Introduction
Osteosarcoma (OS) is the most common type of primary
malignant bone tumor, and demonstrates a high degree of malignancy,
invasion, metastasis and recurrence that significantly affects
patient prognosis; OS accounts for ~5% of cases of childhood cancer
(1–3). Currently, clinical treatment of OS
consists of a combination of chemotherapy, radiation therapy and
surgery, which has demonstrated limited improvement in OS survival
during the last decade. Unfortunately, ~30% of patients with
metastatic or recurrent disease will not survive for >5 years
(3–5). Thus, there remains an urgent
requirement to discover improved therapeutic targets that have the
potential to be developed into alternative treatment strategies
that could improve the survival rate of patients with OS.
Long non-coding RNAs (lncRNAs) are transcripts of
~200 nucleotides and ~10 kb in length (6–9),
which serve numerous biological functions across many life
processes, including cell cycle control, development and
differentiation (10,11). Recently, there has been increased
focus on lncRNAs due to their crucial role in the progression of
multiple types of cancer, including OS (12,13).
For example, lncRNA H19 is aberrantly expressed and induced by
upregulated Hedgehog signaling and Yes-associated protein 1
overexpression, and is responsible for the pathogenesis of
osteoblastic OS (14). The
oncogenic lncRNA metastasis-associated lung adenocarcinoma
transcript 1 has been reported to serve as a competing endogenous
RNA of histone deacetylase 4 by decoying microRNA-140-5p to promote
OS tumor growth (15). However, it
remains relatively unknown whether other lncRNAs have roles in
OS.
Among the wide variety of lncRNAs, long intergenic
non-protein coding RNA 00460 (LINC00460) is of importance. It has
been reported that LINC00460 is involved in the progression of
multiple types of solid carcinoma, such as kidney carcinoma, lung
adenocarcinoma, head and neck squamous cell carcinoma and
epithelial ovarian cancer (16–21).
However, to the best of our knowledge, the role of LINC00460 in OS
has not yet been reported. Through online data analysis using the
Gene Expression Profiling Interactive Analysis (GEPIA) database,
which includes RNA sequencing expression data of 9,736 tumors and
8,587 normal samples from The Cancer Genome Atlas and The
Genotype-Tissue Expression projects, it was observed in the present
study that the overall prognosis of patients with low LINC00460
expression in sarcoma was significantly improved compared with
patients with high LINC00460 expression. This implied that
LINC00460 may be involved in the progression of OS.
In the present study, a series of experiments were
performed to detect the roles of LINC00460 in OS. It was determined
that LINC00460 knockdown with small interfering RNA (siRNA)
inhibited cell viability, and migratory and invasive potential of
OS cells, which may be related to its ability to cause cell cycle
arrest, apoptosis and reduced epithelial-mesenchymal transition
(EMT) of OS cells.
Materials and methods
Chemicals and reagents
DMEM and RPMI-1640 medium were purchased from
HyClone; GE Healthcare Life Sciences, FBS was obtained from Gibco;
Thermo Fisher Scientific, Inc. and Lipofectamine® 2000
from Invitrogen; Thermo Fisher Scientific, Inc. Matrigel was
obtained from BD Biosciences, and siRNA-LINC00460 and the scrambled
siRNA were synthesized by Guangzhou Ribobio Co., Ltd. The following
primary antibodies: Anti-CDK4 (1:1,000; cat. no. 12790S), anti-CDK6
(1:1,000; cat. no. 3136P), anti-vimentin (1:1,000; cat. no. 3878S),
anti-E-cadherin (1:1,000; cat. no. 4065), anti-N-cadherin (1:1,000;
cat. no. 4061P), anti-cyclin D1 (1:1,000; cat. no. 2978P),
anti-Slug (1:1,000; cat. no. 9585S) and anti-GAPDH (1:5,000; cat.
no. 2118L) were purchased from Cell Signaling Technology, Inc. Goat
anti-rabbit or goat anti-mouse horseradish peroxidase
(HRP)-conjugated secondary antibodies (cat. nos. G1215 and G1216)
and ECL reagent were obtained from Wuhan Sanying Biotechnology. The
propidium iodide (PI) was purchased from Beyotime Institute of
Biotechnology, and the Cell Counting Kit (CCK)-8, trypsin,
penicillin, streptomycin sulfate and crystal violet were obtained
from Beijing Solarbio Science & Technology Co., Ltd. Unless
otherwise stated, other reagents were obtained from CoWin
Biosciences Co., Ltd.
Cell lines and culture
OS cell lines MG-63 (TCHu124) and U2-OS (SCSP-5030)
were obtained from the Shanghai Cell Bank of Chinese Academy of
Medical Sciences and cultured in DMEM supplemented with 10%,
penicillin (100 U/ml) and streptomycin sulfate (100 µg/ml). All
cells were maintained in a humidified incubator at 37°C and 5%
CO2. During the logarithmic growth phase, cells were
washed three times in PBS and digested with trypsin to obtain
single cell suspensions. The cells were subsequently plated out and
cultured in 6-well plates for further experimentation.
Cell transfection
OS cells in the logarithmic growth phase were
replenished with fresh DMEM 2 h prior to transfection. The siRNA
sequences against LINC00460 or the negative control (NC) were
designed and synthesized by Guangzhou RiboBio Co., Ltd. siRNA (3
µg/ml) was transfected into 3×104 OS cells at a cell
density of 3×104 cells for 6 h at 37°C using
Lipofectamine® 2000 transfection reagent, according to
the manufacturer's protocol. Subsequently, the medium was replaced
and the cells were cultured in DMEM for 48 h for subsequent
experiments. The sequences of the LINC00460 siRNAs were as follows:
NC, 5′-UUCUCCGAACGUGUCACGUTT-3′; LINC00460-1,
5′-CCAUCCACUUCAAAGUAUUTT-3′; LINC00460-2,
5′-GCCUCUGAAAUGGUGACAATT-3′; LINC00460-3,
5′-GGGAAAGAAGACGCAUUCUTT-3′ and LINC00460-4,
5′-UCACCUUGACUACUGCUAUTT-3′.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from transfected cells using
an UltraPure RNA kit (CoWin Biosciences Co., Ltd.), according to
the manufacturer's protocol. Total RNA was reverse transcribed into
cDNA using a HiFiScript cDNA Synthesis kit (CoWin Biosciences Co.,
Ltd.). The reverse transcription reaction conditions were as
follows: Incubation at 42°C for 50 min, followed by incubation at
85°C for 5 min to terminate the reaction. qPCR was performed to
assess the silencing effects of siRNA using the UltraSYBR mixture
(CoWin Biosciences Co., Ltd.). The following primer pairs were used
for the qPCR: LINC00460 forward, 5′-CAGAAATCCTCCAGCCCTGTTA-3′ and
reverse, 5′-AAGTGTCTTGGGTCATGAGTCC-3′; and β-actin forward,
5′-CCCGAGCCGTGTTTCCT-3′ and reverse, 5′-GTCCCAGTTGGTGACGATGC-3′.
The following thermocycling conditions were used for qPCR: Initial
denaturation at 95°C for 30 sec; 40 cycles of 95°C for 5 sec, 60°C
for 30 sec and one cycle of melting curve at 95°C for 15 sec, 60°C
for 1 min, 95°C for 15 sec and 50°C for 30 sec. Expression levels
were quantified using the 2−ΔΔCq method (22) from three independent experiments
and normalized to the internal reference gene β-actin.
Cell viability assay
Cell viability was determined using a CCK-8 assay.
After 24 h of transfection with siRNA, cells were trypsinized and
resuspended in DMEM; 100 µl of the cell suspension was seeded into
96-well plates at a density of 1×103 cells/well. Next,
10 µl CCK-8 reagent was added into each well and cells were
incubated in a humidified incubator at 37°C and 5% CO2
for 1.5 h. Optical density (OD) values were determined by measuring
the absorbance at a wavelength of 450 nm using a microplate reader.
Cell viability was determined every 24 h.
Colony formation assay
A total of ~800 MG-63 or U2-OS cells/well were
seeded into 6-well culture plates for colony formation assays and
incubated in the culture medium for 14 days. Subsequently, cells
were fixed with 4% methanol for 20 min and stained with 0.1%
crystal violet for 30 min at 37°C. The colonies were visualized and
quantified by counting stained colonies containing ≥50 cells.
Wound healing assay
Cells (1×104 cells/well) were seeded in
6-well plates and cultured until the cells reached 95% confluence.
A sterile 200-µl tip was used to generate the wound in the
monolayer of cells in each well, which was subsequently washed
three times and was cultured with serum-free DMEM for 24 h.
Subsequently, the cells were visualized using an Olympus IX71
inverted microscope (Olympus Corporation; magnification, ×40). The
area of the wound surface was measured with ImageJ software,
version 1.41 (National Institutes of Health). The width of each
wound was measured at five random fields for quantification. All
analysis was performed relative to the starting wound width (0 h
time point).
Transwell invasion and Matrigel
assays
Cell invasion analysis was performed using a
Matrigel-coated Transwell assay. Matrigel was defrosted from −20°C
to 4°C in a refrigerator overnight and the experiment was performed
on ice at all times, with all consumables pre-cooled. A total
volume of 100 µl diluted Matrigel in serum free-cold DMEM was added
to the upper chamber of the Transwell, which was subsequently
inserted into 24-well plates. After incubating the Transwell at
37°C for 4 h, gelled Matrigel was gently washed with warmed
serum-free RPMI-1640 medium. After 24 h of transfection, the cells
were trypsinized and re-suspended in serum-free 1640 medium at a
density of 1×105 cells/ml; 100 µl of this cell
suspension (1×104 cells) was plated in the upper chamber
and 600 µl 1640 medium containing 10% FBS was plated in the lower
chamber. After 24 h of incubation at 37°C, the non-invasive cells
remaining in the upper chamber of the Transwell plate were scraped
off with a cotton swab. The invading cells on the lower surface of
the chamber were fixed with 4% formaldehyde for 15 min at room
temperature and subsequently stained with 0.1% crystal violet for 5
min. After washing with PBS, invasive cells were counted using a
light microscope (magnification, ×100).
Cellular migration was also detected by a Transwell
assay and followed the same protocol as the Matrigel invasion
assay, with the exception that Matrigel was not used to coat the
Transwell plates.
Flow cytometry
After 24 h of transfection, cells were removed from
the culture medium and incubated in serum-free DMEM for 24 h.
Subsequently, 1×106 cells were trypsinized with
EDTA-free-trypsin and harvested by centrifugation (984 × g; 20°C; 5
min). Cells were resuspended with ice-cold PBS and harvested by
centrifugation (same as previous). Cells were fixed overnight with
70% ice-cold ethanol and then collected by centrifugation (984 × g;
20°C; 5 min). Cells were resuspended in 100 µl PBS containing 100
µg/ml RNase and incubated for 5 min at room temperature.
Subsequently, the cell suspension was added to 400 µl PBS
containing 50 µg/ml PI at room temperature for 30 sec. Following
the addition of PI, cells were immediately subjected to FACS
analysis by a BD FACSCalibur™ flow cytometer (BD Biosciences) and
FlowJo software, version 4.5 (Tree Star, Inc.) was used to analyze
the percentage of cells in each phase of the cell cycle (G0/G1, S
or G2/M).
Detection of apoptosis
A 1 mg/ml stock solution of Hoechst 33342 dye
(Beyotime Institute of Biotechnology) and a 0.5 mg/ml stock
solution of PI (Beyotime Institute of Biotechnology) in
H2O were filter-sterilized. Cells incubated in 24-well
plates were washed twice with PBS, then subsequently stained with
10 µg/ml Hoechst 33342 combined with 5 µg/ml PI at 37°C for 15 min,
according to the manufacturer's protocol. After washing twice with
PBS, the cells were visualized and counted under a fluorescence
microscope (magnification, ×100).
Gelatin zymography
The enzymatic activities of matrix metalloproteinase
(MMP)-9 were analyzed by gelatin zymography, according to previous
studies (23–25). A total of 2.5×106 cells
were seeded in 6-cm dishes and transfection was performed as
previously described. After 24 h, cells were washed twice with
serum-free DMEM prior to being cultured in serum-free DMEM for 24
h. The culture supernatant was collected by centrifugation (984 ×
g; 20°C; 5 min) and the pellet was discarded. Gelatin solution was
prepared by dissolving 500 mg gelatin in 50 ml distilled and
deionized H2O for 2 h at room temperature, and was
subsequently placed in a 65°C water bath for 15 min to obtain a
clear gelatin stock solution with a concentration of 10 mg/ml. A
10% acrylamide gel was prepared and gelatin stock solution was
added to obtain a final gelatin concentration of 0.5 mg/ml. 16 µl
of supernatant was loaded into each well and were run for 1.5–2 h
until the indicator dye reached the bottom of the gel. The gel was
washed four times for 15 min with zymogram renaturing buffer at
room temperature, and then the gel was incubated overnight in
incubation buffer at 37°C. Following incubation, the gel was
stained with 0.25% Coomassie Blue R-250 for 4 h at room
temperature, followed by the application of destaining solution
(20% methanol, 10% acetic acid, 70% distilled and deionized
H2O) with gentle agitation until areas of enzyme
activity appeared as transparent bands against the dark blue
background. The mixture was rinsed with H2O until excess
staining solution was removed. PageRuler Prestained Protein Ladder
(Thermo Fisher Scientific, Inc.) was used as a marker. The clear
bands were visualized using Image Scanner III (GE Healthcare) and
the intensity of each band was measured using ImageQuant TL v2003
software (GE Healthcare).
Western blotting
After 48 h of transfection, total protein from cells
transfected with siRNA-NC or siRNA-LINC00460 was extracted using
RIPA lysis buffer containing protease inhibitors at 4°C. Total
protein was quantified using a bicinchoninic acid protein assay and
20 µg protein/lane was separated via SDS-PAGE on a 10% gel. The
separated proteins were transferred onto a PVDF membrane and
blocked for 1 h at room temperature with 5% non-fat milk. The
membranes were incubated with the corresponding primary antibodies
overnight at 4°C. Membranes were washed three times with TBS-0.1%
Tween 20. Following the primary antibody incubation, membranes were
incubated with goat anti-rabbit/mouse HRP-conjugated secondary
antibodies (1:5,000) for 1 h at room temperature. Protein bands
were visualized using a chemiluminescence kit. GAPDH served as the
loading control. The protein bands intensity was analyzed using
Image J software, version 1.41 (National Institutes of Health).
Statistical analysis
Statistical analysis of all results was performed
using SPSS version 20.0 (IBM Corp.) software. Each experiment was
independently conducted ≥3 times and all data are expressed as the
mean ± SD. Significant differences between groups were determined
using Student's t-test or one-way ANOVA with Bonferroni correction
used as the post hoc test. Kaplan Meier analysis was used for
survival analysis, and log-rank test was conducted to determine
P-values. The prognostic value of LINC00460 in OS was analyzed
using the GEPIA database (26).
P<0.05 was considered to indicate a statistically significant
difference.
Results
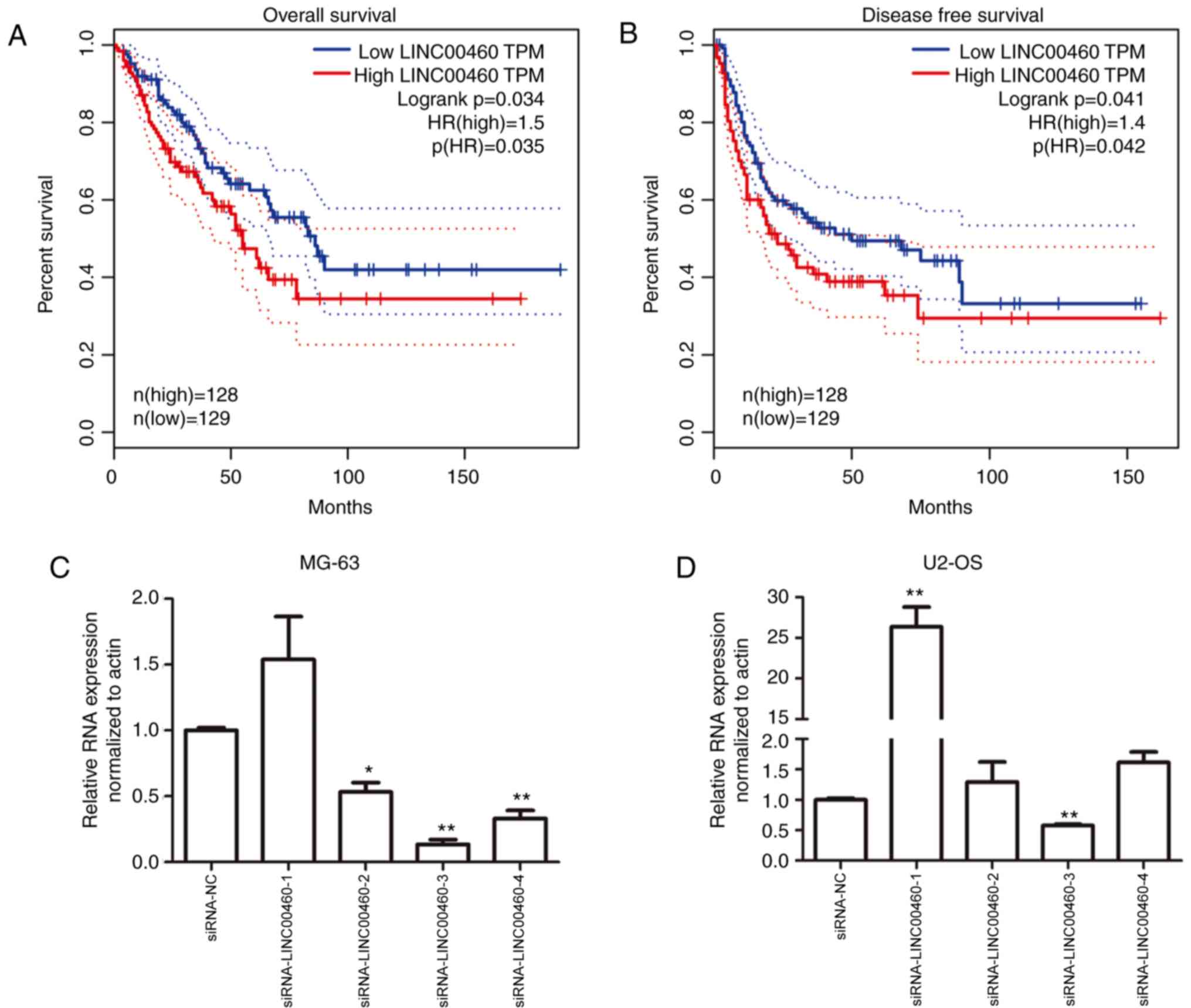
Prognostic value of LINC00460
The overall and disease-free survival of patients
with low LINC00460 expression in OS was significantly improved
compared with patients with high LINC00460 expression (P<0.05;
Fig. 1A and B).
Genetic knockdown of LINC00460 with
siRNA affects OS cell viability
To investigate the function of LINC00460 on OS
progression, siRNA was used to knockdown gene expression. Four
siRNA fragments, siRNA-LINC00460-(1–4),
were designed and synthesized to silence LINC00460 in MG-63 and
U2-OS cell lines. siRNA-LINC00460-3 demonstrated the highest siRNA
transfection efficiency in MG-63 and U2-OS cell lines, providing a
knockdown efficiency of >36.6 and 47.1% compared with NC,
respectively (P<0.01; Fig. 1C and
D). Therefore, siRNA-LINC00460-3 was selected as the siRNA
fragment used to target LINC00460 for subsequent experiments. To
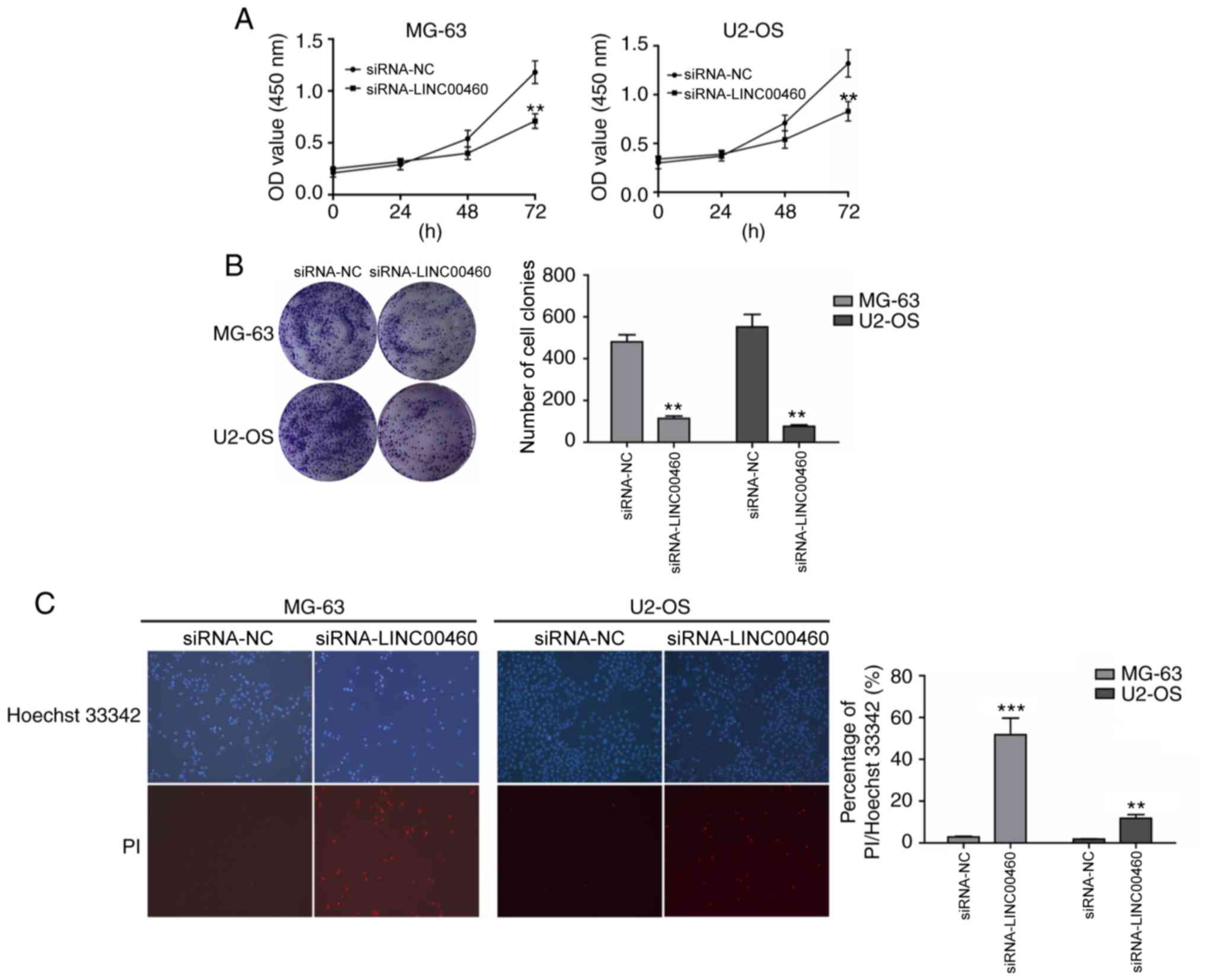
evaluate the effect of LINC00460 on cell viability, CCK-8 and
colony formation assays were conducted to detect the effect of
LINC00460 knockdown on the viability of MG-63 and U2-OS cell lines.
MG-63 and U2-OS cell lines transfected with siRNA-LINC00460-3
demonstrated significantly reduced cell viability by 48–72 h
compared with the NC (P<0.05; Fig.
2A). Similarly, the colony formation assay reported a
significant decrease in the number and size of colonies formed in
both cell lines following LINC00460 gene silencing compared with
the NC group (P<0.01; Fig. 2B).
These results suggested that the genetic knockdown of LINC00460
significantly affected OS cell viability and growth.
Genetic knockdown of LINC00460 induces
apoptosis in OS cells
To further investigate how LINC00460 knockdown
affects OS cell proliferation, the effect of LINC00460 on apoptosis
was evaluated using Hoechst/PI staining. siRNA-LINC00460-
transfected OS cells exhibited significant increases in apoptosis
compared with the siRNA-NC group (MG-63, 52.93±8.25% vs.
2.61±0.20%; U2-OS, 12.33±2.26% vs. 2.16±0.11%; P<0.01; Fig. 2C). These data demonstrated that the
genetic knockdown of LINC00460 increased apoptosis of OS cells.
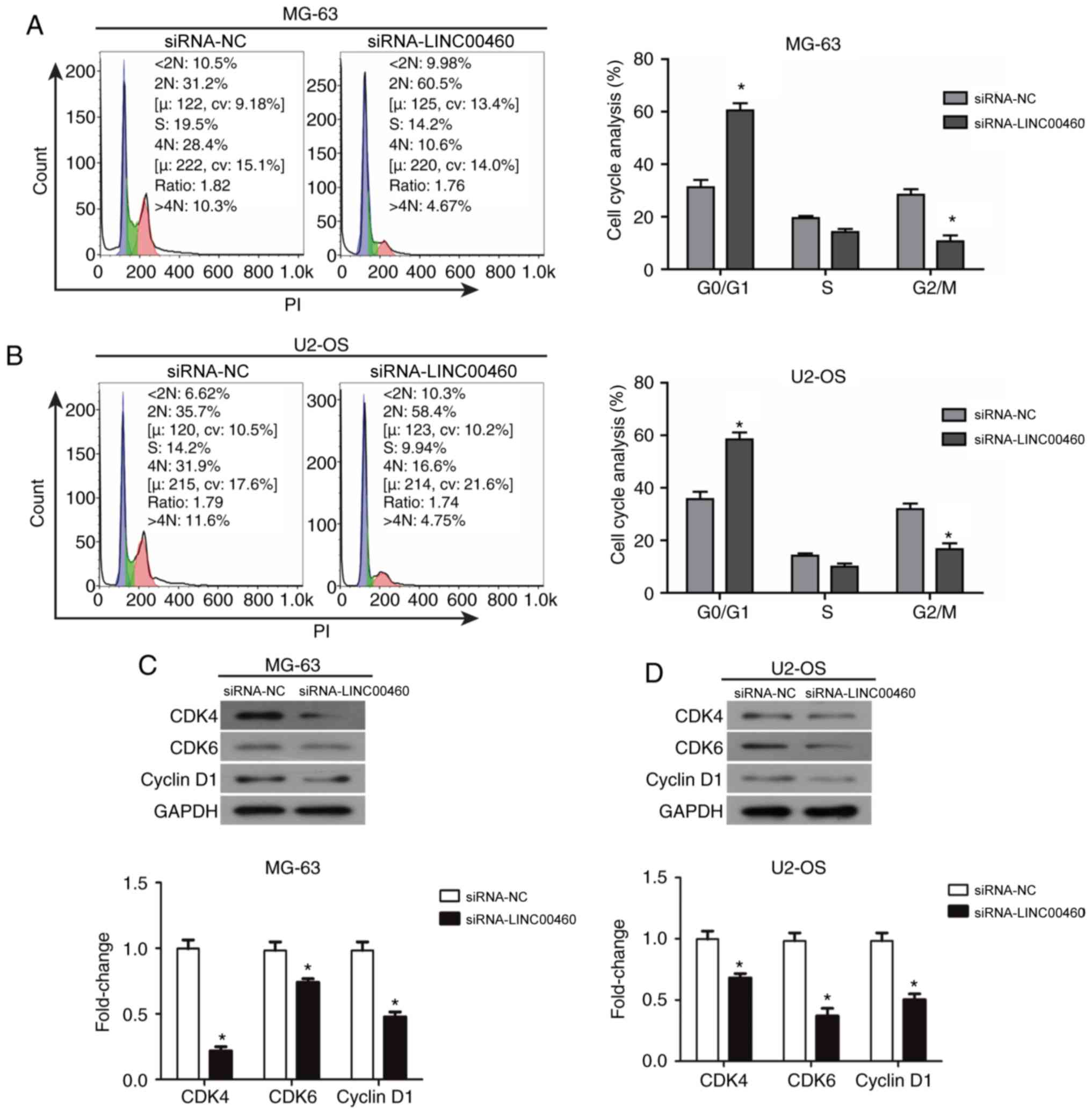
Genetic knockdown of LINC00460 induces
the cell cycle arrest of OS cells
To determine which phase of the cell cycle was
affected by LINC00460 knockdown, flow cytometric analysis was
performed to estimate the distribution of cells at different stages
of the cell cycle. The results indicated that siRNA-LINC00460
transfection induced a significant accumulation of cells in the
G0/G1 phase of the cell cycle in both MG-63 and U2-OS cell lines
compared with the siRNA-NC group (MG-63, 62.1 vs. 32.6%,
respectively; U2-OS, 59.2 vs. 38.1%, respectively; P<0.05;
Fig. 3A and B). This indicated
that siRNA-LINC00460 arrested MG-63 and U2-OS cells in the G0/G1
phase. In addition, the percentage of cells in the G2/M phase
significantly decreased in both cell lines compared with the
siRNA-NC group (MG-63, 28.4±3.21% vs. 10.6±2.20%, respectively;
U2-OS, 31.9±4.37% vs. 16.6±2.78%, respectively; P<0.05; Fig. 3A and B). There were no significant
changes observed in the S phase of either cell line. Based on these
data, it was hypothesized that siRNA-LINC00460 induced G0/G1 phase
arrest and triggered cell apoptosis to reduce cell viability in
OS.
Downregulated expression of cyclin D1
and CDK4/CDK6 may induce cell cycle arrest of OS cells in the G0/G1
phase following transfection with siRNA-LINC00460
To further validate the mechanism underlying the
G0/G1 phase cell cycle arrest following siRNA-LINC00460
transfection in OS cells, western blot analysis was performed to
examine the changes in the protein expression levels of cyclin D1,
CDK4 and CDK6 in MG-63 and U2-OS cell lines (Fig. 3C). Cyclin D1, CDK4 and CDK6
expression levels were significantly decreased following the
genetic knockdown of LINC00460 with siRNA in both cell lines
compared with the siRNA-NC group (P<0.05; Fig. 3C). These data demonstrated that OS
cells were arrested in the G0/G1 phase following transfection with
siRNA-LINC00460, which may be caused by the downregulated
expression of cyclin D1 and CDK4/CDK6.
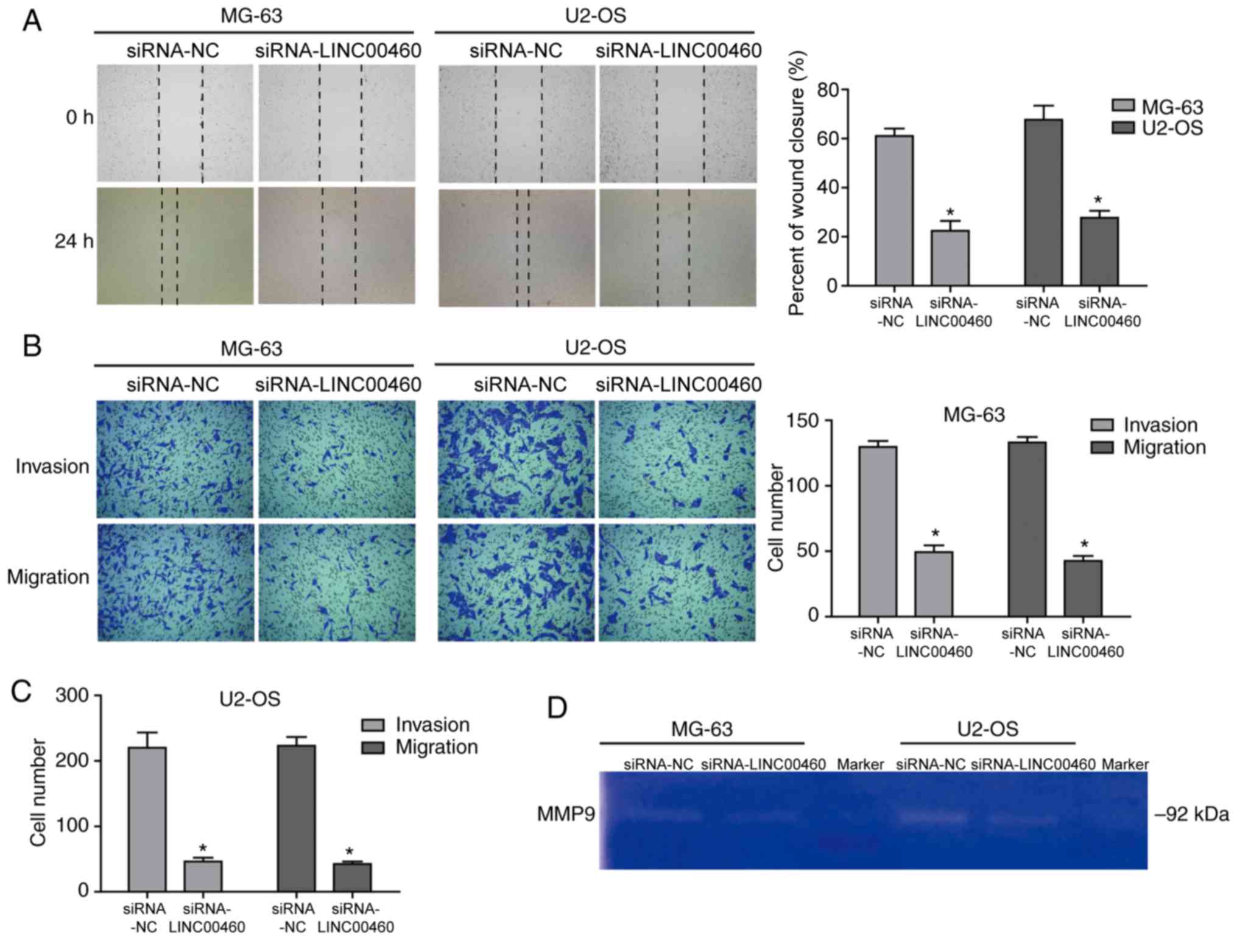
Genetic knockdown of LINC00460
inhibits the migratory and invasive abilities of OS cells
Following siRNA-LINC00460 transfection, MG-63 and
U2-OS cells demonstrated a significantly reduced migratory ability
(0.22±0.05 and 0.28±0.03, respectively) compared with the siRNA-NC
groups (0.62±0.03 and 0.67±0.08, respectively; P<0.05; Fig. 4A). Similar results were observed
using the Transwell migration assay (P<0.05; Fig. 4B and C), which suggested that the
genetic knockdown of LINC00460 significantly decreased the
migration of OS cells. In addition, the invasive ability of MG-63
and U2-OS cells was significantly decreased by ~61.4 and ~83.1%,
respectively, following transfection with siRNA-LINC00460 compared
with their respective siRNA-NCs (Fig.
4B and C). These data indicated that the invasive ability of OS
cells transfected with siRNA-LINC00460 was significantly
inhibited.
Decreased activity of MMP-9 may affect
cell invasion and migration following siRNA-LINC00460
transfection
MMPs are a family of zinc- and calcium-dependent
endopeptidases that selectively degrade components of the
extracellular matrix (27). MMPs
serve crucial roles in tumor cell progression; in particular, MMP-9
is a 92-kDa gelatinase that has been implicated in tumor invasion,
growth and distant metastasis (28). Provided that gelatin is an
important substrate of MMP-9, a gelatin zymography assay was used
to analyze MMP activity to further investigate the effect of
LINC00460 on the invasive and migratory capacity of OS cells.
Weaker gelatinolytic intensity was observed in MG-63 and U2-OS
cells transfected with siRNA-LINC00460 compared with their
respective siRNA-NC groups (Fig.
4D). These findings indicated that MMP-9 activity was decreased
in the siRNA-transfected cells and thus, that the knockdown of
LINC00460 suppressed the OS cell invasive and migratory ability
which may be caused by the decreased activity of MMP-9.
Genetic knockdown of LINC00460
inhibits the EMT process
To further confirm whether the suppression of the
EMT process could represent a mechanism behind the
siRNA-LINC00460-mediated inhibition of cellular migration and
invasion in OS cells, western blot analysis was used to examine the
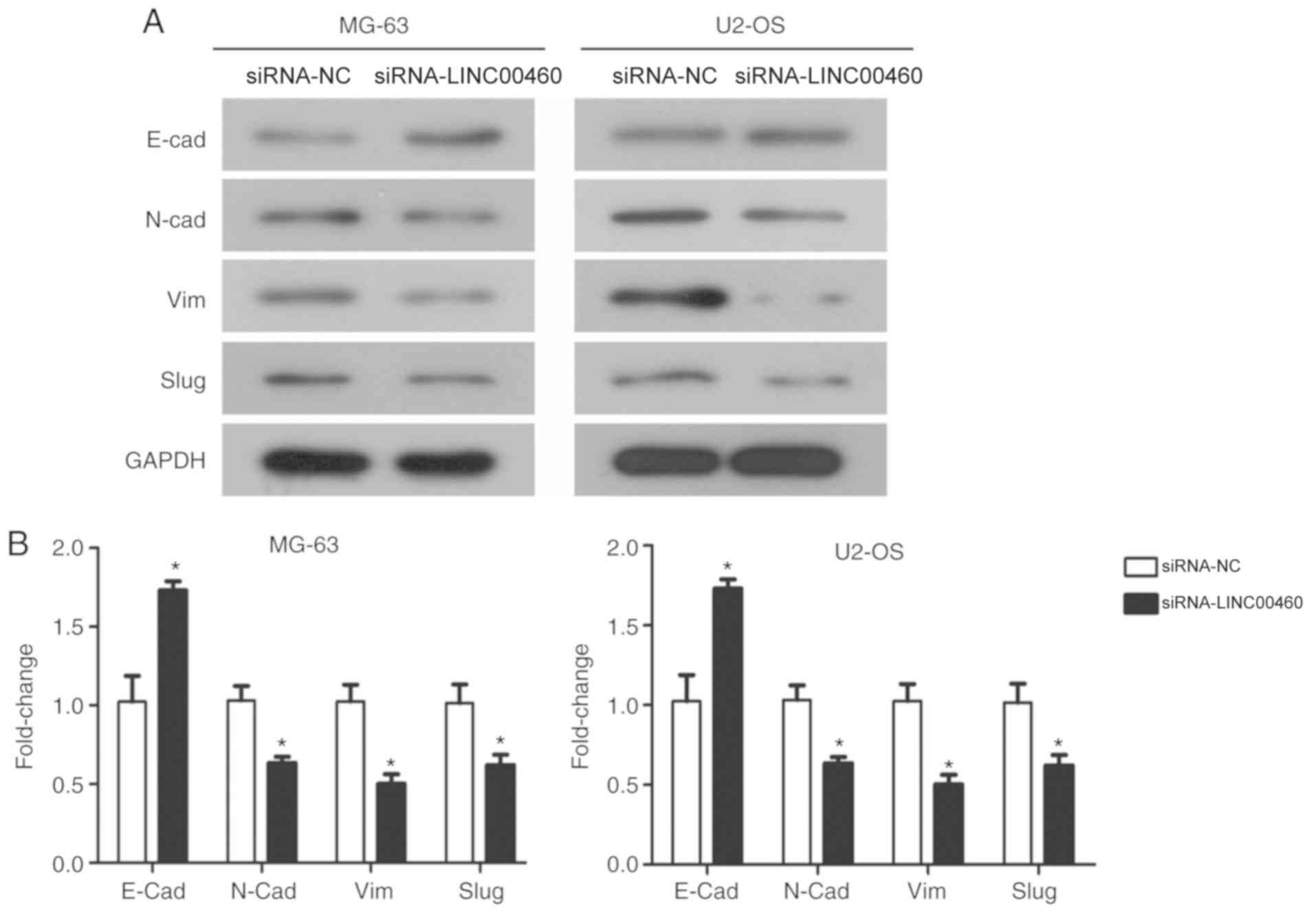
expression of EMT marker proteins. siRNA-LINC00460 transfected
MG-63 and U2-OS cells were observed to have significantly increased
expression levels of E-cadherin, an epithelial marker, and
significantly downregulated expression levels of N-cadherin and
vimentin mesenchymal markers compared with their respective
siRNA-NC groups (Fig. 5A and B).
Furthermore, there was a significant decrease in Slug, a mediator
of EMT, in both cell lines transfected with siRNA-LINC00460
compared with siRNA-NC groups (P<0.05; Fig. 5A and B). Taken together, these
results suggested that the genetic knockdown of LINC00460 inhibited
the migratory and invasive properties of OS cells through
suppressing EMT and inhibiting the expression of MMP-9.
Discussion
To the best of our knowledge, there are few previous
studies reporting the roles and respective mechanism of LINC00460.
In the present study, a siRNA knockdown strategy was used to
silence LINC00460 and investigate the effects of LINC00460 on the
biological function of OS cells. The knockdown of LINC00460
inhibited cellular proliferation through inducing apoptosis and
cell cycle arrest, effectively reducing the cellular viability of
OS cells. The rapid growth of cancer cells is a major factor
contributing to malignancy (29),
thus it is important to explore the mechanism of tumor cell
proliferation to improve cancer treatments. Cellular proliferation
is tightly regulated by different cyclins and catalytic CDKs in
each cell cycle phase (30,31);
for example, cyclin D1 interacts with CDK4 and CDK6 to form a
cyclin/CDK complex that acts as an early regulator to activate
downstream gene cascades and drive cells through the G1/S
checkpoint (32). Therefore,
abnormal levels of cyclin D1, CDK4 and CDK6 are related with a
disordered cell cycle. Using flow cytometric analysis, OS cells
with silenced LINC00460 gene expression were observed to be
arrested in the G0/G1 phase. The underlying mechanisms mediating
this were observed to be, at least partly, owing to the reduced
expression of cyclin D1 and CDK4/CDK6 in siRNA-LINC00460
transfected cells; thus, LINC00460 is suggested to modulate cell
proliferation and/or function in OS partly by upregulating the
levels of cyclin D1 and CDK4/CDK6. Kong et al (33) suggested that silencing LINC00460
suppresses nasopharyngeal carcinoma cell proliferation and growth
in vitro and in vivo through regulating
miR-149-5p/interleukin 6 signaling pathway. Liang et al
(19) also reported that silencing
LINC00460 suppresses esophageal squamous cell carcinoma cell
proliferation and growth through regulating cell cycle and inducing
apoptosis, and LINC00460 was regulated by transcriptional
co-activator CBP/P300 through histone acetylation. The present
study results are consistent with the previous research results.
However, the underlying mechanisms of carcinogenesis, such as the
other molecules involved in this interaction, will require further
exploration.
Furthermore, the genetic knockdown of LINC00460
inhibited the migratory and invasive ability of OS cells through
reducing MMP-9 expression and inhibiting EMT. MMP-9 is a member of
the MMP family, which consists of proteolytic enzymes that
selectively degrade all components of the ECM; it is also a
biological marker for tumor invasion and metastasis (27). In the present study, MMP-9 activity
was observed to be decreased following LINC00460 knockdown with
siRNA in OS cells. In addition, EMT, a pivotal biological process
in which epithelial cells gradually transform into mesenchymal-like
cells through the loss of epithelial markers and the gain of a
mesenchyme-like phenotype, serves a crucial role in the induction
of cancer cell invasion and metastasis. To investigate whether
LINC00460 knockdown decreased the migratory and invasive ability of
OS cells through the EMT pathway, the high expression of epithelial
markers and the low expression of mesenchymal markers in
LINC00460-silenced OS cells was verified. Li et al (20) also demonstrated that LINC00460
promotes cell migration and invasion by inducing EMT in lung cancer
cells by physically interacting with heterogeneous nuclear
ribonucleoprotein K, whereas it has no effect on cell
proliferation. However, this present study has several limitations.
Firstly, cultured cells in vitro are unable to simulate the
tumor microenvironment in vivo, thus this hypothesis
requires further validation using in vivo animal
tumorigenesis experiments. Secondly, the present study primarily
focused on the action mechanism of LINC00460 knockdown in the
progression of OS and additional studies with overexpressed
LINC00460 gene expression are required to validate these
findings.
In conclusion, the present study indicated that
LINC00460 functions as an oncogenic factor in OS that may
facilitate tumor cell growth, migration and invasion, and inhibit
apoptosis. The genetic knockdown of LINC00460 induced cell cycle
arrest in the G0/G1 phase, which may in part, be due to its
inhibition over the expression of cyclin D1, CDK4 and CDK6. In
addition, the knockdown of LINC00460 inhibited the migratory and
invasive potential of OS cells, which may be due to the reduced
MMP-9 expression and suppressed EMT phenotype observed. These data
suggested that LINC00460 upregulation may be a potential risk
factor associated with a poor prognosis in OS. However, the
downstream target molecules of LINC00460 will require further
investigation prior to conclusions being made. Based on these
findings, it is proposed that LINC00460 may serve as a potential
therapeutic target for OS treatment to obtain an improved
prognosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
JJJ and LPH designed the study; JJJ and FCW
performed all the experiments and analyzed the data. LPH wrote the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hansen MF, Seton M and Merchant A:
Osteosarcoma in Paget's disease of bone. J Bone Miner Res. 21
(Suppl 2):S58–S63. 2006. View Article : Google Scholar
|
|
2
|
Ottaviani G and Jaffe N: The etiology of
osteosarcoma. Cancer Treat Res. 152:15–32. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Broadhead ML, Clark JC, Myers DE, Dass CR
and Choong PF: The molecular pathogenesis of osteosarcoma: A
review. Sarcoma. 2011:9592482001.
|
|
5
|
Zhou G, Shi X, Zhang J, Wu S and Zhao J:
MicroRNAs in osteosarcoma: From biological players to clinical
contributors, a review. J Int Med Res. 41:1–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Costa FF: Non-coding RNAs: Meet thy
masters. Bioessays. 32:599–608. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He JH, Han ZP and Li YG: Association
between long non-coding RNA and human rare diseases (Review).
Biomed Rep. 2:19–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chu C, Qu K, Zhong FL, Artandi SE and
Chang HY: Genomic maps of long noncoding RNA occupancy reveal
principles of RNA-chromatin interactions. Mol Cell. 44:667–678.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP and Belgard TG: Transcribed
dark matter: meaning or myth? Hum Mol Genet. 19:R162–R168. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rios-Barrera LD, Gutiérrez-Pérez I,
Dominguez M and Riesgo- Escovar JR: acal is a long non-coding RNA
in JNK signaling in epithelial shape changes during drosophila
dorsal closure. PLoS Genet. 11:e10049272015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun J, Lin Y and Wu J: Long non-coding RNA
expression profiling of mouse testis during postnatal development.
PLoS One. 8:e757502013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chan LH, Wang W, Yeung W, Deng Y, Yuan P
and Mak KK: Hedgehog signaling induces osteosarcoma development
through Yap1 and H19 overexpression. Oncogene. 33:4857–4866. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun Y and Qin B: Long noncoding RNA MALAT1
regulates HDAC4-mediated proliferation and apoptosis via decoying
of miR-140-5p in osteosarcoma cells. Cancer Med. 7:4584–4597. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye JJ, Cheng YL, Deng JJ, Tao WP and Wu L:
LncRNA LINC00460 promotes tumor growth of human lung adenocarcinoma
by targeting miR-302c-5p/FOXA1 axis. Gene. 685:76–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji
T, Chen WT and Zou X: A three-lncRNA signature derived from the
Atlas of ncRNA in cancer (TANRIC) database predicts the survival of
patients with head and neck squamous cell carcinoma. Oral Oncol.
65:94–101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao G, Fu Y, Su Z and Wu R: How long
non-coding RNAs and MicroRNAs mediate the endogenous RNA network of
head and neck squamous cell carcinoma: A comprehensive analysis.
Cell Physiol Biochem. 50:332–341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang Y, Wu Y, Chen X, Zhang S, Wang K,
Guan X, Yang K, Li J and Bai Y: A novel long noncoding RNA
linc00460 up-regulated by CBP/P300 promotes carcinogenesis in
esophageal squamous cell carcinoma. Biosci Rep. 37(pii):
BSR201710192017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y,
Zhou JK, Guo C, Xia Z, Liu L, et al: Long non-coding RNA linc00460
promotes epithelial-mesenchymal transition and cell migration in
lung cancer cells. Cancer Lett. 420:80–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Wen J, Wang H and Wang Y: Long
non-coding RNA LINC00460 promotes epithelial ovarian cancer
progression by regulating microRNA-338-3p. Biomed Pharmacother.
108:1022–1028. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak JK and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lou C, Zhu Z, Zhao Y, Zhu R and Zhao H:
Arctigenin, a lignan from Arctium lappa L., inhibits metastasis of
human breast cancer cells through the downregulation of MMP-2/-9
and heparanase in MDA-MB-231 cells. Oncol Rep. 37:179–184. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Muthukuru M and Cutler CW: Resistance of
MMP9 and TIMP1 to endotoxin tolerance. Pathog Dis. 73(pii):
ftu0032015.PubMed/NCBI
|
|
25
|
Banu SK, Lee J, Starzinski-Powitz A and
Arosh JA: Gene expression profiles and functional characterization
of human immortalized endometriotic epithelial and stromal cells.
Fertil Steril. 90:972–987. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wick W, Platten M and Weller M: Glioma
cell invasion: Regulation of metalloproteinase activity by
TGF-beta. J Neurooncol. 53:177–185. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hua J and Muschel RJ: Inhibition of matrix
metalloproteinase 9 expression by a ribozyme blocks metastasis in a
rat sarcoma model system. Cancer Res. 56:5279–5284. 1996.PubMed/NCBI
|
|
29
|
Zhou JJ, Xie Y, Zhao Y and Li ZX: Neuron
specific enolase gene silencing suppresses proliferation and
promotes apoptosis of lung cancer cells in vitro. Nan Fang Yi Ke Da
Xue Xue Bao. 31:1336–1340. 2011.(In Chinese). PubMed/NCBI
|
|
30
|
Hall M and Peters G: Genetic alterations
of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human
cancer. Adv Cancer Res. 68:67–108. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sherr CJ and Roberts JM: CDK inhibitors:
Positive and negative regulators of G1-phase progression. Genes
Dev. 13:1501–1512. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kong YG, Cui M, Chen SM, Xu Y, Xu Y and
Tao ZZ: LncRNA-LINC00460 facilitates nasopharyngeal carcinoma
tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene.
639:77–84. 2018. View Article : Google Scholar : PubMed/NCBI
|