Introduction
Head and neck cancer (HNC) is a broad spectrum of
disease that encompasses malignancies in the aero-digestive tract,
including the oral cavity, pharynx, larynx, nasal cavity, paranasal
sinuses, salivary glands, thyroid and parathyroid glands (1). HNC is the sixth most common cancer
worldwide. Sri Lanka is considered as one of the high-risk
countries, where HNCs are the leading cancer type in men accounting
for 29% of all cancers (2–4).
Cigarette smoking and alcohol consumption, which are
considered to be the most common risk factors for HNC, are
generally lower in Sri Lankans compared to Western populations
(2,5). However, tobacco chewing and
consumption of areca nut are considered to be the most common
causes of HNC in Sri Lankan males (3,5–7).
Tobacco chewing results in exposure to 28 known carcinogens,
including the non-volatile alkaloid-derived tobacco-specific
N-nitrosamine and N-nitrosamino acids, which are different from the
carcinogens involved in smoking cigarettes (3,8). The
high risk associated with human papilloma virus (HPV) infection for
specific types of HNCs is well established and studied in different
geographical areas (9). However
there is a scarcity of literature available on HPV associated HNC
in the Sri Lankan population.
p53 acts as a ‘guardian of the genome’ to maintain
the balance of cell death and proliferation by regulating the cell
cycle, DNA repair, apoptosis, cellular metabolism and senescence
(10). It is altered in ~50% of
cancers overall and is more frequent in adult vs. childhood
malignancies (11). The efficacy
of many cancer therapeutic approaches is influenced by the
functional status of the p53 tumour suppressor protein. Thus
identification of TP53 mutation status prior to
administration of therapy can predict potential effectiveness of
the treatment and influence treatment selection. Furthermore, the
TP53 mutation spectrum provides information on tumour
origin, cause of mutation, aetiology, molecular pathogenesis,
prediction of patient survival and chances of recurrence (12–15).
There were numerous studies on TP53 variants
in various cancers including head and neck cancer over the last few
decades, particularly in Western populations. But there are only
few studies done considering all subsets of HNC in Asia including
India (16) and Japan (17) excluding Sri Lanka. Since the
frequency of TP53 mutations and the mutation spectra vary in
different geographic areas, according to aetiological factors, life
style, dietary pattern and culture, the present study has focused
on establishing the TP53 mutation spectrum in Sri Lankan HNC
patients. Furthermore we used immunohistochemistry (IHC) to assess
p53 protein expression and correlated immuno-expression of p53 with
TP53 gene mutational status. We also studied HPV infection
in HNC and oesophageal cancer using p16 immuno-expression and HPV
DNA detection, as the latter has reported to be associated with
oral cancer in Sri Lankan patients (18).
Materials and methods
Patient recruitment and sample
processing
Ethical approval was obtained from the Ethics Review
Committee, Faculty of Medicine, University of Colombo, Sri Lanka
(EC/14/160). Patients with HNC (N=44) who had undergone surgical
resection at the National Cancer Institute, Sri Lanka, were
recruited for this study. Written informed consent from the study
participants was obtained prior to recruitment. Socio-demographic
and clinical data were obtained from study participants using
questionnaires and by reviewing their medical reports. The majority
of our patient population represents the Sinhalese ethnicity.
Healthy controls (N=20; 10 males, 10 females) with no
personal/family history of any cancer were recruited for this
study.
Surgically excised tumour tissues were collected and
the close adjacent region of the tissue section was placed in 10%
formalin to prepare Formalin Fixed Paraffin Embedded tissue while
the other section was immediately placed in Allprotect®
Tissue Reagent (cat no. 76405; Qiagen, Hilden, Germany) and stored
at −20°C until processed. The hematoxylin and eosin stained slides
of each tissue were reviewed by a pathologist to confirm the
percentage of tumour region. Studied samples were with >50% area
coverage of tumour in the study, except only two samples had
<10% of tumour cells in the sections.
Genomic DNA was extracted from the excised tumour
tissue of patients and from peripheral venous blood of healthy
controls. Disruption of tissue specimens was done in liquid
nitrogen using a motor and pestle followed by homogenization using
QIAshredder (cat. no. 79654; Qiagen). Tissue DNA was extracted from
homogenized sample using an All prep DNA/RNA/Protein mini kit (cat.
no. 80004; Qiagen) following the manufacturer's protocol and stored
at −20°C until used. Genomic DNA was extracted from blood using the
modified protocol described by Miller et al (19).
Seven sets of primers covering the entire exon 2–11
coding regions and adjacent flanking 5′ and 3′ intronic regions
were designed using the online NCBI/Primer-BLAST software
(https://www.ncbi.nlm.nih.gov/tools/primerblast/index.cgi?ORGANISM=9606&INPUT_SEQUENCE=NM_001618.3).
Polymerase Chain Reaction (PCR) amplification was performed using
each primer set in a final volume of 25 µl containing 100 ng
genomic DNA, 3.5 mM MgCl2, 1X Green GoTaq®
reaction buffer [10 mM Tris-HCl (pH 8.3) and 50 mM KCl], 2.5 mM
dNTPs (Promega Corporation, Madison, WI, USA), 5 pmols of each
primer (IDT Integrated DNA Technologies, Coralville, IA, USA) and 1
unit of GoTaq® Flexi DNA polymerase (Promega
Corporation). PCR conditions: 94°C for 7 min, followed by 33 cycles
of 94°C for 1 min, at the optimized annealing temperature for 1 min
and 72°C for 1 min and a final extension step of 72°C for 10 min
was performed in a thermocycler (Veriti Thermal Cycler; Thermo
Fisher Scientific, Waltham, MA USA). The annealing temperature and
MgCl2 concentration were optimized for each primer set.
The primer nucleotide sequences, amplicon sizes and annealing
temperatures are shown in Table
I.
 | Table I.Nucleotide sequence of primers used
for amplification of TP53, amplicon size and annealing
temperature. |
Table I.
Nucleotide sequence of primers used
for amplification of TP53, amplicon size and annealing
temperature.
| Exon | Primer sequence,
5′-3′ | Amplicon size
(bp) | Annealing
temperature (°C) |
|---|
| 2 and 3 |
CAGCCATTCTTTTCCTGCTC | 497 | 62 |
|
|
GGGGACTGTAGATGGGTGAA |
|
|
| 4 |
CCTGGTCCTCTGACTGCTCT | 361 | 64 |
|
|
GCCAGGCATTGAAGTCTCAT |
|
|
| 5 and 6 |
GTTTCTTTGCTGCCGTCTTC | 500 | 64 |
|
|
CTTAACCCCTCCTCCCAGAG |
|
|
| 7 |
GAGCTTGCAGTGAGCTGAGA | 444 | 63 |
|
|
TCCCAAAGCCAGAGAAAAGA |
|
|
| 8 and 9 |
CAAGGGTGGTTGGGAGTAGA | 532 | 65 |
|
|
TGTCTTTGAGGCATCACTGC |
|
|
| 10 |
TGCATGTTGCTTTTGTACCG | 299 | 56 |
|
|
GAAGGCAGGATGAGAATGGA |
|
|
| 11 |
TGTCATCTCTCCTCCCTGCT | 438 | 61 |
|
|
AAGTGGGCCCCTACCTAGAA |
|
|
PCR products were purified using the
Wizard® SV Gel and PCR Clean-Up kit (Promega
Corporation) and purified products were directly sequenced using
the BigDye® Terminator v3.1 kit (Thermo Fisher
Scientific) and an Applied Biosystems™ 3500Dx Genetic
Analyzer (Thermo Fisher Scientific). Sequence variants detected
were reconfirmed by performing a second PCR and direct
sequencing.
Sequencing results were analysed to identify
variants by alignment with a human TP53 NCBI reference
sequence (GenBank accession number-NC_000017), via Bio
Edit® software and further confirmed by Mutation
Surveyor® v4.0.9 and Alamut® Visual 2.7.2
Documentation. Identified sequence variants were named according to
the Human Genome Variation Society/HGVS nomenclature guidelines
(http://www.hgvs.org/mutnomen/).
Variant analysis
Identified sequence variants were checked for
previous reports in the following databases: Catalogue Of Somatic
Mutations in Cancer (COSMIC) (http://cancer.sanger.ac.uk/cosmic); NCBI (https://www.ncbi.nlm.nih.gov/); IARC TP53 (http://p53.iarc.fr/); Ensembl (https://asia.ensembl.org/index.html); the p53 website
(https://p53.fr/tp53-database).
Pathogenicity of the identified exonic variants was
analysed using five comparative missense prediction programs: Align
GVGD (http://agvgd.hci.utah.edu/agvgd_input.php); SIFT
(http://sift.jcvi.org/www/SIFT_seq_submit2.html);
MutationTaster (http://www.mutationtaster.org/); PolyPhen-2
(http://genetics.bwh.harvard.edu/pph2/); Provean
(http://provean.jcvi.org/seq_submit.php). p53 specific
structural and functional activity (transcriptional activity,
dominant negative effect) data available on IARC TP53 database was
also considered while determining the pathogenicity of the
identified variants (18,19). Human Splicing Finder V3.0
(http://www.umd.be/HSF3/) and splicing window of
Alamut® Visual software, integrating a number of
prediction methods (for splice signal detection: MaxEntScan,
GeneSplicer and ESE/exonic splicing enhancer binding site
detection: ESEFinder, RESCUE-ESE) were used to assess the impact on
gene splicing of identified intronic variants. All variants were
classified according to American College of Medical Genetics
standards and guidelines (20).
Immunohistochemistry for p53
IHC characterization of p53 expression was performed
on twenty four randomly selected representative formalin fixed
paraffin-embedded tumour tissue sections. The primary antibody used
was mouse monoclonal Anti-Human p53 clone DO-7 (Agilent DaKo, Santa
Clara, USA) at 1:100 dilution. Tissue sections of 4 micron
thickness were mounted on microscopic slides and dried at 60°C for
2 h. After dewaxing in xylene, sections were rehydrated in graded
(100, 95, 70 and 60%) alcohol. Microwave heating in citrate (pH 6)
buffer was used for antigen retrieval and endogenous peroxidase was
inhibited by incubating tissue sections in freshly prepared 3%
hydrogen peroxide. Tissue sections were incubated with primary
antibody at room temperature for an hour and then exposed to
horseradish peroxidase (A. Menarini Diagnostics Ltd., Winnersh, UK)
conjugated anti-mouse IgG secondary antibody for 30 min. Universal
probe (A. Menarini Diagnostics Ltd.) was applied for 30 min before
the addition of primary antibody, in order to increase the staining
sensitivity 10 to 40 times for mouse monoclonal antibodies. Then,
the slides were incubated with 3,3′-diaminobenzidine (A. Menarini
Diagnostics Ltd.). Following the incubation, the sections were
washed and counter stained with haematoxylin. Slides that were not
incubated in primary antibody were used as antibody negative
controls to check for specificity of staining.
Images of the stained slides were visualized using
the AperioScanScope® CS System, an automated digital
scanner (Aperio Technologies, Bristol, UK) technology and
Spectrum™ image management software. The slides were
analysed by both automated and manual methods, blinded from the
clinical data.
Human papilloma virus DNA
screening
PCR with GP5+/GP6+ HPV specific primers was
performed for all HNC samples in a final volume of 25 µl containing
100–150 ng genomic DNA, 3.5 mM MgCl2, 1X Green
GoTaq® reaction buffer [10 mM Tris-HCl (pH 8.3) and 50
mM KCl], 2.5 mM dNTPs (Promega Corporation), 50 pmols of each
primer (IDT Integrated DNA Technologies) and 1 unit of
GoTaq® Flexi DNA polymerase (Promega Corporation). PCR
conditions: 94°C for 4 min, followed by 40 cycles of 94°C for 1
min, 40°C for 1 min and 72°C for 1 min and a final extension step
at 72°C for 10 min (21). The
negative control included all reagents except for DNA. A p1203
PML2d HPV-16 plasmid (gift from Peter Howley: Addgene plasmid
#10869) was used as the positive control. Samples positive for HPV
generate a 140-bp-long fragment from the HPV L1 structural
gene.
p16 immunohistochemistry
p16 cyclin-dependent kinase inhibitor protein
expression was detected using IHC at the pathology laboratory of
the Royal Victoria Infirmary, Newcastle upon Tyne using a Ventana
Benchmark XT Automated IHC/In situ hybridization slide
staining system (Ventana Medical Systems, Inc., Tucson, AZ, USA).
The CINtec® p16 Histology (Hoffmann-La Roche AG, Basel,
Switzerland) and UltraView DAB detection kit (Ventana Medical
Systems Inc.) were used for the detection of p16. Images of the
stained slides were visualized and analysed using the same process
as for p53 detection.
Patient follow-up
Data on response to treatment, survival, recurrence
and current status during the follow-up period were collected by
reviewing patient medical records and collecting data directly from
patients/guardians.
Statistical analysis
All categorical data were analysed using Fisher's
exact test to assess the significance of the associations. A
P-value <0.05 was considered as statistically significant.
Results
Baseline characteristics of
participants
Out of the 44 patients, 75% (N=33) were males and
25% (N=11) were females. Mean ± SD age was 59.03±11.68 years for
males and 53.27±19.04 years for females. Healthy controls were
younger (males 33.2±4.92; females 33.1±5.84 years). Table II summarizes the characteristics
of the patients, tumour type and possible risk factors the patients
were exposed to.
 | Table II.Baseline characteristics of the
patients, tumour type and possible risk factors the patients were
exposed to. |
Table II.
Baseline characteristics of the
patients, tumour type and possible risk factors the patients were
exposed to.
|
Characteristics | Total number of
patients | Patients with
wild-type TP53, no. | Patients with
mutated TP53, no. | Total number of
healthy controls | Two-tailed P-value
between patient and healthy controlsa |
|---|
| Sex |
|
Male | 33 | 23 | 10 | 10 | 0.1176 |
|
Female | 11 | 7 | 4 | 10 |
|
| Ethnicity |
|
Sinhalese | 35 | 23 | 12 | 13 | 0.0888 |
| Sri
Lankan Tamil | 3 | 3 | 0 | 5 |
|
| Indian
Tamil | 3 | 1 | 2 | 1 |
|
|
Muslims | 3 | 3 | 0 | 0 |
|
|
Burghers | 0 | 0 | 0 | 1 |
|
| Age at study
entry |
| <30
years | 2 | 2 | 0 | 2 | 0.0007 |
| 30–60
years | 24 | 14 | 10 | 18 |
|
| >61
years | 18 | 14 | 4 | 0 |
|
| Tumour type |
|
Squamous cell carcinoma | 25 | 13 | 12 | N/A | N/A |
|
Adenocarcinoma | 4 | 3 | 1 | N/A |
|
|
Papillary carcinoma | 4 | 4 | 0 | N/A |
|
|
Others | 11 | 10 | 1 | N/A |
|
| Smoking
history |
|
Yes | 17 | 11 | 6 | 0 | 0.0005 |
| No | 25 | 17 | 8 | 20 |
|
|
Unknown | 2 | 2 | 0 | 0 |
|
| Alcohol
consumption |
|
Yes | 13 | 8 | 5 | 1 | 0.0251 |
| No | 29 | 20 | 9 | 19 |
|
|
Unknown | 2 | 2 | 0 | 0 |
|
| Betel-quid
chewing |
| Yes
with tobacco | 21 | 12 | 9 | 0 | <0.0001 |
| Yes
without tobacco | 2 | 2 | 0 | 0 |
|
| No | 19 | 14 | 5 | 20 |
|
|
Unknown | 2 | 2 | 0 | 0 |
|
| Roofing |
|
Asbestos | 21 | 14 | 7 | 1 | <0.0001 |
|
Tile | 15 | 11 | 4 | 5 |
|
|
Concrete | 1 | 1 | 0 | 12 |
|
|
Metal | 3 | 2 | 1 | 0 |
|
|
Unknown | 4 | 2 | 2 | 2 |
|
| Codon 72
polymorphism |
|
|
|
| 0.6225 |
|
Arginine | 10 | 6 | 4 | 7 |
|
|
Proline | 19 | 12 | 7 | 7 |
|
|
Arginine/proline | 15 | 12 | 3 | 6 |
|
Analysis of TP53 sequence variants and
epidemiological/clinical correlations
A total of 36 sequence variants (18 translated
exonic variants, 12 intronic and 2 non-translated exonic variants)
were found in 44 patients and 4 additional sequence variants were
found only in healthy controls (two exonic silent and two novel
intronic variants). Table III
illustrates the characteristics, In silico and functional
analysis of each variant. All the exonic variants were in the DNA
binding domain of the p53 protein (codons 94–292).
 | Table III.In silico and functional
analysis of identified variants. |
Table III.
In silico and functional
analysis of identified variants.
|
| HGVS
Nomenclature |
|
| No. of carriers in
the study cohort |
| In silico
prediction |
| Prediction of
functional activity |
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|---|
| No. | cDNA | Protein | Location | Mutation type | Patients
(n=44) | Controls
(n=20) | Code reported in
databases | Splice site | CpG site | Align GVGD | Mutation
taster | Provean | SIFT | Poly Phen 2 | NCBI | Transcriptional
activity [21] | Functional activity
prediction [22] | Dominant negative
effect | Conclusion |
|---|
| 1 | c.298delC |
p.Gln100Argfs*23 | E4 | FS | 1 | 0 | Novel | – | – | – | DC | – | – | – | – | – | – | – | Path |
| 2 | c.383delC |
p.Pro128Leufs*42 | E5 | FS | 1 | 0 | COSM5198771 | – | – | – | DC | – | – | – | – | – | – | – | Path |
| 3 | c.626_637del |
p.Asn210_Arg213 | E6 | IF | 1 | 0 | Novel | – | – | – | DC | – | – | – | – | – | – | – | Path |
|
| GAAACAC | del |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| TTTTC |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 4 | c.493C>T | p.Gln165* | E 5 | NS | 1 | 0 | rs730882001,
COSM43632 | – | – | – | DC | D | – | – | Path | – | – | – | Path |
| 5 | c.637C>T | p.Arg213* | E 6 | NS | 1 | 0 | rs397516436 | – | Yes | – | – | – | – | – | Path | – | – | – | Path |
| 6 | c.422G>T | p.Cys141Phe | E 5 | M | 1 | 0 | COSM44911 | – | – | C65 | DC | D | D | PRB | – | PF | F | – | Path |
| 7 | c.455C>T | p.Pro152Leu | E5 | M | 1 | 0 | rs587782705 | – | Yes | C65 | DC | D | D | POB | Path | NF | NF | ND | Path |
| 8 | c.524G>A | p.Arg175His | E 5 | M | 2 | 0 | rs28934578,
COSM10648 | – | Yes | C25 | DC | D | D | POB | Path | NF | NF | DN | Path |
| 9 | c.578A>G | p.His193Arg | E 6 | M | 1 | 0 | rs786201838,
COSM10742 | – | – | C25 | DC | D | D | PRB | LP | NF | NF | MD | Path |
| 10 | c.583A>T | p.Ile195Phe | E6 | M | 1 | 0 | COSM44633 | – | – | C0 | DC | D | D | PRB | – | NF | NF | – | Path |
| 11 | c.646G>C | p.Val216Leu | E 6 | M | 1 | 0 | rs730882025 | – | – | C25 | DC | – | D | – | LP | NF | NF | – | Path |
| 12 | c.659A>G | p.Tyr220Cys | E6 | M | 1 | 0 | rs121912666,
COSM10758 | – | – | C65 | DC | D | D | PRB | Path | NF | NF | MD | Path |
| 13 | c.747G>T | p.Arg249Ser | E7 | M | 1 | 0 | rs28934571,
COSM10817 | – | – | C65 | DC | D | D | PRB | Path | NF | NF | DN | Path |
| 14 | c.818G>A | p.Arg273His | E 8 | M | 1 | 0 | rs28934576,
COSM10660 | – | Yes | C25 | DC | – | D | – | Path | NF | NF | DN | Path |
| 15 | c.844C>T | p.Arg282Trp | E 8 | M | 1 | 0 | rs28934574,
COSM10704 | – | Yes | C65 | DC | D | D | PRB | Path | NF | NF | MD | Path |
| 16 | c.467G>A | p.Arg156His | E 5 | M | 1 | 0 | rs371524413,
COSM43739 | – | Yes | C0 | P | N | T | POB | LP | PF | NF | ND | LP |
| 17 | c.576G>C | p.Gln192His | E 6 | M | 2 | 0 | COSM44554 | – | – | C0 | DC | N | T | B | – | F | F | – | LB |
| 18 | c.648G>C | p.Val216Val | E 6 | S | 1 | 0 | rs199693249 | – | – | – | – | – | – | – | – | – | – | – | LB |
| 19 | c.63C>T | p.Asp21Asp | E 2 | S | 0 | 1 | rs1800369 | – | – | – | – | – | – | – | – | – | – | – | LB |
| 20 | c.459C>T | p.Pro153Pro | E 5 | S | 0 | 1 | rs72661116 | – | – | – | – | – | – | – | LB | – | – | – | LB |
| 21 | c.-140G>A | – | E 1 | 3′UTR | 4 | 0 | Novel | – | – | – | – | – | – | – | – | – | – | – | LB |
| 22 | c.-159delT | – | E 2 | 3′UTR | 1 | 0 | Novel | – | – | – | – | – | – | – | – | – | – | – | LB |
| 23 | c.97-29C>A | – | I 3 | I | 4 | 2 | rs17883323 | Al | – | – | – | – | – | – | – | – | – | – | US |
| 24 | c.994-95 delT | – | I 9 | I | 4 | 0 | Novel | Al | – | – | – | – | – | – | – | – | – | – | US |
| 25 | c.74+16G>C | – | I 2 | I | 2 | 0 | Novel | – | – | – | – | – | – | – | – | – | – | – | LB |
| 26 | c.74+38C>G | – | I 2 | I | 25 | 12 | rs1642785 | – | – | – | – | – | – | – | – | – | – | – | LB |
| 27 | c.96+41_96+56 | – | I 3 | I | 39 | 19 | rs59758982 | – | – | – | – | – | – | – | – | – | – | – | LB |
|
| delACCTGG |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| AGGGCTG |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| GGG |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 28 | c.97-52G>A | – | I 3 | I | 1 | 0 | rs540683791 | – | – | – | – | – | – | – | – | – | – | – | LB |
| 29 | c.375+37C>A | – | I 4 | I | 1 | 0 | Novel | – | – | – | – | – | – | – | – | – | – | – | LB |
| 30 | c.782+72C>T | – | I 7 | I | 22 | 9 | rs12947788 | – | – | – | – | – | – | – | – | – | – | – | LB |
| 31 | c.782+92T>G | – | I 7 | I | 22 | 9 | rs12951053 | – | – | – | – | – | – | – | – | – | – | – | LB |
| 32 | c.993+12T>C | – | I 9 | I | 1 | 0 | rs1800899 | – | – | – | – | – | – | – | – | – | – | – | LB |
| 33 |
c.1100+76T>A | – | I 10 | I | 2 | 0 | Novel | – | – | – | – | – | – | – | – | – | – | – | LB |
| 34 | c.75-42G>A | – | I 2 | I | 0 | 1 | Novel | – | – | – | – | – | – | – | – | – | – | – | LB |
| 35 | c.782+79C>T | – | I 7 | I | 0 | 1 | Novel | – | – | – | – | – | – | – | – | – | – | – | LB |
| 36 | c.673-36G>C | – | I 6 | I | 2 | 0 | rs17880604 | – | – | – | – | – | – | – | B | – | – | – | B |
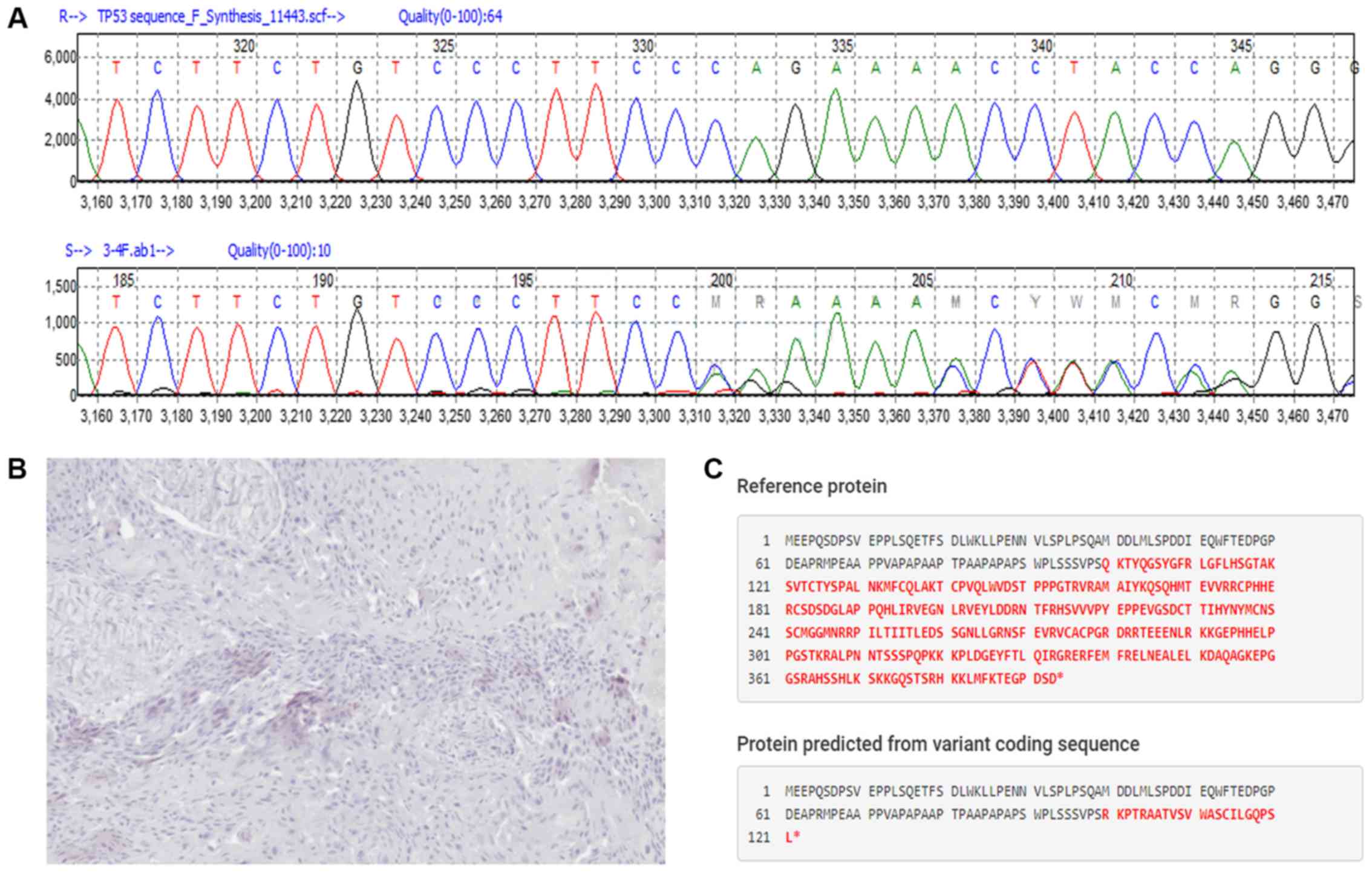
c.298delC/p.Gln100Argfs*23, a novel heterozygous
frameshift variant in exon 4 that created a stop codon at position
123, was found in a male patient with upper oesophageal cancer
(Fig. 1) diagnosed at 51 years of
age. A c.383delC/p.Pro128Leufs*42 frameshift variant in exon 5 that
created a stop codon at position 170 was also found in a male
patient with cancer recurrence in the vocal cord.
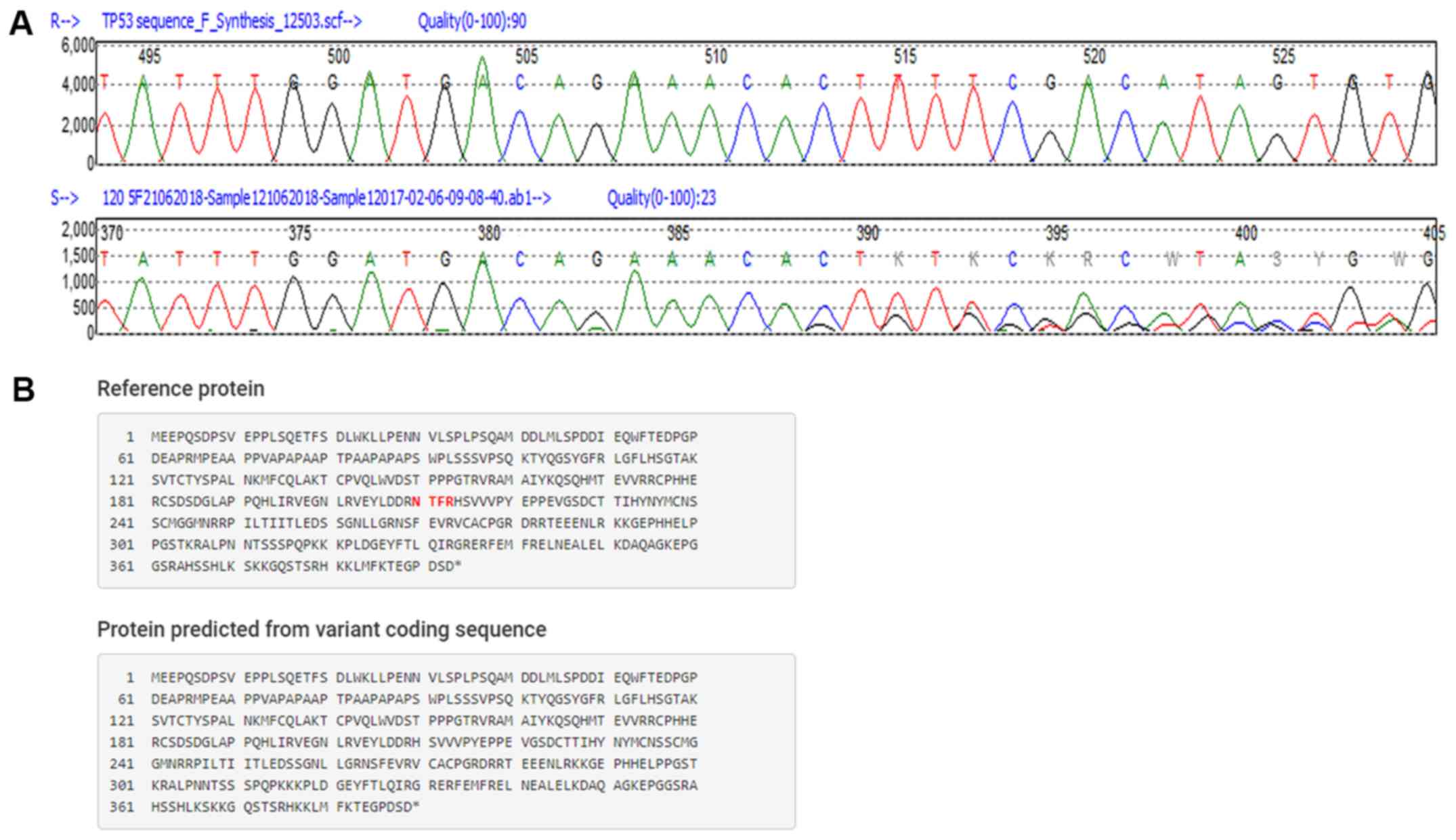
A novel 12 base pair in-frame deletion in exon 6
(c.626_637delGAAACACTTTTC/p.Asn210_Arg213del), that would results
in the loss of 4 amino acid residues from 210 to 213, producing a
389 amino acid protein (Fig. 2)
was detected in the tumour DNA of a 61-year-old male patient with
oesophageal cancer.
Another male patient with recurrent cheek cancer had
one nonsense pathogenic variant, c.493C>T (p.Gln165*) in exon 5,
creating a stop codon at position 165, together with a pathogenic
missense variant (c.578A>G/p.His193Arg) in exon 6 and survived
only two years and eight months following recurrence. A c.637C>T
(p.Arg213*) a nonsense pathogenic variant in exon 6, creating a
stop codon at 213 position was observed in a 72-year-old male
patient with tumour of the mandible.
A 59-year-old female patient with a malignancy in
the cheek carried two variants in exon 5; a pathogenic missense
variant (c.422G>T/p.Cys141Phe) and a likely-pathogenic missense
variant (c.467G>A/p.Arg156His). She survived for one year and
eight months following diagnosis.
Another 59-year-old male patient had a cancer
diagnosed in 2010 in the retro molar region which recurred in the
mandible after 5 years. He had a single pathogenic missense
variant, c.455C>T/p.Pro152Leu in exon 5. Another pathogenic
variant, c.524G>A/p.Arg175His in exon 5 was observed in a male
patient with left antral malignancy and in a female patient with
oesophageal cancer.
Four pathogenic missense variants were detected in
exon 6 (c.583A>T, c.659A>G, c.646G>C, c.578A>G). One
male patient aged 59 diagnosed with upper oesophageal cancer had a
c.583A>T/p.Ile195Phe variant. A c.659A>G/p.Tyr220Cys variant
was found in a 55-year-old male patient with malignancy in the
retro molar region. This variant co-existed with another pathogenic
variant, c.818G>A/p.Arg273His, in exon 8. Co-existence of
c.646G>C and c.648G>C (p.Val216Leu) variants were identified
in the malignant cheek tumour of a 60-year-old female patient.
c.576G>C/p.Gln192His, a likely-benign variant was
identified in exon 6 in a 68-year-old female patient with
oesophageal cancer and in a male patient with arytenoid cartilage
cancer. The only pathogenic missense variant
c.747G>T/p.Arg249Ser in exon 7 was observed in a male patient
aged 68 years with cancer in the cheek.
A c.844C>T/p.Arg282Trp variant in exon 8 was
found in a 60-year-old female patient with a mandible cancer who
survived only 1 year and 5 months following diagnosis.
Among the 14 patients who had pathogenic variants,
eight had a history of betel quid-tobacco chewing, five in the
absence of smoking or alcoholism and three in the presence of
smoking and alcoholism. Six of the patients with pathogenic
variants did not have a history of betel quid-tobacco chewing, but
three of them were smokers and had a history of alcoholism.
Among the intronic variants, c.994-95delT, a novel
deletion in intron 9 was identified in 4 patients but not in
controls and c.97-29C>A, a reported variant substitution in
intron 3, which was identified in four patients and in two controls
were categorized as variants with uncertain significance as both
alter the WT branch point and potentially alter the splicing
according to Human Splicing finder.
Two novel variants were found in the 5′UTR of the
gene; c.-140G>A (N=4) and c.-159delT (N=1) in patients but not
in healthy controls. Three more novel intronic variants were found
in patients [c.74+16G>C (N=2), c.375+37C>A (N=1),
c.1100+76T>A (N=2)] in introns 2, 4 and 10 respectively but not
in any healthy controls. In contrast, two other novel variants
[c.75-42G>A (intron 2) and c.782+79C>T (intron 7)] were
observed in healthy controls but not in patients.
TP53 gene status and expression of p53
protein
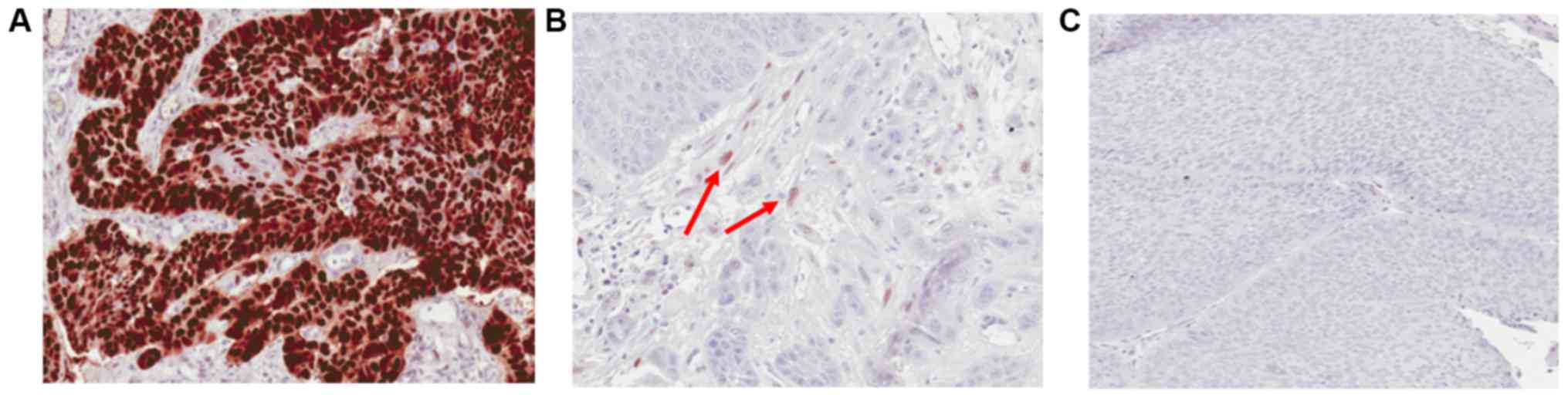
Twenty four samples were randomly chosen for IHC
analysis. The H-Score for each slide was calculated by multiplying
the intensity of staining (no staining, 0; weak, 1; intermediate,
2; and strong, 3) by the percentage of cells at each staining
intensity level. Scores ranged from 0 to 300. Scores were grouped
into three categories; from 0 to <1 was considered negative
(Pattern-C), score ranging from 1 to 20 (Pattern-B) and all the
scores >20 (Pattern-A) (Fig.
3).
IHC staining for p53 showed positive
immuno-reactivity in 13/24 (54.17%) cases. Nine of these tumours
showed widespread IHC positive tumour nuclear staining involving
either the entire tissue section or a segment of it (Pattern-A)
while 4 showed rare/scattered p53 positive single tumour nuclei
(Pattern-B). All cases with a missense variant in TP53 gene
showed Pattern-A IHC staining. However, vice versa is not true
because 2 cases showed IHC positivity with Pattern-A in the absence
of detectable pathogenic TP53 gene variants. These had only
~10% of positively stained tumour cells in the respective tissue
sections. One case with both missense and nonsense variants showed
Pattern-A IHC staining. All 4 cases with Pattern-B IHC staining had
no detectable pathogenic TP53 variants. Among the 11 cases
with immuno-negativity (Pattern-C), two had frameshift variants and
one had a nonsense variant. The remaining eight patients who showed
immuno-negativity had wild-type TP53.
Prevalence of HPV
Sensitivity of the GP5+/GP6+ primer pair was at the
level of femtograms for HPV genotypes which match strongly with the
primers and at the level of picograms for HPV genotypes having four
or more mismatches to the primers (21). Absence of positive results in our
study cohort indicated either absence of HPV DNA or the presence of
HPV DNA at a concentration below the level of sensitivity.
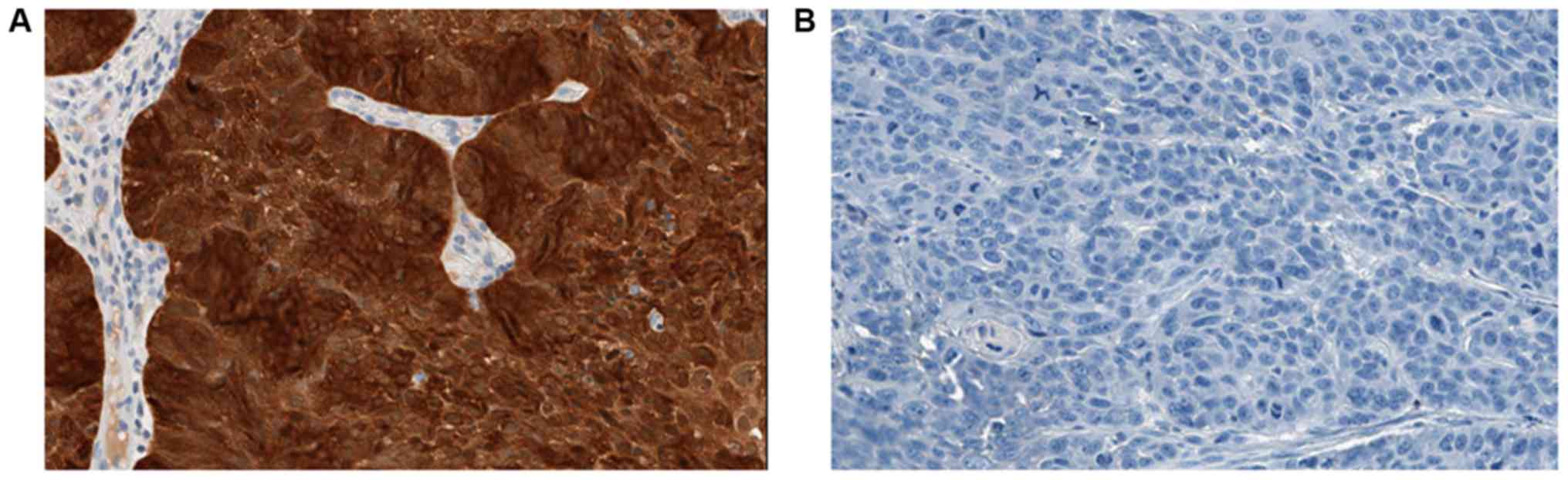
The IHC staining for p16 is defined as positive if
there is strong and diffuse nuclear and cytoplasmic staining
pattern in >70% of the tumour specimen (Fig. 4). Regardless of cancer type,
TP53 mutation status and p53 protein expression, all HNC
samples in the Sri Lankan study cohort were negative for p16
staining, despite clear positive staining with the control
sample.
Discussion
This is a preliminary study on Sri Lankan HNC
patients, focused on the mutation spectrum of the TP53 gene,
expression of p53 protein and relationship to possible risk
factors. In the present study, we also included upper oesophageal
cancer, as HNCs are often associated with oesophageal cancer and
both tumour types share common risk factors (22).
Identification of hotspot regions of TP53
mutations in the Sri Lankan context is useful to prioritize
screening of such regions rather screening the entire gene, prior
to treatment. In the current study, 70% (11 out of 15) of
pathogenic mutations reported were detected in the exon 5 and 6
which is covered by a single set of primer. Therefore it is worth
to understand the hotspot region in TP53 for initiation of
screening of TP53 in a resource limited setting such as Sri
Lanka. Hotspot regions of TP53 identified in the present
study are similar to those reported for other ethnic groups. This
is also supported by the data in the ‘IARC TP53 database’
(http://p53.iarc.fr/TP53SomaticMutations.aspx) and
‘cBioPortal for cancer genomics’ (http://www.cbioportal.org/).
Among the pathogenic variants found, the frameshift
variant c.298delC and in-frame variant c.626_637delGAAACACTTTTC
have not been previously reported. The c.646G>C somatic missense
variant is reported only in the IARC TP53 database, but the details
are not provided. c.383delC and c.576G>C are novel variants in
HNCs, as they have been reported only in the COSMIC database for
breast and stomach tumours respectively. Although c.422G>T,
c.455C>T, c.467G>A, c.747G>T and c.818G>A have been
commonly reported for many types of cancer, these variants have not
been reported in HNC or oesophageal cancer.
Some mutations of TP53 such as frame shift
deletions or insertions and nonsense or splice site mutations are
recessive and require loss of the remaining normal allele for cells
to lose p53 tumour suppressor function. However, some point
missense mutant forms can have a dominant-negative effect (DNE)
because they are no longer recognised by their negative regulator
MDM2 and accumulate in the cell to a much greater level than the
normal wild-type TP53 allele. These so-called
‘dominant-negative’ forms of mutant p53 nevertheless have lost
their normal transcriptional activity. Loss of function of each
identified variant in the current study was assessed based on the
reported loss of transcriptional activity data available in the
IARC and other databases. Overall transcriptional activity of 2314
distinct mutant proteins has been experimentally measured on a
panel of p53-response elements of promoters of downstream
transcriptional target genes such as CDKN1A (p21),
MDM2 and BAX (23–25).
In addition sequence variants have also been evaluated using a
computational geometry approach called Delaunay Tessellations to
predict the functional impact (26). DNE of sequence variants has been
assessed based on studies carried out on promoters of
p21WAF, Ribosomal Gene Clusters, etc. (24). Out of ten pathogenic missense
variants reported in our study cohort, nine (c.455C>T,
c.524G>A, c.578A>G, c.583A>T, c.659A>G, c.646G>C,
c.747G>T, c.818G>A and c.844C>T) are predicted to involve
complete loss of transcriptional function based on databases
compiled from both yeast assay and computational method.
Codon 72 variant (p.Arg72Pro) in exon 4 is a
well-known polymorphism present in the normal human population. The
implication of this polymorphism in cancer risk and prognosis is
controversial, with some earlier studies reporting that p53 protein
containing the codon 72 Arginine form has a greater apoptotic
potential (27–31) while others have failed to replicate
these findings (32,33). In the present study neither the
p.Arg72Pro alleles was significantly associated with cancer.
One previous study of TP53 carried out in
1998 on 23 oral squamous cell carcinoma from Sri Lankan patients
has been published. This reported nine sequence variants: Missense
variant in codons 135, 164, 176, 245, 248; deletions in codons 130,
144–148, 172–187 and one insertion in codon 250 (34). None of these variants were found in
the current study, which included 19 patients with oral squamous
cell carcinoma. However studies done in all subset of HNC in India
and Japan showed 21 and 11.6% of pathogenic TP53 variant and
69 and 13.7% of HPV positivity respectively (16,17).
Generally, for head and neck cancers without HPV
infection has a higher prevalence of TP53 mutation. However,
the lower percentage of TP53 mutations was observed in the
current study is due to the inclusion of thyroid cancer in the
studied cohort as it is considered under head and neck cancer in
Sri Lankan context. In the current study, out of 44 patients, 9
patients were with thyroid carcinoma and none of them carried any
pathogenic TP53 variants, which may be the possible reason
for a decreased percentage of TP53 mutation prevalence
(35,36).
TP53 mutations can be classified as
disruptive and non-disruptive based on the location and type of
mutations. Disruptive mutations are missense mutations located in
the DNA-binding domain, or stop codons in any region which create
truncated proteins. Non-disruptive mutations are missense mutations
located outside the DNA binding domain. Disruptive mutations are
closely associated with poor prognosis of head and neck cancer
(37) and all the reported
pathogenic somatic TP53 mutations in the current study are
associated with poor prognosis.
However, we also analysed the pathogenic missense
mutations with a computational approach called evolutionary action
or EAp53, which has been validated both in vivo and in
vitro, for TP53 mutation classification (http://mammoth.bcm.tmc.edu/EAp53/). This approach
assigns an evolutionary action score and classifies each p53
missense mutation as either high- or low-risk. According to Neskey
et al (38), patients with
high risk TP53 mutations show poor survival with high invasive and
aggressive tumour behaviour. According to the EAp53 analysis
c.422G>T, c.455C>T, c.524G>A, c.578A>G, c.747G>T
were categorized as high risk mutations and c.583A>T,
c.646G>C, c.659A>G, c.818G>A, c.844C>T as low risk
mutations. In addition all truncating mutations are also considered
to have poor prognosis.
Generally wild-type p53 protein has very short
half-life in the cells under normal conditions due to constant
degradation by the MDM2-mediated negative feedback loop (39). In the case of mutant p53 protein,
MDM2 protein cannot be induced, thus MDM2-mediated negative
feedback is absent leading to accumulation of mutant p53 (40). It is also reported that a
contribution to the stabilization of mutant p53 is due to impaired
ubiquitination (41). All the
missense variations identified in the current study showed the
accumulation of p53 protein with the Pattern-A strong
immunostaining. However the converse was not true, as there were
two samples with Pattern-A IHC which showed no detectable
variations in sequence. But these two samples have only <10% of
tumour cells in the sections used for immunostaining which would
have been below the sensitivity of detection by sanger
sequencing.
All the truncated proteins that resulted either from
frameshift or nonsense variations showed immuno-negativity. The
antibody used to detect the expression of p53 protein is DO-7 which
binds to the p53 protein between amino acid 1–45 (42). Even though all four truncated
protein are longer than 123 amino acids, they remained
undetectable. This may be due to nonsense-mediated decay of mRNA
with premature stop codon may have resulted in less stable
truncated proteins.
All the samples (N=4) with Pattern-B IHC were
TP53 wild-type and, there were 57% (8/14) of the wild-type
samples that showed no positive immuno-reactivity. Thus IHC
analysis cannot be used as a standalone method to detect the
alteration in p53 as it cannot differentiate between truncated p53
protein and wild-type p53 protein as both show an immuno-negative
IHC pattern.
Cigarette smoking and alcoholism are well known
triggers of HNCs. The awareness of these risk factors is higher
among Sri Lankans. But despite a reduction in cigarette smoking,
the burden of HNC has increased constantly over the years, which
prompted us to search for other risk factors.
Betel-quid chewing with tobacco is a common habit
among Sri Lankans especially those who are engaged in hard physical
work for long hour. Furthermore, there is poor awareness of the
harmfulness of this habit. 47.7% of the patients (N=21) studied
were addicted to betel-quid chewing with tobacco and out of them,
nine have TP53 sequence variants conferring an altered
protein structure.
HPVs are non-enveloped DNA viruses, containing a
double stranded circular DNA which consists of an upstream
regulatory region (URR), an early region (E) encoding viral
regulatory proteins and a late region (L) encoding viral capsid
proteins (43). HPV is an
identified cause of HNCs and it is reported that HPV-positive HNCs
have a better prognosis (44–46).
The high-risk HPV proteins E6, E7 target tumour suppressor proteins
p53 and RB respectively. Disruption of the function of p53 and pRB
lead to uncontrolled cell division (47). Here we studied the expression of
p16 protein which is used as a surrogate marker to identify
functionally active HPV infection as it is reciprocally
overexpressed due to functional inactivation of pRB by E7 protein
(48,49). Several previous studies in other
countries reported p16 overexpression in HNC. The highest (38%) and
lowest (7.8%) level of p16 expression are reported in the USA,
while intermediate levels are reported from India (20%) and Kenya
(14.6%) (50–53). All samples in our study cohort were
negative for p16 staining which may suggest the absence of
functionally active HPV infection (54).
In a previous study from Sri Lanka 37.2% of tumour
samples from oral cancer patients were reported to be infected when
analysed using HPV DNA typing (18). However in the current study, none
of the patients were positive on DNA typing which also indicates
the absence or presence of very low concentration of HPV DNA in the
tumour samples.
It has also been reported that p53 protein with the
Arginine72 polymorphism is efficiently degraded by the E6 protein
of HPV 16, thus increasing susceptibility to cancer (55). Nevertheless, there are studies that
show no correlation between p.Arg72Pro and HPV related cervical
cancers (56,57). Our cohort of HNC patients also
showed no correlation between p.Arg72Pro and HPV infection.
Exposure to asbestos due to living in houses with
asbestos roofing and exposure to rubber by occupations may also
have contributed to mutagenesis of TP53 and carcinogenesis
in some patients in the present cohort.
In conclusion, we examined all exons and splicing
sites of the TP53 gene in HNC and oesophageal cancers from a
cohort of Sri Lankan patients and found a high occurrence of gene
alterations including several novel variants. All p53 protein
altering variants found were positioned between exons 4–8. Only the
point missense variants were associated with strong
immuno-positivity, thus limiting the use of IHC in detecting
mutation status of p53. Our cohort of patients did not appear to
carry significant levels of HPV infection.
Acknowledgements
The authors would like to thank Ms Anoma Jayasoma
(senior technical officer of the Institute of Biochemistry,
Molecular Biology and Biotechnology, University of Colombo,
Colombo, Sri Lanka), Ms Chameera Helani (library information
assistant of the Institute of Biochemistry, Molecular Biology and
Biotechnology, University of Colombo) for their assistance in
conducting research at the institute and collecting details of the
patients. Ms Gayani Kokila Wijesinghe and Ms Yamuna Wickramasinghe
Jayasekera (staff technical officers of Department of Pathology,
Faculty of Medicine, University of Colombo) for their assistance in
making formalin-fixed paraffin-embedded blocks. Dr Calum Kirk
(research technician of Northern Institute for Cancer Research,
Newcastle University, Newcastle, UK) for assisting in the
immunohistochemistry.
Funding
The present study was funded by National Research
Council, Sri Lanka (grant no. 15-33) and Commonwealth Scholarship
Commission of the UK (grant no. LKCN-2015-100).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
VM performed the experiments, contributed to the
analysis and interpretation of the data, and drafted the
manuscript. EHK, KHT and SDS conceived and designed the study, were
involved in the molecular genetic studies, data analysis and
interpretation, and the revision of the manuscript. KDS and PA
provided clinical expertise, recruited the study participants, and
supervised the clinical data and histopathological
characterization. JL designed the immunohistochemical and HPV
studies, and contributed to the analysis of the sequencing data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics Review
Committee, Faculty of Medicine, University of Colombo, Sri Lanka
(EC/14/160). Written informed consent form at the time of enrolment
was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DNE
|
dominant negative effect
|
|
HNC
|
head and neck cancer
|
|
HPV
|
human papilloma virus
|
|
IHC
|
immunohistochemistry
|
|
PCR
|
polymerase chain reaction
|
|
UTR
|
untranslated region
|
References
|
1
|
Shah JP and Lydiatt W: Treatment of cancer
of the head and neck. CA Cancer J Clin. 45:352–368. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Joshi P, Dutta S, Chaturvedi P and Nair S:
Head and neck cancers in developing countries. Rambam Maimonides
Med J. 5:e00092014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khan Z, Tönnies J and Müller S: Smokeless
tobacco and oral cancer in South Asia: A systematic review with
meta-analysis. J Cancer Epidemiol. 2014:3946962014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
National Cancer Control Programme (NCCP),
. Cancer Incidence Data Sri Lanka 2010. (12). NCCP. (Colombo).
1–82. 2016.
|
|
5
|
Amarasinghe HK, Usgodaarachchi US, Johnson
NW, Lalloo R and Warnakulasuriya S: Betel-quid chewing with or
without tobacco is a major risk factor for oral potentially
malignant disorders in Sri Lanka: A case-control study. Oral Oncol.
46:297–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
No authors listed, . Sri Lankan
declaration on oral cancer, smokeless tobacco and areca nut -
Towards a society free of oral cancer. Sri Lanka Dental Journal.
45:27–29. 2015.
|
|
7
|
Somatunga LC, Sinha DN, Sumanasekera P,
Galapatti K, Rinchen S, Kahandaliyanage A, Mehta FR and Nishirani
Lanka JD: Smokeless tobacco use in Sri Lanka. Indian J Cancer.
49:357–363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
World Health Organization - International
Agency for Research on Cancer (IARC), . IARC Monographs on the
Evaluation of Carcinogenic Risks to Humans. 89:IARC. (Lyon). 1–593.
2017.
|
|
9
|
Kreimer AR, Clifford GM, Boyle P and
Franceschi S: Human papillomavirus types in head and neck squamous
cell carcinomas worldwide: A systematic review. Cancer Epidemiol
Biomarkers Prev. 14:467–475. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deb SP and Deb S: Mutant p53 and MDM2 in
cancer. Springer. 2014. View Article : Google Scholar
|
|
11
|
Hollstein M, Sidransky D, Vogelstein B and
Harris CC: p53 mutations in human cancers. Science. 253:49–53.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen MB, Wu XY, Yu R, Li C, Wang LQ, Shen
W and Lu PH: P53 status as a predictive biomarker for patients
receiving neoadjuvant radiation-based treatment: A meta-analysis in
rectal cancer. PLoS One. 7:e453882012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu HL, Shao L, Wang Q, Jia T, Li M and
Yang DP: A systematic review of p53 as a biomarker of survival in
patients with osteosarcoma. Tumour Biol. 34:3817–3821. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Greenblatt MS, Bennett WP, Hollstein M and
Harris CC: Mutations in the p53 tumor suppressor gene: Clues to
cancer etiology and molecular pathogenesis. Cancer Res.
54:4855–4878. 1994.PubMed/NCBI
|
|
15
|
Lane DP: On the expression of the p53
protein in human cancer. Mol Biol Rep. 19:23–29. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mitra S, Banerjee S, Misra C, Singh RK,
Roy A, Sengupta A, Panda CK and Roychoudhury S: Interplay between
human papilloma virus infection and p53 gene alterations in head
and neck squamous cell carcinoma of an Indian patient population. J
Clin Pathol. 60:1040–1047. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Maruyama H, Yasui T, Ishikawa-Fujiwara T,
Morii E, Yamamoto Y, Yoshii T, Takenaka Y, Nakahara S, Todo T,
Hongyo T and Inohara H: Human papillomavirus and p53 mutations in
head and neck squamous cell carcinoma among Japanese population.
Cancer Sci. 105:409–417. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jayasooriya PR, Kurose K, Terai M,
Sivagnanam K, Siriwardana S, Tilakaratne WM, Tagami J and Takagi M:
Human papillomavirus in oral cancer from Sri Lanka: Prevalence and
relationship with clinico-pathological parameters. Oral Med Pathol.
8:45–50. 2003. View Article : Google Scholar
|
|
19
|
Miller SA, Dykes DD and Polesky HF: A
simple salting out procedure for extracting DNA from human
nucleated cells. Nucleic Acids Res. 16:12151988. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Richards S, Aziz N, Bale S, Bick D, Das S,
Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al:
Standards and guidelines for the interpretation of sequence
variants: A joint consensus recommendation of the American college
of medical genetics and genomics and the association for molecular
pathology. Genet Med. 17:405–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
de Roda Husman AM, Walboomers JM, van den
Brule AJ, Meijer CJ and Snijders PJ: The use of general primers GP5
and GP6 elongated at their 3′ ends with adjacent highly conserved
sequences improves human papillomavirus detection by PCR. J Gen
Virol. 76:1057–1062. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim JS and Kim BW: Esophageal cancer and
head and neck cancer: The earlier, the better. Gut Liver.
9:131–132. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bouaoun L, Sonkin D, Ardin M, Hollstein M,
Byrnes G, Zavadil J and Olivier M: TP53 variations in human
cancers: New lessons from the IARC TP53 database and genomics data.
Hum Mutat. 37:865–876. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Petitjean A, Mathe E, Kato S, Ishioka C,
Tavtigian SV, Hainaut P and Olivier M: Impact of mutant p53
functional properties on TP53 mutation patterns and tumor
phenotype: Lessons from recent developments in the IARC TP53
database. Hum Mutat. 28:622–629. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kato S, Han SY, Liu W, Otsuka K, Shibata
H, Kanamaru R and Ishioka C: Understanding the function-structure
and function-mutation relationships of p53 tumor suppressor protein
by high-resolution missense mutation analysis. Proc Natl Acad Sci
USA. 100:8424–8429. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mathe E, Olivier M, Kato S, Ishioka C,
Vaisman I and Hainaut P: Predicting the transactivation activity of
p53 missense mutants using a four-body potential score derived from
Delaunay tessellations. Hum Mutat. 27:163–172. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dumont P, Leu JI, Della Pietra AC III,
George DL and Murphy M: The codon 72 polymorphic variants of p53
have markedly different apoptotic potential. Nat Genet. 33:357–365.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Toyama T, Zhang Z, Nishio M, Hamaguchi M,
Kondo N, Iwase H, Iwata H, Takahashi S, Yamashita H and Fujii Y:
Association of TP53 codon 72 polymorphism and the outcome of
adjuvant therapy in breast cancer patients. Breast Cancer Res.
9:R342007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Damin AP, Frazzon AP, Damin DC, Roehe A,
Hermes V, Zettler C and Alexandre CO: Evidence for an association
of TP53 codon 72 polymorphism with breast cancer risk. Cancer
Detect Prev. 30:523–529. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li T, Lu ZM, Guo M, Wu QJ, Chen KN, Xing
HP, Mei Q and Ke Y: p53 codon 72 polymorphism (C/G) and the risk of
human papillomavirus-associated carcinomas in China. Cancer.
95:2571–2576. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan R, Wu MT, Miller D, Wain JC, Kelsey
KT, Wiencke JK and Christiani DC: The p53 codon 72 polymorphism and
lung cancer risk. Cancer Epidemiol Biomarkers Prev. 9:1037–1042.
2000.PubMed/NCBI
|
|
32
|
Baynes C, Healey CS, Pooley KA, Scollen S,
Luben RN, Thompson DJ, Pharoah PD, Easton DF, Ponder BA and Dunning
AM; SEARCH breast cancer study, : Common variants in the ATM,
BRCA1, BRCA2, CHEK2 and TP53 cancer susceptibility genes are
unlikely to increase breast cancer risk. Breast Cancer Res.
9:R272007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mabrouk I, Baccouche S, El-Abed R,
Mokdad-Gargouri R, Mosbah A, Saïd S, Daoud J, Frikha M, Jlidi R and
Gargouri A: No evidence of correlation between p53 codon 72
polymorphism and risk of bladder or breast carcinoma in Tunisian
patients. Ann NY Acad Sci. 1010:764–770. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chiba I, Muthumala M, Yamazaki Y, Uz Zaman
A, Iizuka T, Amemiya A, Shibata T, Kashiwazaki H, Sugiura C and
Fukuda H: Characteristics of mutations in the p53 gene of oral
squamous-cell carcinomas associated with betel-quid chewing in Sri
Lanka. Int J Cancer. 77:839–842. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Poeta ML, Manola J, Goldwasser MA,
Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D,
Saunders J, et al: TP53 mutations and survival in squamous-cell
carcinoma of the head and neck. N Engl J Med. 357:2552–2561. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Neskey DM, Osman AA, Ow TJ, Katsonis P,
McDonald T, Hicks SC, Hsu TK, Pickering CR, Ward A, Patel A, et al:
Evolutionary action score of TP53 identifies high-risk mutations
associated with decreased survival and increased distant metastases
in head and neck cancer. Cancer Res. 75:1527–1536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Reich NC, Oren M and Levine A: Two
distinct mechanisms regulate the levels of a cellular tumor
antigen, p53. Mol Cell Biol. 3:2143–2150. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Blagosklonny MV: Loss of function and p53
protein stabilization. Oncogene. 15:1889–1893. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nagata Y, Anan T, Yoshida T, Mizukami T,
Taya Y, Fujiwara T, Kato H, Saya H and Nakao M: The stabilization
mechanism of mutant-type p53 by impaired ubiquitination: The loss
of wild-type p53 function and the hsp90 association. Oncogene.
18:6037–6049. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vojtĕsek B, Bártek J, Midgley CA and Lane
DP: An immunochemical analysis of the human nuclear phosphoprotein
p53. New monoclonal antibodies and epitope mapping using
recombinant p53. J Immunol Methods. 151:237–244. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Harden ME and Munger K: Human
papillomavirus molecular biology. Mutat Res Rev Mutat Res.
772:3–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ang KK, Harris J, Wheeler R, Weber R,
Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C,
et al: Human papillomavirus and survival of patients with
oropharyngeal cancer. N Engl J Med. 363:24–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lassen P, Eriksen JG, Hamilton-Dutoit S,
Tramm T, Alsner J and Overgaard J: Effect of HPV-associated
p16INK4A expression on response to radiotherapy and survival in
squamous cell carcinoma of the head and neck. J Clin Oncol.
27:1992–1998. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fakhry C, Westra WH, Li S, Cmelak A, Ridge
JA, Pinto H, Forastiere A and Gillison ML: Improved survival of
patients with human papillomavirus-positive head and neck squamous
cell carcinoma in a prospective clinical trial. J Natl Cancer Inst.
100:261–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dok R and Nuyts S: HPV positive head and
neck cancers: Molecular pathogenesis and evolving treatment
strategies. Cancers (Basel). 8:E412016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Grønhøj Larsen C, Gyldenløve M, Jensen DH,
Therkildsen MH, Kiss K, Norrild B, Konge L and Von Buchwald C:
Correlation between human papillomavirus and p16 overexpression in
oropharyngeal tumours: A systematic review. Br J Cancer.
110:1587–1594. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lewis JS Jr, Thorstad WL, Chernock RD,
Haughey BH, Yip JH, Zhang Q and El-Mofty SK: p16 positive
oropharyngeal squamous cell carcinoma: An entity with a favorable
prognosis regardless of tumor HPV status. Am J Surg Pathol.
34:1088–1096. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Smith EM, Rubenstein LM, Hoffman H, Haugen
TH and Turek LP: Human papillomavirus, p16 and p53 expression
associated with survival of head and neck cancer. Infect Agent
Cancer. 5:42010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Murthy V, Swain M, Teni T, Pawar S, Kalkar
P, Patil A, Chande A, Ghonge S, Laskar SG, Gupta T, et al: Human
papillomavirus/p16 positive head and neck cancer in India:
Prevalence, clinical impact, and influence of tobacco use. Indian J
Cancer. 53:387–393. 2016.PubMed/NCBI
|
|
52
|
Githaiga BK, Muchiri LW and Rogena EA: P16
expression in subsets of head and neck squamous cell carcinoma
reported in Kenyatta National Hospital Nairobi, Kenya. Pathol.
46:S962014. View Article : Google Scholar
|
|
53
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
54
|
Sritippho T, Pongsiriwet S,
Lertprasertsuke N, Buddhachat K, Sastraruji T and Iamaroon A: p16-a
possible surrogate marker for high-risk human papillomaviruses in
oral cancer? Asian Pac J Cancer Prev. 17:4049–4057. 2016.PubMed/NCBI
|
|
55
|
Storey A, Thomas M, Kalita A, Harwood C,
Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G and
Banks L: Role of a p53 polymorphism in the development of human
papillomavirus-associated cancer. Nature. 393:229–234. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Klug SJ, Wilmotte R, Santos C, Almonte M,
Herrero R, Guerrero I, Caceres E, Peixoto-Guimaraes D, Lenoir G,
Hainaut P, et al: TP53 polymorphism, HPV infection, and risk of
cervical cancer. Cancer Epidemiol Biomarkers Prev. 10:1009–1012.
2001.PubMed/NCBI
|
|
57
|
Andersson S, Rylander E, Strand A,
Sällström J and Wilander E: The significance of p53 codon 72
polymorphism for the development of cervical adenocarcinomas. Br J
Cancer. 85:1153–1156. 2001. View Article : Google Scholar : PubMed/NCBI
|