Introduction
Tissue factor (TF) is a 47-kD transmembrane
cell-surface glycoprotein, which is primarily known as the
initiator of the blood coagulation cascade (1). Human TF is genetically encoded by the
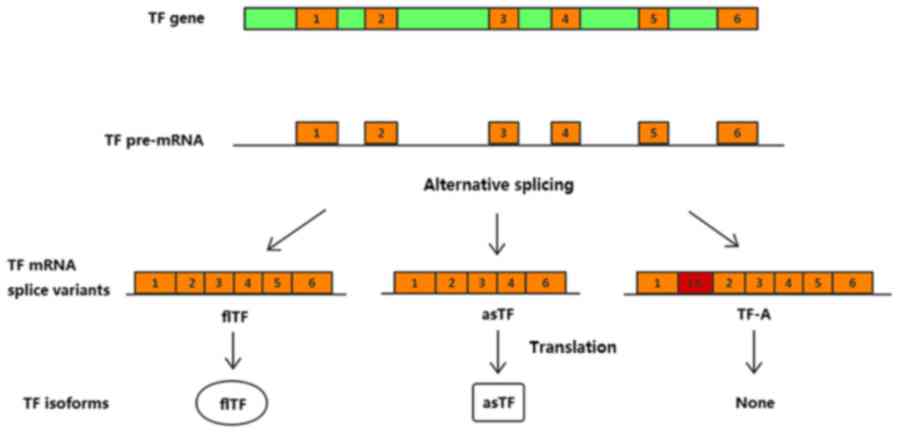
TF gene, which is transcribed to TF premature mRNA. Alternative
splicing of TF results in three naturally occurring protein
isoforms: Full-length (fl)TF, alternatively spliced (as)TF and
TF-A. asTF and flTF serve important and distinct roles in various
biological processes that involve vessel formation and maturation,
and initiation of the blood coagulation cascade (2). asTF, which arises from exclusion of
the fifth exon of the primary TF transcript, exhibits low
prothrombogenic potential but is more closely associated with tumor
growth, angiogenesis, metastasis and cell growth (3–5).
TF-A, another splice variant, is only expressed at the mRNA level
in a number of cancer cell lines and in endothelial cells (Fig. 1). To the best of our knowledge, the
biological function of TF-A mRNA is currently unknown; therefore,
this splice variant was not taken into account in the present study
(6–8).
Few studies regarding TF expression have
discriminated between flTF and asTF; therefore, it is necessary to
investigate the expression status of these two isoforms in
different diseases. Bile acids are strong signaling molecules that
are capable of influencing various biological processes, including
inflammation, apoptosis, cancer progression and atherosclerosis.
Chenodeoxycholic acid (CDCA) is a bile acid that has been
demonstrated to enhance ectopic vessel formation (9). Similarly, apolipoprotein M (apoM),
which was discovered by Xu and Dahlbäck in 1999 (10), is mainly located in high-density
lipoprotein in the blood and has been demonstrated to be associated
with tumor growth, atherosclerosis and thrombosis (11–13).
In order to investigate the association between TF variants and
CDCA/apoM, the difference in the expression of TF variants in
various cell strains and tissues was examined in the present study.
The results of this study may contribute to further studies on the
function and mechanism of TF in associated diseases.
Materials and methods
Cell lines and cell culture
Human cervical cancer cell lines [C-33A, human
papilloma virus (HPV)-negative; HeLa, HPV18-positive; and SiHa,
HPV16-positive], human breast cancer cell lines (ZR-75-1, luminal A
subtype; MCF-7, luminal A subtype; BT-474, luminal B subtype;
MDA-MB-468, basal-like subtype; and MDA-MB-231, basal-like
subtype), a human hepatoblastoma cell line (HepG2), a human
colorectal cancer cell line (Caco-2) and a human umbilical vein
cell line (EA.hy926) were purchased from the Cell Bank of Type
Culture Collection of the Chinese Academy of Sciences (Shanghai,
China). All cells were cultured according to their respective
conditions and maintained in the Comprehensive Laboratory of The
Third Affiliated Hospital of Soochow University (Changzhou,
China).
The C-33A, HeLa, SiHa, Caco-2 and HepG2 cells were
cultured in minimum essential medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and the MCF-7 and EA.hy926
cells were maintained and cultured in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific, Inc.). The BT-474 and
ZR-75-1 cells were cultured in RPMI-1640 media (Gibco; Thermo
Fisher Scientific, Inc.), whereas the MDA-MB-468 and MDA-MB-231
cells were cultured in Leibovitz's L-15 medium (Gibco; Thermo
Fisher Scientific, Inc.). All of the complete media were
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin, and 10%
non-essential amino acid solution was added for the culture of
cervical cancer cell lines. All cells were incubated at 37°C in an
atmosphere containing 5% CO2, with the exception of the
MDA-MB-468 and MDA-MB-231 cells, which were cultured at 37°C in a
humidified atmosphere containing 100% air.
The HepG2 and EA.hy926 cells were seeded in 6-well
plates with the concentration of 2×105 cells/well, and
allowed to grow to 80–90% confluence. Subsequently, they were
washed and incubated in serum-free medium containing 50 µM CDCA
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) with 0.1% ethanol
for 24 h at 37°C. Total RNA was then extracted.
The apoM coding sequence was obtained by polymerase
chain reaction (PCR) amplification from human genomic DNA (forward
primer, 5′-GAGGATCCCCGGGTACCGGTCGCCACCATGTTCCACCAAATTTGGGCAGC-3′;
reverse primer, 5′-TCCTTGTAGTCCATACCGTTATTGGACAGCTCACAGGCCTC-3′)
and was inserted into the Ubi-MCS-3FLAG-CMVEGFP vector (cat. no.
GV365; Shanghai Genechem Co., Ltd., Shanghai, China). Empty
lentiviral vectors with green fluorescent protein (GFP) and
lentivirus-mediated human apoM overexpression vectors with GFP were
prepared by Shanghai GeneChem Co., Ltd. In brief, 20 µg/ml of GV365
vector, 15 µg/ml of pHelper 1.0 and 10 µg/ml of pHelper 2.0
(Shanghai Genechem Co., Ltd.) were cotransfected into 293T cells
(Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences, Shanghai, China) with enhanced infection solution and
polybrene (Shanghai Genechem Co., Ltd.,), cultured at 37°C for 48 h
at 70% confluence. Lentiviral particles [1×109
transducing units (TU)/ml] were obtained from supernatants
following centrifugation at a speed of 75,000 × g for 2 h at 4°C.
Subsequently, Caco-2 and EA.hy926 cells at 50% confluence were
incubated with the lentiviral vector (multiplicity of infection,
50; 3.75×108 TU) in dishes. After 12 h, culture medium
containing the lentivirus was aspirated from the wells and fresh
complete medium was added. The expression intensity of GFP was
observed 3 days later and apoM overexpression was confirmed using
reverse transcription-quantitative PCR (RT-qPCR). Caco-2 and
EA.hy926 cells transfected with empty vectors (multiplicity of
infection=50) were set up as the corresponding negative control
(NC) groups.
Tissue sample collection
Placenta tissue specimens from 20 women (age, 24–33
years); esophageal cancer tissue specimens from 18 men and two
women (age, 56–72 years); 14 breast cancer tissue specimens (age,
46–74); 34 cervical cancer specimens (age, 35–75 years) and 16
normal cervical control samples (age, 38–65 years); and non-small
cell lung cancer (NSCLC) tissues and adjacent/normal tissues from
seven men and nine women (age, 48–76 years) were collected at the
Third Affiliated Hospital of Soochow University between July 2014
and September 2016. The patients had not received preoperative
radiotherapy and/or chemotherapy. The experimental protocols were
approved by the Institutional Ethics Committee of the Third
Affiliated Hospital of Soochow University and all patients provided
written informed consent for this study. All patients had undergone
modified radical operations. All tissue samples including placental
tissues, esophageal cancer tissues, breast cancer tissues, cervical
cancer tissues, normal cervical tissues, NSCLC tissues and their
adjacent/normal tissues were excised and quickly frozen in liquid
nitrogen until subsequent analysis.
RNA isolation and RT-qPCR
Total RNA was extracted from cells and tissues using
the total RNA purification kit (Shenergy Biocolor Biological
Science & Technology Company, Shanghai, China), according to
the manufacturer's protocol. cDNA was synthesized, according to the
manufacturer's protocol, using the RevertAid First Strand cDNA
Synthesis kit (Thermo Fisher Scientific, Inc.). Primers and TaqMan
probes (labeled with carboxyfluorescein) for human flTF and asTF
were designed using Primer Premier version 5.0 (Premier Biosoft
International, Palo Alto, CA, USA) and were synthesized by Sangon
Biotech Co., Ltd. (Shanghai, China) (Table I). The mRNA expression levels of
flTF and asTF were quantified relative to the mRNA expression
levels of GAPDH, and quantification was performed using a
LightCycler® 480 Instrument II (Roche Applied Science,
Penzberg, Germany) in a final volume of 25 µl. PCR reactions,
purchased from the Shenergy Biocolor BioScience and Technology
Company (Shanghai, China), were performed using the following
mixture: 2.5 µl MgCl2 (25 mM; with 4 µl MgCl2 in the asTF PCR
reactions), 2.5 µl PCR buffer (10X), 0.5 µl 4X dNTP (10 mM), 0.25
µl Taq DNA polymerase (5 units), 0.04 µl each primer and probe (100
µM), 2 µl cDNA template (replaced by water in no template controls)
and nuclease-free water to a final volume of 25 µl. Thermal cycling
was performed under the following conditions: 3 min of initial
denaturation at 95°C, followed by 40 cycles at 95°C for 5 sec
(temperature transition rate 4.4°C/sec) and 60°C extension for 25
sec. Samples were amplified simultaneously in triplicate in a
single assay run. mRNA expression levels are presented as a ratio
between the target gene and GAPDH gene expression; the fold-change
was calculated using 2−ΔΔCq (14).
 | Table I.Primers and probes for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers and probes for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Probe (5′FAM,
3′BHQ-1) |
|---|
| flTF |
TGATGTGGATAAAGGAGAAAACTACTGT |
CTACCGGGCTGTCTGTACTCTTC |
TTCAAGCAGTGATTCCCTCCCGAACA |
| asTF |
ATCTTCAAGTTCAGGAAAGAAATATTCTAC |
GCTCTGCCCCACTCCTGCC |
TTGGAGCTGTGGTATTTGTGGTCATCATC |
| GAPDH |
CAGGGCTGCTTTTAACTCTGGT |
CATGGGTGGAATCATATTGGAAC |
TGGATATTGTTGCCATCAATGA CCCCT |
Statistical analysis
Statistical analysis was performed using GraphPad
Prism software version 5.0 (GraphPad Software, Inc., La Jolla, CA,
USA). All data are expressed as the means ± standard deviation or
standard error of the mean. Data were analyzed using Student's
t-test or one-way analysis of variance (ANOVA). Tukey's multiple
comparison test was conducted following ANOVA to compare multiple
groupse (NSCLC tissues and their adjacent/normal tissues).
P<0.05 was considered to indicate a statistically significant
difference.
Results
mRNA expression levels of TF variants
in human cell lines and tissues
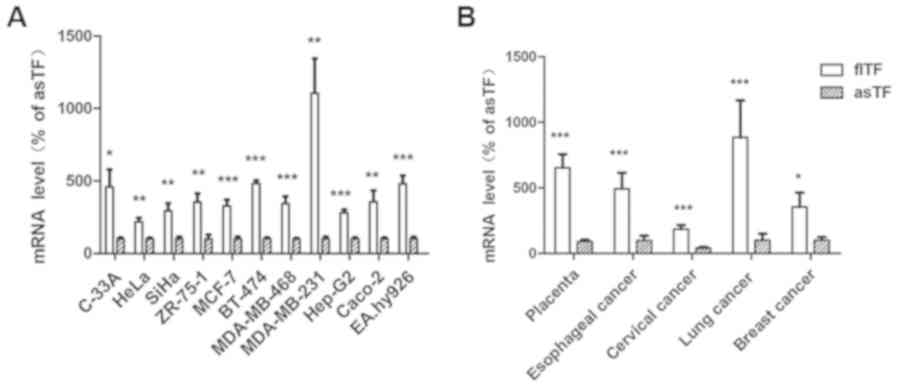
As shown in Fig.
2A, the mRNA expression levels flTF were compared with those of
asTF in 11 human cell lines, including human cervical cancer
(C-33A, HeLa and SiHa), breast cancer (ZR-75-1, MCF-7, BT-474,
MDA-MB-468 and MDA-MB-231), hepatoblastoma (HepG2), colorectal
cancer (Caco-2) and umbilical vein (EA.hy926) cells. The expression
levels were also compared in five types of tissue specimen,
including placenta, esophageal cancer, cervical cancer, lung cancer
and breast cancer tissues (Fig.
2B). The results demonstrated that flTF and asTF exist in a
wide range of human tissues and cells, and the mRNA expression
levels of flTF were significantly higher compared with asTF in all
samples tested (P<0.05).
Effects of CDCA on the mRNA expression
levels of TF variants in HepG2 and EA.hy926 cells
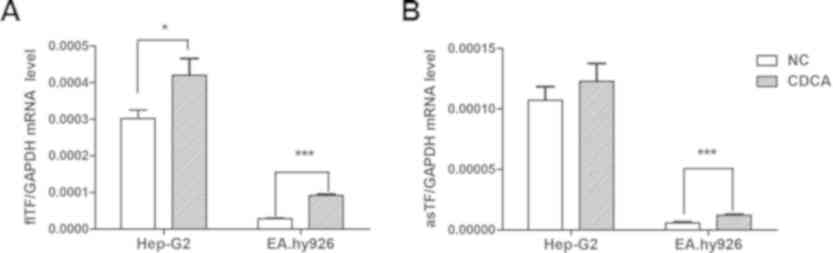
There was a significant increase in the expression
levels of flTF in the HepG2 and EA.hy926 cells treated with CDCA
(Fig. 3A). CDCA also promoted the
expression of asTF in EA.hy926 cells, but had no significant effect
on HepG2 cells (Fig. 3B).
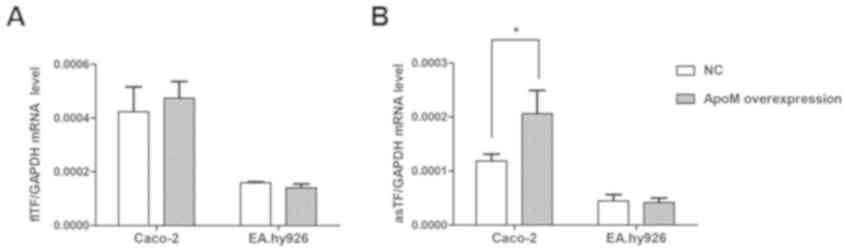
Effects of apoM overexpression on the
mRNA expression levels of TF variants in Caco-2 and EA.hy926
cells
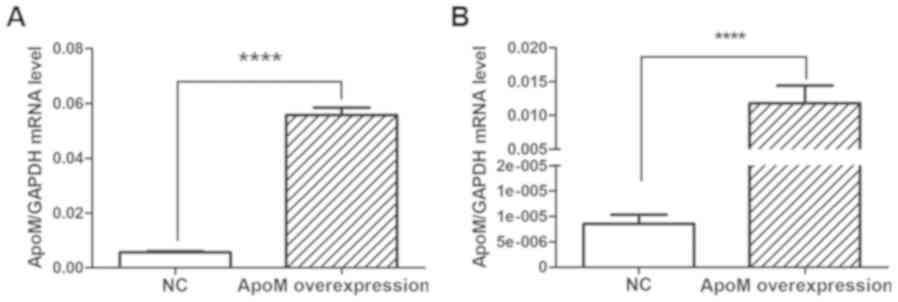
Compared with in the NC (empty vector control)
group, apoM lentivirus transduction increased apoM expression by
9.95-fold (P<0.0001) in Caco-2 cells (Fig. 4A) and 1,379.07-fold (P<0.0001)
in EA.hy926 cells (Fig. 4B). As
shown in Fig. 5, the results
demonstrated that the mRNA expression levels of asTF were increased
in Caco-2 cells overexpressing apoM compared with in the NC group
(P<0.05); however, no significant effect was observed on flTF
expression in those cells. In addition, apoM overexpression had no
significant effect on flTF and asTF in EA.hy926 cells.
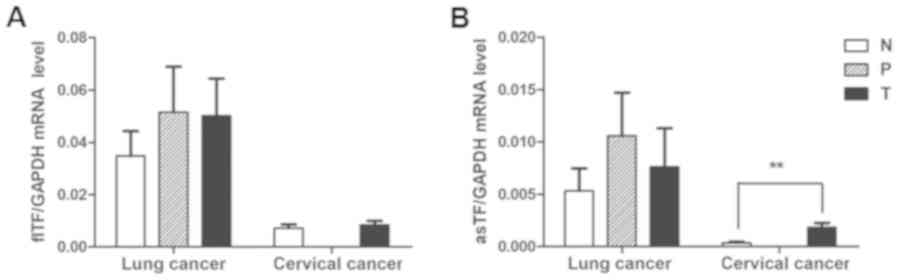
mRNA expression levels of TF variants
in tumor, normal and adjacent tissues
To investigate whether the expression of individual
splice variants differed during the tumor process, the expression
levels of flTF and asTF were investigated in cervical and lung
cancer specimens. As shown in Fig.
6, there was no significant difference in the mRNA expression
levels of flTF and asTF splice variants among any of the lung
cancer tissues. There was also no significant difference in the
mRNA expression levels of flTF between cervical cancer and normal
cervical tissues; however, asTF was significantly increased in
cervical cancer tissues compared with in normal cervical tissues
(P<0.01).
Summary of the negative results
Overexpression of apoM had little effect on the mRNA
expression levels of flTF in the Caco-2 and EA.hy926 cells. In
addition, there was no significant difference between the mRNA
expression levels of flTF in the adjacent/normal and tumor NSCLC
and cervical cancer tissues (Table
II). With regards to asTF mRNA expression, CDCA had little
effect on HepG2 cells and apoM overexpression did not affect the
levels of asTF in the EA.hy926 cells. Similarly, there was no
statistically significant difference in asTF mRNA levels between
the adjacent and tumor NSCLC tissues.
 | Table II.Expression levels of flTF and asTF in
cell lines and tissues. |
Table II.
Expression levels of flTF and asTF in
cell lines and tissues.
|
|
| flTF | asTF |
|---|
|
|
|
|
|
|---|
| Cell lines and
tissues | Group | Expression
levels | P-value | Expression
levels | P-value |
|---|
| Caco-2 | ApoM OE group |
4.73×10−4±6.22×10−5 | 0.66 |
2.29×10−4±4.53×10−5 | 0.03a |
|
| Control group |
4.23×10−4±9.22×10−5 |
|
1.19×10−4±1.27×10−5 |
|
| EA.hy926 | ApoM OE group |
1.40×10−4±1.43×10−5 | 0.24 |
4.17×10−5±8.43×10−6 | 0.82 |
|
| Control group |
1.59×10−4±3.86×10−6 |
|
4.50×10−5±1.14×10−5 |
|
| HepG2 cells | CDCA group |
4.20×10−4±4.53×10−5 | 0.04a |
1.23×10−4±1.48×10−5 | 0.42 |
|
| Control group |
3.02×10−4±2.34×10−5 |
|
1.07×10−4±1.11×10−5 |
|
| NSCLC | Normal tissues | 0.034±0.034 | 0.97 | 0.005±0.008 | 0.54 |
|
| Adjacent
tissues | 0.052±0.067 |
| 0.011±0.016 |
|
|
| Tumor tissues | 0.040±0.041 |
| 0.008±0.015 |
|
| Cervical
cancer | Normal tissues | 0.007±0.006 | 0.62 | 0.0003±0.0004 | 0.001a |
|
| Tumor tissues | 0.007±0.007 |
| 0.002±0.002 |
|
Discussion
Since asTF and flTF serve important and often
distinct roles in various biological processes, it is appropriate
to study them separately. However, as shown in Table III (15–52),
total TF, including asTF and flTF, or flTF alone, has been
investigated in studies over the last decade. Few studies have been
conducted that have detected asTF and flTF levels separately,
according to the primer sequences that have been designed.
Furthermore, the primers used in three of these previous studies
(27,29,48)
are not completely matched with the TF sequence and therefore
cannot be used to amplify asTF or flTF.
 | Table III.Information regarding the TF primers
and amplified products in studies over the last decade
(2008–2017). |
Table III.
Information regarding the TF primers
and amplified products in studies over the last decade
(2008–2017).
| Author, year | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Products | (Refs.) |
|---|
| Rossiello MR et
al, 2008 |
AAACCCGTCAATCAAGTCTAC |
GAAAGTGTTGTTCCTTCTGAC | flTF and asTF | (15) |
| Sovershaev MA et
al, 2008 |
CCCCAGAGTTCACACCTTACCT |
CACTTTTGTTCCCACCTGTTCA | flTF and asTF | (16) |
| Regina S et
al, 2008 |
CCAAACCCGTCAATCAAGT |
TGCTTCACATCCTTCACAATCTCG | flTF and asTF | (17) |
| Plotkowski MC et
al, 2008 | CCCGAACAGTTAACC
GGAAGA |
GCTCCAATGATGTAGAATATTTCTCTGA | flTF | (18) |
| Liu F et al,
2008 |
GCACTAAGTCAGGAGATTGG |
CTGTCTGTACTCTTCCGGTT | flTF | (19) |
| Usui M et
al, 2009 |
CAGTGCAATATAGCATTTGCAGTAGC |
CTACTGTTTCAGTGTTCAAGCAGTGA | flTF | (20) |
| Lockwood CJ et
al, 2009 |
GAAGCAGACGTACTTGGCACGG |
CCGAGGTTTGTCTCCAGGTA | flTF and asTF | (21) |
| Di Stefano R et
al, 2009 |
TGAATGTGACCGTAGAAGATGA |
GGAGTTCTCCTTCCAGCTCT | flTF and asTF | (22) |
| Wang HJ et
al, 2009 |
TCCCGAACAGTTAACCGGAA |
GACCACAAATACCACAGCTCCA | flTF | (23) |
| Wang HJ et
al, 2010 |
CAGGGAATGTGGAGAGCAC |
GGCTGTCCGAGGTTTGTC | flTF and asTF | (24) |
| Teng Y et
al, 2010 |
CCGAACAGTTAACCGGAAGA |
TCAGTGGGGAGTTCTCCTTC | flTF | (25) |
| Valsami S et
al, 2011 |
AATGTGGAGAGCACCGGTTCT |
CGTTCATCTTCTACGGTCACATTC | flTF and asTF | (26) |
| Blank M et
al, 2011 |
TTCACACCTTACCTGGAGACAAAC |
AACATCCCGGAGGAGGCTTAGGA | -- | (27) |
| Ben-Hadj-Khalifa S
et al, 2011 |
CCGACGAGATTGTGAAGGATGT |
AGAGGCTCCCCAGCAGAAC | flTF and asTF | (28) |
| Gerotziafas GT
et al, 2012 |
AATGTGGAGACCGGTTCT |
TCCGTTCATCTTCTACGGTCAC | -- | (29) |
| Wang JG et
al, 2012 |
AATGTGGAGAGCACCGGTTC |
CGTTCATCTTCTACGGTCACATTC | flTF and asTF | (30) |
| Gebhard C et
al, 2012 |
TCCCCAGAGTTCACACCTTACC |
TGACCACAAATACCACAGCTCC | flTF and asTF | (31) |
| Lin M et al,
2012 |
AGAGTACAGACAGCCCGGTAGAGTG |
GCCAGGATGATGACAAGGATGA | flTF | (32) |
| Maruyama K et
al, 2012 |
CTCGGACAGCCAACAATTCAGAGT |
TGTTCGGGAGGGAATCACTGCTTGAACACT | flTF | (33) |
| Sun L et al,
2013 |
GCCAGGAGAAAGGGGAAT |
CAGTGCAATATAGCATTTGCAGTAGC | flTF | (34) |
| Yang HP et
al, 2013 |
GAACCCAAACCCGTCAAT |
TCTCATACAGAGGCTCCC | flTF and asTF | (35) |
| Eisenreich A et
al, 2013 |
CTCGGACAGCCAACAATTCAG |
CGGGCTGTCTGTACTCTTCC | flTF | (36) |
| Carneiro-Lobo TC
et al, 2014 |
CAGGCACTACAAATACTGTGG |
TGTAGACTTGATTGACGGGTT | flTF and asTF | (37) |
| Chen C et
al, 2014 |
GTGATTCCCTCCCGAACAGTT |
CTGGCCCATACACTCTACCG | flTF | (38) |
| Balia C et
al, 2014 |
TTGGCAAGGACTTAATTTATACAC |
CTGTTCGGGAGGGAATCAC | flTF | (39) |
| Liu LX et
al, 2014 |
ACGCTCCTGCTCGGCTGGGT |
CGTCTGCTTCACATCCTTCA | flTF and asTF | (40) |
| Bravo ML et
al, 2015 |
CCAAACCCGTCAATCAAGTC |
ACAATCTCGTCGGTGAGGTC | flTF and asTF | (41) |
| Sovershaev TA et
al, 2015 |
GAATGTGACCGTAGAAGATG |
CACTGAAACAGTAGTTTTCTCC | flTF | (42) |
| Wang B et
al, 2015 |
CAAACCCGTCAATCAAGTCTAC |
CTTCACATCCTTCACAATCTCG | flTF and asTF | (43) |
| Wang R et
al, 2015 |
CACCGACGAGATTGTGAAGG |
CGGAGGCTTAGGAAAGTGTTG | flTF and asTF | (44) |
| Orellana R et
al, 2015 |
CCAAACCCGTCAATCAAGTC |
ACAATCTCGTCGGTGAGGT | flTF and asTF | (45) |
| Jacobsen C et
al, 2015 |
GCCAGGAGAAAGGGGAAT |
CAGTGCAATATAGCATTTGCAG | flTF | (46) |
| Dong R et
al, 2015 |
TGACCTCACCGACGAGATTGTGAA |
TCTGAATTGTTGGCTGTCCGAGGT | flTF and asTF | (47) |
| Li W et al,
2016 |
AAGCAGTGATTCCCTCTCG |
AACACAGCATTGGCAGCAG | -- | (48) |
| Scalise V et
al, 2016 |
TTGGCAAGGACTTAATTTATACAC |
CTGTTCGGGAGGGAATCAC | flTF | (49) |
| Krychtiuk KA et
al, 2017 |
CAGACAGCCCGGTAGAGTGT |
CCACAGCTCCAATGATGTAGAA | flTF | (50) |
| Gao H et al,
2017 |
GGCGCTTCAGGCACTACAA |
TTGATTGACGGGTTTGGGTTC | flTF and asTF | (51) |
| Brambilla M et
al, 2018 |
TGATGTGGATAAAGGAGAAAACTACTGT |
TCTACCGGGCTGTCTGTACTCTT | flTF | (52) |
In terms of expression, as the major form of TF, the
mRNA expression levels of flTF were higher than those of asTF in
all specimens tested in the present study. Furthermore, in terms of
function, CDCA increased the expression levels of flTF in HepG2
cells, whereas those of asTF were not affected. Therefore, it was
hypothesized that flTF may contribute to the onset of liver cancer.
The present study also demonstrated that CDCA increased the
expression levels of flTF and asTF in EA.hy926 cells, which
indicated that CDCA may be associated with vasoconstriction.
Furthermore, Kundu et al (9) demonstrated that CDCA is capable of
promoting vessel formation. In a previous study, both variants were
revealed to mediate various physiological and pathological
functions, including angiogenesis (2). Therefore, CDCA may promote vessel
formation through the upregulation of these two variants; however,
the possible mechanism requires further investigation.
asTF is associated with the development of numerous
types of cancer. A previous study demonstrated that the mRNA
expression levels of apoM in colorectal cancer tissues were
significantly increased in patients with lymph node metastasis
(53). The present study revealed
that the expression levels of asTF were increased in Caco-2 cells
that overexpressed apoM compared with the control cells.
Conversely, overexpression of apoM had little effect on flTF.
Consistent with this result, Yu et al (54) demonstrated that flTF expression in
colorectal cancer has no influence on cell proliferation in
vitro; however, asTF has been reported to promote cell
proliferation in vitro and tumor growth in vivo
(1,36,55).
It may therefore be hypothesized that asTF, not flTF, contributes
to the onset of cancer.
Venteclef et al (56) demonstrated that bile acids suppress
apoM expression in vitro and in vivo. The results of
the present study demonstrated that as one of the bile acids, CDCA
increased the expression levels of the two TF variants in the
EA.hy926 cells and apoM increased as TF expression in Caco-2 cells.
These data suggested that apoM may be involved in the regulation of
TF expression induced by CDCA, but this requires further
investigation.
The mRNA expression levels of flTF were not
significantly different between the cervical cancer and normal
tissues; however, those of asTF were markedly increased in cancer
tissues. Previous studies regarding the coagulant properties of
asTF have been fairly inconclusive (57,58),
but they are likely to be essential during angiogenesis associated
with the development of cancer (59,60).
The present study demonstrated that the measurement of asTF mRNA
may be associated with cervical cancer risk. The expression levels
of flTF and asTF in lung cancer and paracancerous tissues were
higher compared with in normal control tissues; however, the
differences were not statistically significant. Rollin et al
(61) demonstrated that the
relative amount of asTF is low. This previous study also analyzed
the levels of asTF in NSCLC tumors according to clinicopathological
features; the results revealed that there is no association between
asTF and sex, age, stage and differentiation grade, yet patients
with high asTF tumor mRNA expression had a poorer prognosis.
Therefore, further studies are required to investigate the
correlation between TF and NSCLC tumors.
A limitation of the present study is that control or
paired normal esophageal and breast cancer tissues samples were not
analyzed. Therefore, the role of flTF and asTF in these two types
of cancer could not be clearly identified.
In conclusion, TF isoforms are able to activate
distinct signaling pathways (2),
leading to the modulation of cancer-associated biological processes
and nonhemostatic pathophysiological processes, including
thrombosis, angiogenesis, tumor growth and metastasis. A previous
study demonstrated that flTF mediates cell signaling via protease
activated receptor 2 and downstream signaling proteins, including
protein kinase C and extracellular signal-regulated kinase 1 and 2,
whereas asTF exhibits activity via integrin ligation (2,62).
To the best of our knowledge, the present study is the first to
reveal that flTF expression may be increased compared with asTF in
all tissue specimens tested, and suggested that overexpression of
apoM and CDCA may affect the mRNA expression levels of the two
variants. Furthermore, the expression levels of the two variants
may be different in the same cancer tissues. These results provide
further information regarding the TF system and emphasize the
significance of flTF and asTF expression in tumor progression and
other types of disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Changzhou
High-Level Medical Talents Training Project (grant no.
2016ZCLJ002).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
GL and NX contributed to the design of the study and
revision of the manuscript. LP performed the experiments, analyzed
the data and contributed to writing the manuscript. YY and QM
performed experiments. MY and SY were responsible for experiments
and statistical analysis.
Ethics approval and consent to
participate
The experimental protocols were approved by the
Institutional Ethics Committee of the Third Affiliated Hospital of
Soochow University and all patients provided written informed
consent for this study.
Patient consent for publication
Written informed consent was obtained from all
patients.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Eisenreich A, Boltzen U, Malz R,
Schultheiss HP and Rauch U: Overexpression of alternatively spliced
tissue factor induces the pro-angiogenic properties of murine
cardiomyocytic HL-1 cells. Circ J. 75:1235–1242. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leppert U and Eisenreich A: The role of
tissue factor isoforms in cancer biology. Int J Cancer.
137:497–503. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Boltzen U, Eisenreich A, Antoniak S,
Weithaeuser A, Fechner H, Poller W, Schultheiss HP, Mackman N and
Rauch U: Alternatively spliced tissue factor and full-length tissue
factor protect cardiomyocytes against TNF-α-induced apoptosis. J
Mol Cell Cardiol. 52:1056–1065. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eisenreich A: Regulation of vascular
function on posttranscriptional level. Thrombosis 2013.
9487652013.
|
|
5
|
van den Berg YW, van den Hengel LG, Myers
HR, Ayachi O, Jordanova E, Ruf W, Spek CA, Reitsma PH, Bogdanov VY
and Versteeg HH: Alternatively spliced tissue factor induces
angiogenesis through integrin ligation. Proc Natl Acad Sci USA.
106:19497–19502. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chand HS, Ness SA and Kisiel W:
Identification of a novel human tissue factor splice variant that
is upregulated in tumor cells. Int J Cancer. 118:1713–1720. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eisenreich A, Bogdanov VY, Zakrzewicz A,
Pries A, Antoniak S, Poller W, Schultheiss HP and Rauch U:
Cdc2-like kinases and DNA topoisomerase I regulate alternative
splicing of tissue factor in human endothelial cells. Circ Res.
104:589–599. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han X, Guo B, Li Y and Zhu B: Tissue
factor in tumor microenvironment: A systematic review. J Hematol
Oncol. 7:542014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kundu S, Bansal S, Muthukumarasamy KM,
Sachidanandan C, Motiani RK and Bajaj A: Deciphering the role of
hydrophobic and hydrophilic bile acids in angiogenesis using in
vitro and in vivo model systems. Medchemcomm. 8:2248–2257. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu N and Dahlbäck B: A novel human
apolipoprotein (apoM). J Biol Chem. 274:31286–31290. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ahmad A, Sundquist K, Zöller B, Dahlbäck
B, Svensson PJ, Sundquist J and Memon AA: Identification of
polymorphisms in apolipoprotein M gene and their relationship with
risk of recurrent venous thromboembolism. Thromb Haemost.
116:432–441. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nádró B, Juhász L, Szentpéteri A, Páll D,
Paragh G and Harangi M: The role of apolipoprotein M and
sphingosine 1-phosphate axis in the prevention of atherosclerosis.
Orv Hetil. 159:168–175. 2018.(In Hungarian). View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu Y, Luo G, Jiang B, Yu M, Feng Y, Wang
M, Xu N and Zhang X: Apolipoprotein M promotes proliferation and
invasion in non-small cell lung cancers via upregulating S1PR1 and
activating the ERK1/2 and PI3K/AKT signaling pathways. Biochem
Biophys Res Commun. 501:520–526. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rossiello MR, Rotunno C, Coluccia A,
Carratù MR, Di Santo A, Evangelista V, Semeraro N and Colucci M:
Ochratoxin A inhibits the production of tissue factor and
plasminogen activator inhibitor-2 by human blood mononuclear cells:
Another potential mechanism of immune-suppression. Toxicol Appl
Pharmacol. 229:227–231. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sovershaev MA, Lind KF, Devold H,
Jørgensen TØ, Hansen JB, Østerud B and Egorina EM: No evidence for
the presence of tissue factor in high-purity preparations of
immunologically isolated eosinophils. J Thromb Haemost.
6:1742–1749. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Regina S, Rollin J, Bléchet C, Iochmann S,
Reverdiau P and Gruel Y: Tissue factor expression in non-small cell
lung cancer: Relationship with vascular endothelial growth factor
expression, microvascular density, and K-ras mutation. J Thorac
Oncol. 3:689–697. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Plotkowski MC, Feliciano LF, Machado GB,
Cunha LG Jr, Freitas C, Saliba AM and de Assis MC: ExoU-induced
procoagulant activity in pseudomonas aeruginosa-infected airway
cells. Eur Respir J. 32:1591–1598. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu F, Huang R, Yao J, Wei W, Hu Y, Song S
and Li J: Homocysteine-induced enhanced expression of tissue factor
in human vascular smooth muscle cells. J Huazhong Univ Sci
Technolog Med Sci. 28:520–524. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Usui M, Kuriyama N, Kisawada M, Hamada T,
Mizuno S, Sakurai H, Tabata M, Imai H, Okamoto K, Uemoto S and
Isaji S: Tissue factor expression demonstrates severe sinusoidal
endothelial cell damage during rejection after living-donor liver
transplantation. J Hepatobiliary Pancreat Surg. 16:513–520. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lockwood CJ, Murk W, Kayisli UA,
Buchwalder LF, Huang ST, Funai EF, Krikun G and Schatz F: Progestin
and thrombin regulate tissue factor expression in human term
decidual cells. J Clin Endocrinol Metab. 94:2164–2170. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Di Stefano R, Barsotti MC, Armani C,
Santoni T, Lorenzet R, Balbarini A and Celi A: Human peripheral
blood endothelial progenitor cells synthesize and express
functionally active tissue factor. Thromb Res. 123:925–930. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang HJ, Huang H, Chuang YC and Huang HC:
Paclitaxel induces up-regulation of tissue factor in human aortic
endothelial cells. Int Immunopharmacol. 9:144–147. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang HJ, Lo WY, Lu TL and Huang H:
(−)-Epigallocatechin-3-gallate decreases
thrombin/paclitaxel-induced endothelial tissue factor expression
via the inhibition of c-Jun terminal NH2 kinase phosphorylation.
Biochem Biophys Res Commun. 391:716–721. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Teng Y, Jiang R, Lin Q, Ding C and Ye Z:
The relationship between plasma and placental tissue factor, and
tissue factor pathway inhibitors in severe pre-eclampsia patients.
Thromb Res. 126:e41–45. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Valsami S, Ruf W, Leikauf MS, Madon J,
Kaech A and Asmis LM: Immunomodulatory drugs increase endothelial
tissue factor expression in vitro. Thromb Res. 127:264–271. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Blank M, Baraam L, Eisenstein M, Fridkin
M, Dardik R, Heldman Y, Katchalski-Katzir E and Shoenfeld Y:
β2-Glycoprotein-I based peptide regulate endothelial-cells
tissue-factor expression via negative regulation of pGSK3β
expression and reduces experimental-antiphospholipid-syndrome. J
Autoimmun. 37:8–17. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ben-Hadj-Khalifa S, Hézard N, Almawi WY,
Lakbakbi S, Macé C, Cornillet-Lefebvre P, Mahjoub T and Nguyen P:
IL-10 modulates fondaparinux inhibition of monocyte-induced
thrombin generation. J Thromb Thrombolysis. 32:311–317. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gerotziafas GT, Galea V, Mbemba E,
Khaterchi A, Sassi M, Baccouche H, Prengel C, van Dreden P, Hatmi
M, Bernaudin JF and Elalamy I: Tissue factor over-expression by
human pancreatic cancer cells BXPC3 is related to higher
prothrombotic potential as compared to breast cancer cells MCF7.
Thromb Res. 129:779–786. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang JG, Geddings JE, Aleman MM, Cardenas
JC, Chantrathammachart P, Williams JC, Kirchhofer D, Bogdanov VY,
Bach RR, Rak J, et al: Tumor-derived tissue factor activates
coagulation and enhances thrombosis in a mouse xenograft model of
human pancreatic cancer. Blood. 119:5543–5552. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gebhard C, Holy EW, Camici GG, Akhmedov A,
Stämpfli SF, Stähli BE, von Rickenbach B, Breitenstein A, Greutert
H, Yang Z, et al: Caffeine induces endothelial tissue factor
expression via phosphatidylinositol 3-kinase inhibition. Thromb
Haemost. 107:884–894. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin M, Weng H, Wang X, Zhou B, Yu P and
Wang Y: The role of tissue factor and protease-activated receptor 2
in endometriosis. Am J Reprod Immunol. 68:251–257. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maruyama K, Morishita E, Yuno T, Sekiya A,
Asakura H, Ohtake S and Yachie A: Carbon monoxide (CO)-releasing
molecule-derived CO regulates tissue factor and plasminogen
activator inhibitor type 1 in human endothelial cells. Thromb Res.
130:e188–193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun L, Liu Y, Lin S, Shang J, Liu J, Li J,
Yuan S and Zhang L: Early growth response gene-1 and
hypoxia-inducible factor-1α affect tumor metastasis via regulation
of tissue factor. Acta Oncol. 52:842–851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang HP, Yue L, Jiang WW, Liu Q, Kou JP
and Yu BY: Diosgenin inhibits tumor necrosis factor-induced tissue
factor activity and expression in THP-1 cells via down-regulation
of the NF-κB, Akt, and MAPK signaling pathways. Chin J Nat Med.
11:608–615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Eisenreich A, Zakrzewicz A, Huber K,
Thierbach H, Pepke W, Goldin-Lang P, Schultheiss HP, Pries A and
Rauch U: Regulation of pro-angiogenic tissue factor expression in
hypoxia-induced human lung cancer cells. Oncol Rep. 30:462–470.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Carneiro-Lobo TC, Lima MT,
Mariano-Oliveira A, Dutra-Oliveira A, Oba-Shinjo SM, Marie SK,
Sogayar MC and Monteiro RQ: Expression of tissue factor signaling
pathway elements correlates with the production of vascular
endothelial growth factor and interleukin-8 in human astrocytoma
patients. Oncol Rep. 31:679–686. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen C, Liao D, Wang J, Liang Z and Yao Q:
Anti-human protein S antibody induces tissue factor expression
through a direct interaction with platelet phosphofructokinase.
Thromb Res. 133:222–228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Balia C, Petrini S, Scalise V, Neri T,
Carnicelli V, Cianchetti S, Zucchi R, Celi A and Pedrinelli R:
Compound 21, a selective angiotensin II type 2 receptor agonist,
downregulates lipopolysaccharide-stimulated tissue factor
expression in human peripheral blood mononuclear cells. Blood
Coagul Fibrinolysis. 25:501–506. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu LX, Zeng H, Liu EY and Chen FP: Tissue
factor expression and methylation regulation in differentiation of
embryonic stem cells into trophoblast. Asian Pac J Trop Med.
7:557–561. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bravo ML, Pinto MP, Gonzalez I, Oliva B,
Kato S, Cuello MA, Lange CA and Owen GI: Progesterone regulation of
tissue factor depends on MEK1/2 activation and requires the
proline-rich site on progesterone receptor. Endocrine. 48:309–320.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sovershaev TA, Egorina EM, Unruh D,
Bogdanov VY, Hansen JB and Sovershaev MA: BMP-7 induces TF
expression in human monocytes by increasing F3 transcriptional
activity. Thromb Res. 135:398–403. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang B, Xiong S, Hua Q, Chen C, Liao H,
Chen L, Yao W, Wu D and Tao Z: Tissue factor is strongly expressed
in pericarcinomatous tissue in patients with laryngeal carcinoma.
Int J Clin Exp Pathol. 8:13719–13724. 2015.PubMed/NCBI
|
|
44
|
Wang R, Ma YT, Ma J, Chen D, Wu B, Wen BZ
and He SL: Simultaneous detection of peripheral mononuclear cell
and plasma tissue factor expression for prevention and treatment of
ischemic cardiocerebrovascular diseases. Int J Clin Exp Pathol.
8:5674–5680. 2015.PubMed/NCBI
|
|
45
|
Orellana R, Kato S, Erices R, Bravo ML,
Gonzalez P, Oliva B, Cubillos S, Valdivia A, Ibañez C, Brañes J, et
al: Platelets enhance tissue factor protein and metastasis
initiating cell markers, and act as chemoattractants increasing the
migration of ovarian cancer cells. BMC Cancer. 15:2902015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jacobsen C, Oechsle K, Hauschild J,
Steinemann G, Spath B, Bokemeyer C, Ruf W, Honecker F and Langer F:
Regulation of tissue factor in NT2 germ cell tumor cells by
cisplatin chemotherapy. Thromb Res. 136:673–681. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dong R, Chen W, Feng W, Xia C, Hu D, Zhang
Y, Yang Y, Wang DW, Xu X and Tu L: Exogenous bradykinin inhibits
tissue factor induction and deep vein thrombosis via activating the
eNOS/phosphoinositide 3-kinase/Akt signaling pathway. Cell Physiol
Biochem. 37:1592–1606. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li W, Chen W, Xie M, Huang H, Su H, Han H,
Zhang D, Zhang Y, Yang X, Xu W, et al: Fasudil inhibits tissue
factor and plasminogen activator inhibitor-1 secretion by
peripheral blood mononuclear cells in CAPD patients. Ren Fail.
38:1359–1363. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Scalise V, Balia C, Cianchetti S, Neri T,
Carnicelli V, Zucchi R, Franzini M, Corti A, Paolicchi A, Celi A
and Pedrinelli R: Non enzymatic upregulation of tissue factor
expression by gamma-glutamyl transferase in human peripheral blood
mononuclear cells. Thromb J. 14:452016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Krychtiuk KA, Kaun C, Hohensinner PJ,
Stojkovic S, Seigner J, Kastl SP, Zuckermann A, Eppel W, Rauscher
S, de Martin R, et al: Anti-thrombotic and pro-fibrinolytic effects
of levosimendan in human endothelial cells in vitro. Vascul
Pharmacol. 90:44–50. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao H, Liu L, Zhao Y, Hara H, Chen P, Xu
J, Tang J, Wei L, Li Z, Cooper DKC, et al: Human IL-6, IL-17,
IL-1β, and TNF-α differently regulate the expression of
pro-inflammatory related genes, tissue factor, and swine leukocyte
antigen class I in porcine aortic endothelial cells.
Xenotransplantation. doi.org/10.1111/xen.12291.
|
|
52
|
Brambilla M, Rossetti L, Zara C, Canzano
P, Giesen PLA, Tremoli E and Camera M: Do methodological
differences account for the current controversy on tissue factor
expression in platelets? Platelets. 29:406–414. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Luo G, Zhang X, Mu Q, Chen L, Zheng L, Wei
J, Berggren-Söderlund M, Nilsson-Ehle P and Xu N: Expression and
localization of apolipoprotein M in human colorectal tissues.
Lipids Health Dis. 9:1022010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yu JL, May L, Lhotak V, Shahrzad S,
Shirasawa S, Weitz JI, Coomber BL, Mackman N and Rak JW: Oncogenic
events regulate tissue factor expression in colorectal cancer
cells: Implications for tumor progression and angiogenesis. Blood.
105:1734–1741. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kocatürk B, Van den Berg YW, Tieken C,
Mieog JS, de Kruijf EM, Engels CC, van der Ent MA, Kuppen PJ, Van
de Velde CJ, Ruf W, et al: Alternatively spliced tissue factor
promotes breast cancer growth in a β1 integrin-dependent manner.
Proc Natl Acad Sci USA. 110:11517–11522. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Venteclef N, Haroniti A, Tousaint JJ,
Talianidis I and Delerive P: Regulation of anti-atherogenic
apolipoprotein M gene expression by the orphan nuclear receptor
LRH-1. J Biol Chem. 283:3694–3701. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Censarek P, Bobbe A, Grandoch M, Schrör K
and Weber AA: Alternatively spliced human tissue factor (asHTF) is
not pro-coagulant. Thromb Haemost. 97:11–14. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Szotowski B, Antoniak S, Poller W,
Schultheiss HP and Rauch U: Procoagulant soluble tissue factor is
released from endothelial cells in response to inflammatory
cytokines. Circ Res. 96:1233–1239. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mackman N and Davis GE: Blood coagulation
and blood vessel development: Is tissue factor the missing link?
Arterioscler Thromb Vasc Biol. 31:2364–2366. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Unruh D, Turner K, Srinivasan R, Kocatürk
B, Qi X, Chu Z, Aronow BJ, Plas DR, Gallo CA, Kalthoff H, et al:
Alternatively spliced tissue factor contributes to tumor spread and
activation of coagulation in pancreatic ductal adenocarcinoma. Int
J Cancer. 134:9–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Rollin J, Regina S and Gruel Y: Tumor
expression of alternatively spliced tissue factor is a prognostic
marker in non-small cell lung cancer. J Thromb Haemost. 8:607–610.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Giannarelli C, Alique M, Rodriguez DT,
Yang DK, Jeong D, Calcagno C, Hutter R, Millon A, Kovacic JC, Weber
T, et al: Alternatively spliced tissue factor promotes plaque
angiogenesis through the activation of hypoxia-inducible factor-1α
and vascular endothelial growth factor signaling. Circulation.
130:1274–1286. 2014. View Article : Google Scholar : PubMed/NCBI
|