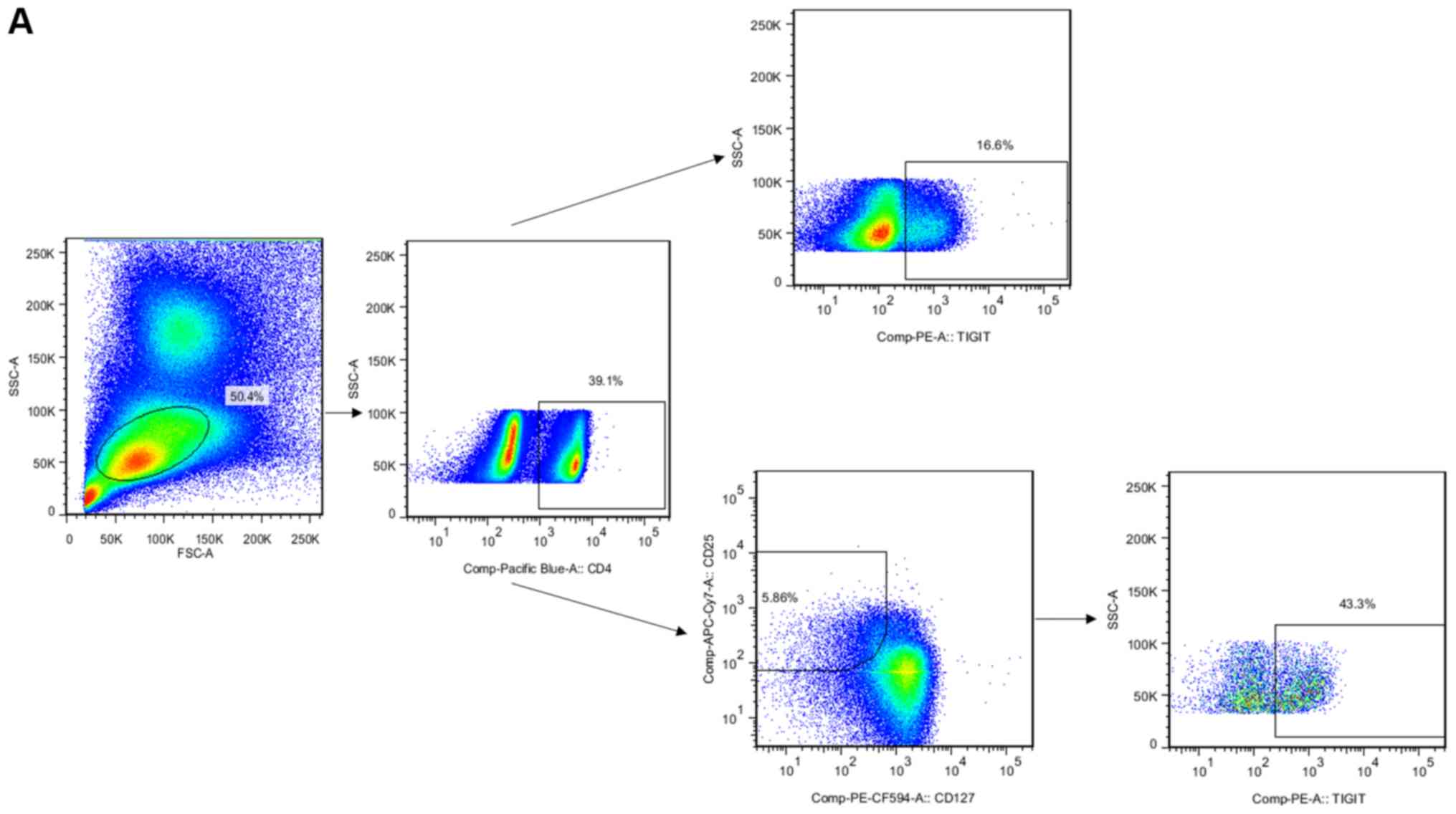
The gated strategy of flow cytometry is shown in Fig. 3A. The frequency of CD4 cells in
patients with HCC was higher than that in the normal control group
and decreased following surgery (Fig.
3B). The frequency of TIGIT+ CD4+ T cells
was increased in patients with HCC following surgery (Fig. 3B). Another important finding was
that the frequency of TIGIT+ Treg cells was higher in
patients with HCC than that in the normal control group and was
decreased following surgery (Fig.
3B). The secretion of cytokines from different T cell
subpopulations was then analyzed. The results showed that the
expression of the pro-inflammatory cytokine IL-17a was increased
(P=0.047; Fig. 3C), the expression
of IL-6 was increased (P=0.043; Fig.
3C), and the expression of the anti-inflammatory factor IL-10
was decreased (P=0.034; Fig. 3D)
following surgery. These results indicate that the frequency and
functional changes of TIGIT+ CD4+ T cells and
TIGIT+ Treg cells are involved in the pathogenesis of
HCC. In addition, when the balance between subsets of T cells was
disrupted, the functional changes of T cell subsets mediated the
regulation of tumor development.
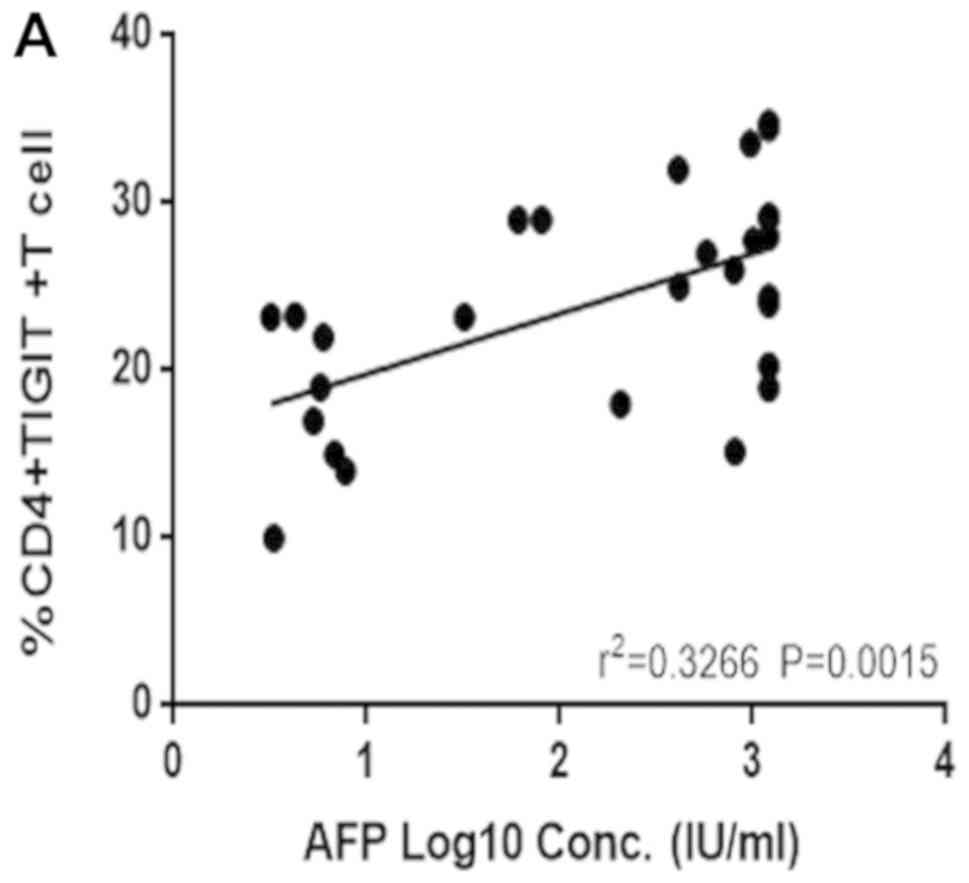
Correlation between TIGIT+
CD4+ T cells and TIGIT+ Treg cell frequency
and AFP
AFP is the most common clinical index used in the
diagnosis of liver cancer. Therefore, a possible correlation
between the expression of TIGIT+ CD4+ T and
TIGIT+ Treg cells and the clinical indicator, AFP, was
examined. The results showed that the frequency of
TIGIT+ CD4+ T cells (Fig. 4A) and TIGIT+ Treg cells
(Fig. 4B) in the peripheral blood
correlated positively with AFP, which may contribute to an early
diagnosis of HCC.
Discussion
The incidence of HCC has been increasing, which is a
concern due to HCC being a disease with a high mortality rate
(24). Liver cancer is one example
of an inflammation-related type of cancer and >90% of cases of
liver cancer are caused by liver damage and infection (25), which contributes to the formation,
progression, escape, proliferation, invasion, angiogenesis and
metastasis of tumor cells. Among inflammatory aspects, the
activation of nuclear transcription factor (NF)-κB contributes to
inflammation-induced tumors. In addition, tumor suppressor genes
with specific mutations or abnormal expression can activate NF-κB
and increase the possibility of immune evasion, which can promote
the development of HCC (26).
Therefore, the liver remains in an inflammatory environment for a
long period of time, causing necrosis that develops into cirrhosis
and eventually leads to liver cancer (27). At present, there is no optimal
diagnostic test or therapy for liver cancer. Therefore, identifying
novel methods to diagnose and treat liver cancer is vital. The
co-inhibitory receptor pathway has attracted widespread attention
as a potential target of immune regulation. Significant success has
been achieved with the use of co-inhibitory receptor pathways, such
as cytotoxic T lymphocyte antigen 4 (CTLA-4) and programmed death-1
(PD-1), in the treatment of cancer (28–30).
TIGIT is a recently identified co-inhibitory receptor that is
expressed on activated T cells, Treg cells, NK cells and tumor
cells (8,11,19,31).
Similar to CTLA-4 and CD28, TIGIT competes with its costimulatory
counterpart, CD226, for the same ligands (CD155 and CD112) and
mediates immune suppression in tumors and chronic infections
(32). In the present study, as
the degree of malignancy increased in cancerous tissues, the number
of CD4+ cells gradually increased, indicating that the
interaction between tumor cells and immune cells determines the
prognosis of HCC. Previous studies have shown that HCC can evade
host immune surveillance and has the ability of self-protection
(33,34). In addition, HCC cells avoid immune
attack through the secretion of immunosuppressive cytokines,
abnormal expression of antigens and changes to the local immune
microenvironment (34). Evidence
has also demonstrated that immunosuppressive factors expressed by
tumor cells, which inhibit APC or T cell function, suppress antigen
presentation and immune response and facilitate the immune evasion
of tumor cells (35,36). A number of studies have also found
that the overexpression of TIGIT reduces the function of
CD4+ T cells in rheumatoid arthritis (20). The expression of TIGIT on
CD4+ T cells was found to be significantly increased in
patients with systemic lupus erythematosus and correlated with the
activity of the disease (37).
CD155/TIGIT signaling has also been found to regulate
CD8+ T cell metabolism and promote tumor progression in
human gastric cancer (8). A
noteworthy finding in the present study was that TIGIT and its
ligand CD155 were upregulated in cancerous tissues from patients
with HCC as the degree of cancerous differentiation reduced,
suggesting that the TIGIT/CD155 pathway is involved in the
pathogenesis of HCC. Other co-inhibitory receptors have also been
reported, such as PD-1 marking dysfunctional regulatory T cells in
malignant gliomas (38). These
results suggest that the TIGIT/CD155 pathway serves an important
role in HCC.
As is well known, the tumor immune microenvironment
is produced by the interaction among tumor cells, immune cells and
the tumor stroma. In addition to effector T cells, the existence of
Treg cells, which negatively regulate immune responses, inhibits
effector T cells. The expression of TIGIT on CD4+ T and
Treg cells was detected by flow cytometry in peripheral blood from
patients with HCC prior to and following surgery. The frequency of
TIGIT+ CD4+ T cells increased prior to
surgery, and the frequency of TIGIT+ Treg cells
decreased following surgery, compared with the frequency in healthy
controls. This indicates that these cells may be involved in tumor
immune evasion and that the expression of TIGIT affects the
functions of CD4+ effector cells and Treg cells, which
mediates HCC immune evasion. The immune environment in the body is
disordered, and the balance of cytokines secreted by immune
effector cells alters with the development of HCC tumors. In the
present study, the levels of IL-17a and IL-6 in patients with HCC
increased following surgery, indicating that antitumor immunity was
reversed following surgery and that immune defense in the body was
restored. IL-10 is one of the immunosuppressive cytokines secreted
by Treg cells to suppress the immune response. The decreased level
of IL-10 in patients postoperatively suggests that the immune
response had recovered.
AFP is the most common tumor marker for the
diagnosis of primary liver cancer. The positive rate and
specificity of AFP in liver cancer is ~70%. In the present study,
TIGIT+ CD4+ T and TIGIT+ Treg were
positively correlated with AFP, suggesting that TIGIT+ T
cells are of potential clinical value in the diagnosis of HCC.
Further investigations based on medical evidence are required to
examine this application.
Taken together, the human immune system is
disordered in liver cancer. The number and function of immune cells
are altered due to the high expression of TIGIT, particularly on
effector T cells and Treg cells, which serve important roles in the
tumor microenvironment. The interaction between TIGIT+
tumor cells and TIGIT+ T cells appears to determine the
severity and prognosis of HCC. In addition, the expression of TIGIT
on CD4+ T and Treg cells was positively correlated with
AFP and may be a potential indicator for HCC detection. Therefore,
the expression of TIGIT may serve as a key molecule for the
diagnosis and treatment of HCC in the future.
Acknowledgements
The authors would like to thank Richard Ho Lye Yin
(Ph.D) and Ms Margaret Vera Trelfa from Ningxia Medical University
for making corrections to the text.
Funding
The present study received financial support from
the National Natural Science Foundation of China (grant nos.
81560466,81760437, 31860695 and 81560504), the Natural Science
Foundation Project of Ningxia (grant no. NZ16055), the Innovation
and Entrepreneurship for Returned Students Project of Ningxia,
China (2017) (grant no. YXW2017084) and the Construction of
First-class Disciplines in Ningxia University (Basic Medicine in
Western China) grant no. NXYLXK2017B07).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XD and CS contributed to study design, drafted the
manuscript and gave final approval of the manuscript; JL, JC and QZ
contributed to study design and data analysis, and gave final
approval of the manuscript; XY and BM contributed to experimental
operation and gave final approval of the manuscript; ZL and YD
acquired the data and revised the manuscript for important
intellectual content. All authors gave final approval of the
version to be published and agreed to be accountable for all
aspects of the work.
Ethics approval and consent to
participate
The tissues and peripheral blood of patients with
HCC were collected in accordance with a protocol approved by the
Ethics Committee for the Conduct of Human Research at Ningxia
Medical University (Ningxia, China; no. 2015-074). Investigators
obtained informed consent before enrolling participants in clinical
trials.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mazzanti R, Gramantieri L and Bolondi L:
Hepatocellular carcinoma: Epidemiology and clinical aspects. Mol
Aspects Med. 29:130–143. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB: Epidemiology of
hepatocellular carcinoma in USA. Hepatol Res. 37 (Suppl 2):S88–S94.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Greten TF, Duffy AG and Korangy F:
Hepatocellular Carcinoma from an immunologic perspective. Clin
Cancer Res. 19:6678–6685. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boles KS, Vermi W, Facchetti F, Fuchs A,
Wilson TJ, Diacovo TG, Cella M and Colonna M: A novel molecular
interaction for the adhesion of follicular CD4 T cells to
follicular DC. Eur J Immunol. 39:695–703. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He W, Zhang H, Han F, Chen X, Lin R, Wang
W, Qiu H, Zhuang Z, Liao Q, Zhang W, et al: CD155T/TIGIT signaling
regulates CD8(+) T-cell metabolism and promotes tumor progression
in human gastric cancer. Cancer Res. 77:6375–6388. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Joller N, Hafler JP, Brynedal B, Kassam N,
Spoerl S, Levin SD, Sharpe AH and Kuchroo VK: Cutting Edge: TIGIT
Has T cell-intrinsic inhibitory functions. J Immunol.
186:1338–1342. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu X, Harden K, Gonzalez LC, Francesco M,
Chiang E, Irving B, Tom I, Ivelja S, Refino CJ, Clark H, et al: The
surface protein TIGIT suppresses T cell activation by promoting the
generation of mature immunoregulatory dendritic cells. Nat Immunol.
10:48–57. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Levin SD, Taft DW, Brandt CS, Bucher C,
Howard ED, Chadwick EM, Johnston J, Hammond A, Bontadelli K,
Ardourel D, et al: Vstm3 is a member of the CD28 family and an
important modulator of T-cell function. Eur J Immunol. 41:902–915.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Joller N, Lozano E, Burkett PR, Patel B,
Xiao S, Zhu C, Xia J, Tan TG, Sefik E, Yajnik V, et al: Treg cells
expressing the coinhibitory molecule TIGIT selectively inhibit
proinflammatory Th1 and Th17 cell responses. Immunity. 40:569–581.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kurtulus S, Sakuishi K, Ngiow SF, Joller
N, Tan DJ, Teng MW, Smyth MJ, Kuchroo VK and Anderson AC: TIGIT
predominantly regulates the immune response via regulatory T cells.
J Clin Invest. 125:4053–4062. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Trehanpati N and Vyas AK: Immune
regulation by T regulatory cells in hepatitis B virus-related
inflammation and cancer. Scand J Immunol. 85:175–181. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kurose K, Ohue Y, Sato E, Yamauchi A,
Eikawa S, Isobe M, Nishio Y, Uenaka A, Oka M and Nakayama E:
Increase in activated treg in til in lung cancer and in vitro
depletion of treg by ADCC using an antihuman CCR4 mAb (KM2760). J
Thorac Oncol. 10:74–83. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Godefroy E, Zhong H, Pham P, Friedman D
and Yazdanbakhsh K: TIGIT-positive circulating follicular helper T
cells display robust B-cell help functions: Potential role in
sickle cell alloimmunization. Haematologica. 100:1415–1425. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Goding SR, Wilson KA, Xie Y, Harris KM,
Baxi A, Akpinarli A, Fulton A, Tamada K, Strome SE and Antony PA:
Restoring immune function of tumor-specific CD4+ T cells during
recurrence of melanoma. J Immunol. 190:4899–4909. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stengel KF, Harden-Bowles K, Yu X, Rouge
L, Yin J, Comps-Agrar L, Wiesmann C, Bazan JF, Eaton DL and Grogan
JL: Structure of TIGIT immunoreceptor bound to poliovirus receptor
reveals a cell-cell adhesion and signaling mechanism that requires
cis-trans receptor clustering. Proc Natl Acad Sci USA.
109:5399–5404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stanietsky N, Simic H, Arapovic J, Toporik
A, Levy O, Novik A, Levine Z, Beiman M, Dassa L, Achdout H, et al:
The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell
cytotoxicity. Proc Natl Acad Sci USA. 106:17858–17863. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao W, Dong Y, Wu C, Ma Y, Jin Y and Ji
Y: TIGIT overexpression diminishes the function of CD4 T cells and
ameliorates the severity of rheumatoid arthritis in mouse models.
Exp Cell Res. 340:132–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chauvin JM, Pagliano O, Fourcade J, Sun Z,
Wang H, Sander C, Kirkwood JM, Chen TH, Maurer M, Korman AJ and
Zarour HM: TIGIT and PD-1 impair tumor antigen-specific CD8(+) T
cells in melanoma patients. J Clin Invest. 125:2046–2058. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song Y, Wang B, Song R, Hao Y, Wang D, Li
Y, Jiang Y, Xu L, Ma Y, Zheng H, et al: T-cell Immunoglobulin and
ITIM Domain Contributes to CD8(+) T-cell Immunosenescence. Aging
Cell. 17:2018. View Article : Google Scholar
|
|
24
|
Imbeaud S, Ladeiro Y and Zucman-Rossi J:
Identification of novel oncogenes and tumor suppressors in
hepatocellular carcinoma. Semin Liver Dis. 30:75–86. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Matsuzaki K, Murata M, Yoshida K, Sekimoto
G, Uemura Y, Sakaida N, Kaibori M, Kamiyama Y, Nishizawa M,
Fujisawa J, et al: Chronic inflammation associated with hepatitis C
virus infection perturbs hepatic transforming growth factor beta
signaling, promoting cirrhosis and hepatocellular carcinoma.
Hepatology. 46:48–57. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang L and Karin M: Roles of tumor
suppressors in regulating tumor-associated inflammation. Cell Death
Differ. 21:1677–1686. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sanz-Cameno P, Trapero-Marugán M, Chaparro
M, Jones EA and Moreno-Otero R: Angiogenesis: From chronic liver
inflammation to hepatocellular carcinoma. J Oncol. 2010:2721702010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pedroza-Gonzalez A, Zhou G, Vargas-Mendez
E, Boor PP, Mancham S, Verhoef C, Polak WG, Grünhagen D, Pan Q,
Janssen H, et al: Tumor-infiltrating plasmacytoid dendritic cells
promote immunosuppression by Tr1 cells in human liver tumors.
Oncoimmunology. 4:e10083552015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Callahan MK, Postow MA and Wolchok JD:
CTLA-4 and PD-1 pathway blockade: Combinations in the clinic. Front
Oncol. 4:3852014.PubMed/NCBI
|
|
30
|
Hamid O, Robert C, Daud A, Hodi FS, Hwu
WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, et al:
Safety and tumor responses with lambrolizumab (anti-PD-1) in
melanoma. N Engl J Med. 369:134–144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stanietsky N, Rovis TL, Glasner A, Seidel
E, Tsukerman P, Yamin R, Enk J, Jonjic S and Mandelboim O: Mouse
TIGIT inhibits NK-cell cytotoxicity upon interaction with PVR. Eur
J Immunol. 43:2138–2150. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Johnston RJ, Comps-Agrar L, Hackney J, Yu
X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B, et al:
The immunoreceptor TIGIT regulates antitumor and antiviral CD8+ T
cell effector function. Cancer Cell. 26:923–937. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jenne CN and Kubes P: Immune surveillance
by the liver. Nat Immunol. 14:996–1006. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nemeth E, Baird AW and O'Farrelly C:
Microanatomy of the liver immune system. Semin Immunopathol.
31:333–343. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhong Z, Carroll KD, Policarpio D, Osborn
C, Gregory M, Bassi R, Jimenez X, Prewett M, Liebisch G, Persaud K,
et al: Anti-transforming growth factor beta receptor II antibody
has therapeutic efficacy against primary tumor growth and
metastasis through multieffects on cancer, stroma, and immune
cells. Clin Cancer Res. 16:1191–1205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zakrzewski PK, Cygankiewicz AI,
Mokrosiński J, Nowacka-Zawisza M, Semczuk A, Rechberger T and
Krajewska WM: Expression of endoglin in primary endometrial cancer.
Oncology. 81:243–250. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mao L, Hou H, Wu S, Zhou Y, Wang J, Yu J,
Wu X, Lu Y, Mao L, Bosco MJ, et al: TIGIT signalling pathway
negatively regulates CD4+ T-cell responses in systemic
lupus erythematosus. Immunology. 151:280–290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lowther DE, Goods BA, Lucca LE, Lerner BA,
Raddassi K, van Dijk D, Hernandez AL, Duan X, Gunel M, Coric V, et
al: PD-1 marks dysfunctional regulatory T cells in malignant
gliomas. JCI Insight. 1(pii): e859352016.PubMed/NCBI
|