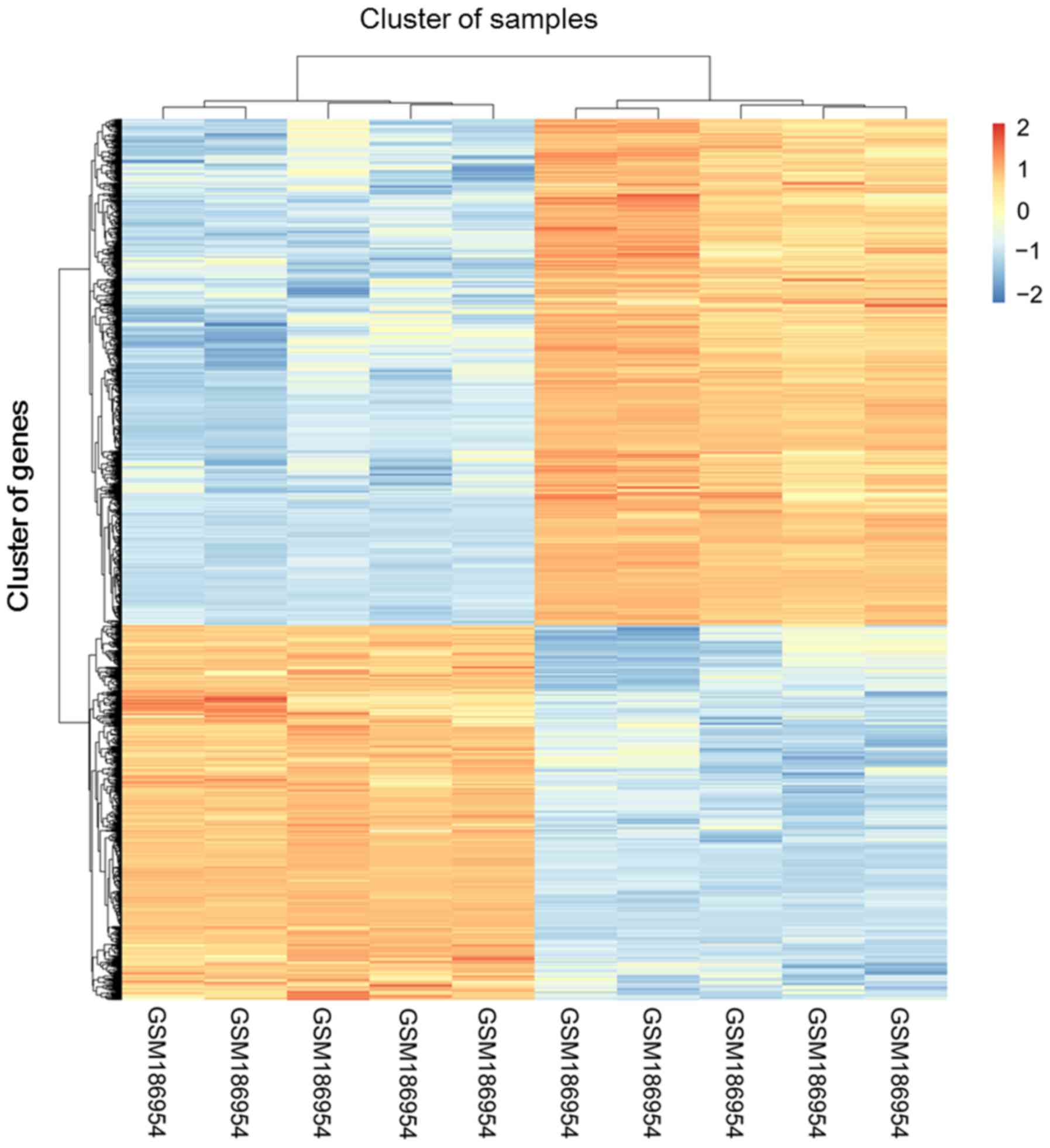
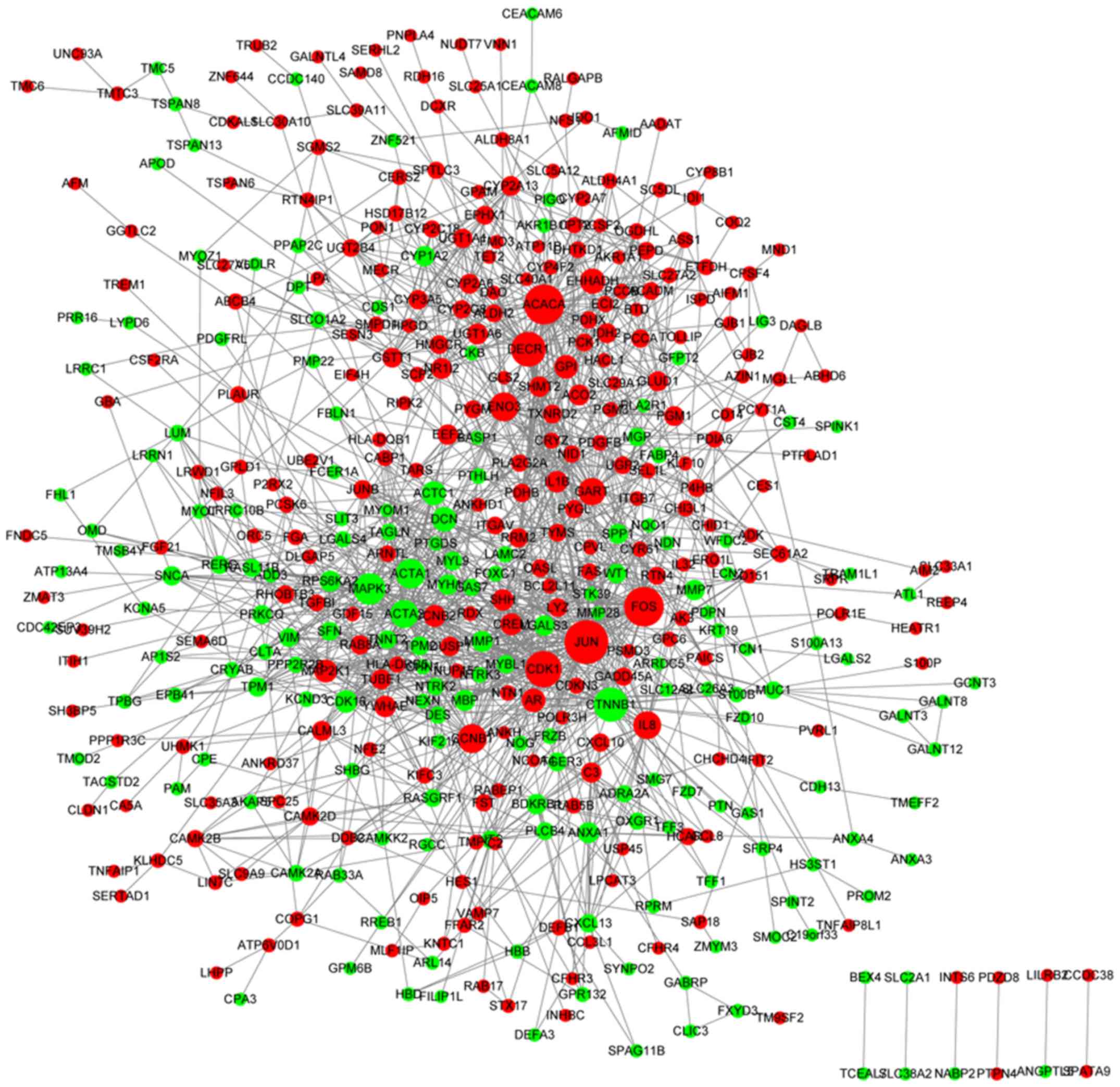
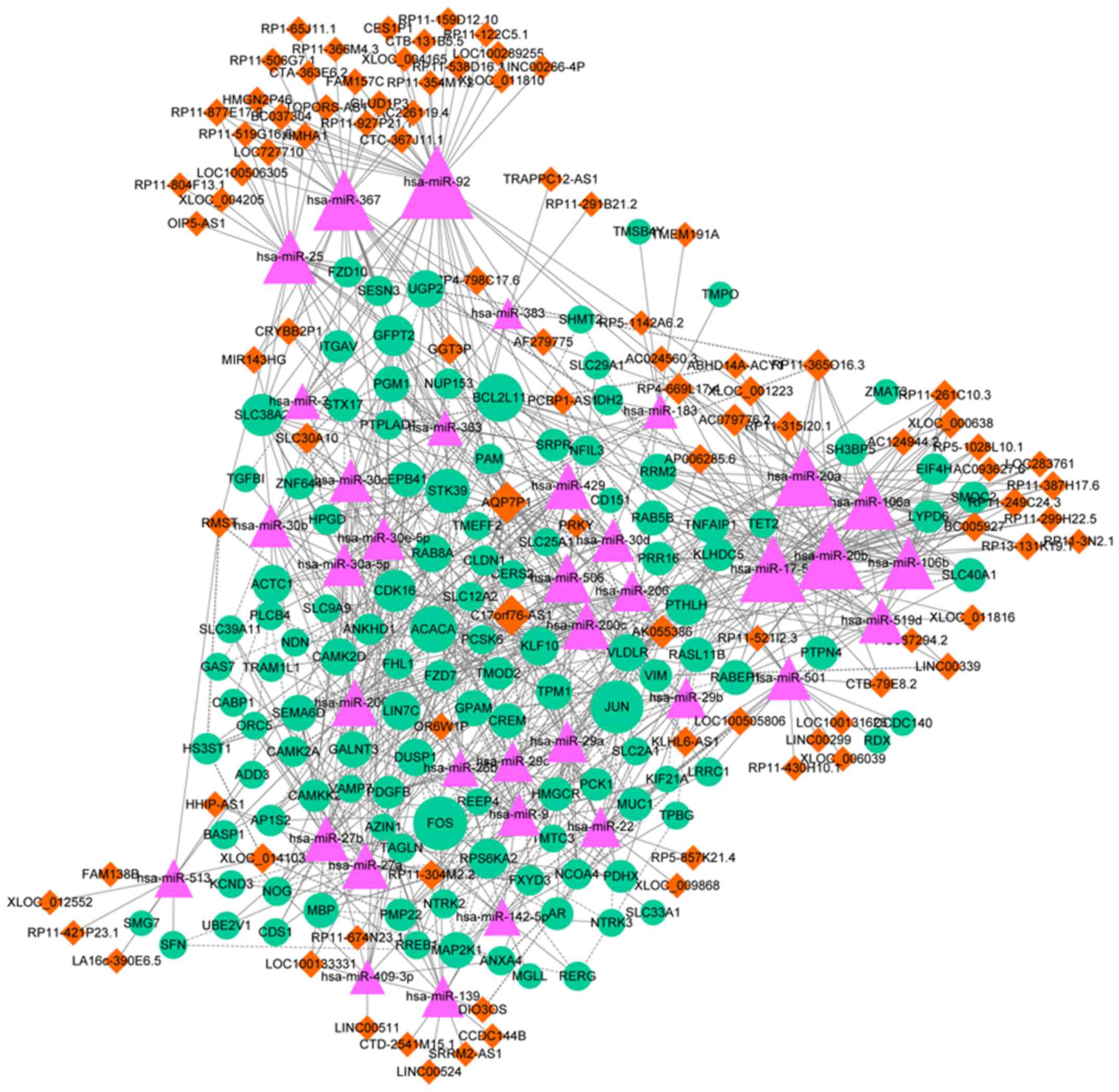
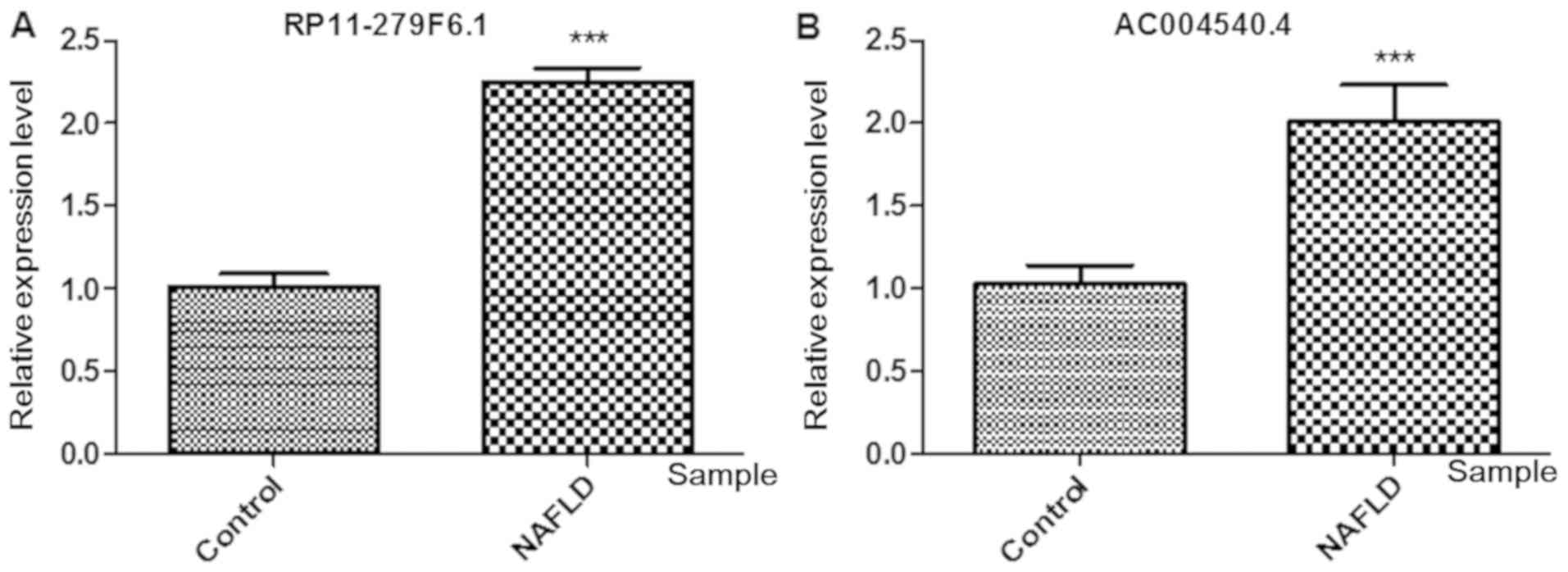
|
1
|
Shaker M, Tabbaa A, Albeldawi M and
Alkhouri N: Liver transplantation for nonalcoholic fatty liver
disease: New challenges and new opportunities. World J
Gastroenterol. 20:5320–5330. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rinella ME: Nonalcoholic fatty liver
disease: A systematic review. JAMA. 313:22632015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tolman KG and Dalpiaz AS: Treatment of
non-alcoholic fatty liver disease. Ther Clin Risk Manag.
3:1153–1163. 2007.PubMed/NCBI
|
|
4
|
Clark JM and Diehl AM: Nonalcoholic fatty
liver disease: An underrecognized cause of cryptogenic cirrhosis.
JAMA. 289:3000–3004. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Omagari K, Kadokawa Y, Masuda J, Egawa I,
Sawa T, Hazama H, Ohba K, Isomoto H, Mizuta Y, Hayashida K, et al:
Fatty liver in non-alcoholic non-overweight Japanese adults:
Incidence and clinical characteristics. J Gastroenterol Hepatol.
17:1098–1105. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen L, Fan JG, Shao Y, Zeng MD, Wang JR,
Luo GH, Li JQ and Chen SY: Prevalence of nonalcoholic fatty liver
among administrative officers in Shanghai: An epidemiological
survey. World J Gastroenterol. 9:1106–1110. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lazo M, Hernaez R, Bonekamp S, Kamel IR,
Brancati FL, Guallar E and Clark JM: Non-alcoholic fatty liver
disease and mortality among US adults: Prospective cohort study.
BMJ. 343:d68912011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Doege H, Grimm D, Falcon A, Tsang B, Storm
TA, Xu H, Ortegon AM, Kazantzis M, Kay MA and Stahl A: Silencing of
hepatic fatty acid transporter protein 5 in vivo reverses
diet-induced non-alcoholic fatty liver disease and improves
hyperglycemia. J Biol Chem. 283:22186–22192. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hoekstra M, Li ZS, Kruijt JK, Van Eck M,
Van Berkel TJ and Kuiper J: The expression level of non-alcoholic
fatty liver disease-related gene PNPLA3 in hepatocytes is highly
influenced by hepatic lipid status. J Hepatol. 52:244–251. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin YC, Chang PF, Hu FC, Yang WS, Chang MH
and Ni YH: A common variant in the PNPLA3 gene is a risk factor for
non-alcoholic fatty liver disease in obese Taiwanese children. J
Pediatr. 158:740–744. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang Y, Bian Z, Zhao L, Liu Y, Liang S,
Wang Q, Han X, Peng Y, Chen X, Shen L, et al: Interleukin-17
exacerbates hepatic steatosis and inflammation in non-alcoholic
fatty liver disease. Clin Exp Immunol. 166:281–290. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamada H, Suzuki K, Ichino N, Ando Y,
Sawada A, Osakabe K, Sugimoto K, Ohashi K, Teradaira R, Inoue T, et
al: Associations between circulating microRNAs (miR-21, miR-34a,
miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta.
424:99–103. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun C, Huang F, Liu X, Xiao X, Yang M, Hu
G, Liu H and Liao L: miR-21 regulates triglyceride and cholesterol
metabolism in non-alcoholic fatty liver disease by targeting HMGCR.
Int J Mol Med. 35:847–853. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rui EC, Ferreira DM, Afonso MB, Borralho
PM, Machado MV, Cortez-Pinto H and Rodrigues CM: miR-34a/SIRT1/p53
is suppressed by ursodeoxycholic acid in the rat liver and
activated by disease severity in human non-alcoholic fatty liver
disease. J Hepatol. 58:119–125. 2012.PubMed/NCBI
|
|
15
|
Sun C, Liu X, Yi Z, Xiao X, Yang M, Hu G,
Liu H, Liao L and Huang F: Genome-wide analysis of long noncoding
RNA expression profiles in patients with non-alcoholic fatty liver
disease. IUBMB Life. 67:847–852. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK: Limma: Linear models for
microarray data. In: Bioinformatics and Computational Biology
Solutions Using R and Bioconductor. Gentleman R, Carey VJ, Huber W,
Irizarry RA and Dudoit S: Springer New York. (New York, NY).
397–420. 2005.
|
|
17
|
Kolde R and Kolde MR: Package ‘pheatmap’.
https://cran.r-project.org/web/packages/pheatmap/October
12–2015
|
|
18
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tweedie S, Ashburner M, Falls K, Leyland
P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D,
Schroeder A, Seal R, et al: FlyBase: Enhancing drosophila gene
ontology annotations. Nucleic Acids Res 37 (Database Issue).
D555–D559. 2009. View Article : Google Scholar
|
|
20
|
Altermann E and Klaenhammer TR:
PathwayVoyager: Pathway mapping using the Kyoto encyclopedia of
genes and genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res 43
(Database Issue). D447–D452. 2015. View Article : Google Scholar
|
|
22
|
Saito R, Smoot ME, Ono K, Ruscheinski J,
Wang PL, Lotia S, Pico AR, Bader GD and Ideker T: A travel guide to
Cytoscape plugins. Nat Methods. 9:1069–1076. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of biological networks. Biosystems. 127:67–72. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Du Y, Gao C, Chen X, Hu Y, Sadiq R and
Deng Y: A new closeness centrality measure via effective distance
in complex networks. Chaos. 25:0331122015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Opsahl T, Agneessens F and Skvoretz J:
Node centrality in weighted networks: Generalizing degree and
shortest paths. Social Networks. 32:245–251. 2010. View Article : Google Scholar
|
|
26
|
Cukierski WJ and Foran DJ: Using
betweenness centrality to identify manifold Shortcuts. Proc IEEE
Int Conf Data Min. 2008:949–958. 2008.PubMed/NCBI
|
|
27
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mukaka MM: Statistics corner: A guide to
appropriate use of correlation coefficient in medical research.
Malawi Med J. 24:69–71. 2012.PubMed/NCBI
|
|
29
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013.
Nucleic Acids Res. 41:W77–W83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res 42 (Database Issue). D68–D73. 2014. View Article : Google Scholar
|
|
32
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Correction: Human MicroRNA targets. PLoS
Biol. 3:e2642005. View Article : Google Scholar :
|
|
33
|
Krüger J and Rehmsmeier M: RNAhybrid:
microRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34:W451–W454. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dorn C, Engelmann JC, Saugspier M, Koch A,
Hartmann A, Müller M, Spang R, Bosserhoff A and Hellerbrand C:
Increased expression of c-Jun in nonalcoholic fatty liver disease.
Lab Invest. 94:394–408. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Singh R, Wang YJ, Xiang YQ, Tanaka KE,
Gaarde WA and Czaja MJ: Differential effects of JNK1 and JNK2
inhibition on murine steatohepatitis and insulin resistance.
Hepatology. 49:87–96. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karin M and Gallagher E: From JNK to pay
dirt: Jun kinases, their biochemistry, physiology and clinical
importance. IUBMB Life. 57:283–295. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen
X, Chen L, Scheuch H, Zheng H, Qin L, et al: Liver cancer
initiation is controlled by AP-1 through SIRT6-dependent inhibition
of survivin. Nat Cell Biol. 15:1203–1211. 2013. View Article : Google Scholar
|
|
39
|
Machida K, Tsukamoto H, Liu JC, Han YP,
Govindarajan S, Lai MM, Akira S and Ou JH: c-Jun mediates hepatitis
C virus hepatocarcinogenesis through signal transducer and
activator of transcription 3 and nitric oxide-dependent impairment
of oxidative DNA repair. Hepatology. 52:480–492. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Trierweiler C, Hockenjos B, Zatloukal K,
Thimme R, Blum HE, Wagner EF and Hasselblatt P: The transcription
factor c-JUN/AP-1 promotes HBV-related liver tumorigenesis in mice.
Cell Death Differ. 23:576–582. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shrivastava S, Petrone J, Steele R, Lauer
GM, Di Bisceglie AM and Ray RB: Up-regulation of circulating
miR-20a is correlated with hepatitis C virus-mediated liver disease
progression. Hepatology. 58:863–871. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Panasiuk A, Dzieciol J, Panasiuk B and
Prokopowicz D: Expression of p53, Bax and Bcl-2 proteins in
hepatocytes in non-alcoholic fatty liver disease. World J
Gastroenterol. 12:6198–6202. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Selzner M, Rüdiger HA, Selzner N, Thomas
DW, Sindram D and Clavien PA: Transgenic mice overexpressing human
B cl-2 are resistant to hepatic ischemia and reperfusion. J
Hepatol. 36:218–225. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
An F, Gong B, Wang H, Yu D, Zhao G, Lin L,
Tang W, Yu H, Bao S and Xie Q: miR-15b and miR-16 regulate TNF
mediated hepatocyte apoptosis via BCL2 in acute liver failure.
Apoptosis. 17:702–716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fort A, Borel C, Migliavacca E,
Antonarakis SE, Fish RJ and Neerman-Arbez M: Regulation of
fibrinogen production by microRNAs. Blood. 116:2608–2615. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tryndyak VP, Marrone AK, Latendresse JR,
Muskhelishvili L, Beland FA and Pogribny IP: MicroRNA changes,
activation of progenitor cells and severity of liver injury in mice
induced by choline and folate deficiency. J Nutr Biochem. 28:83–90.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang Y, Yu T, Jiang S, Zhang Y, Li M, Tang
N, Ponnusamy M, Wang JX and Li PF: miRNAs as potential therapeutic
targets and diagnostic biomarkers for cardiovascular disease with a
particular focus on WO2010091204. Expert Opin Ther Pat.
27:1021–1029. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wong CC, Wong CM, Tung EK, Au SL, Lee JM,
Poon RT, Man K and Ng IO: The microRNA miR-139 suppresses
metastasis and progression of hepatocellular carcinoma by
down-regulating Rho-kinase 2. Gastroenterology. 140:322–331. 2011.
View Article : Google Scholar : PubMed/NCBI
|