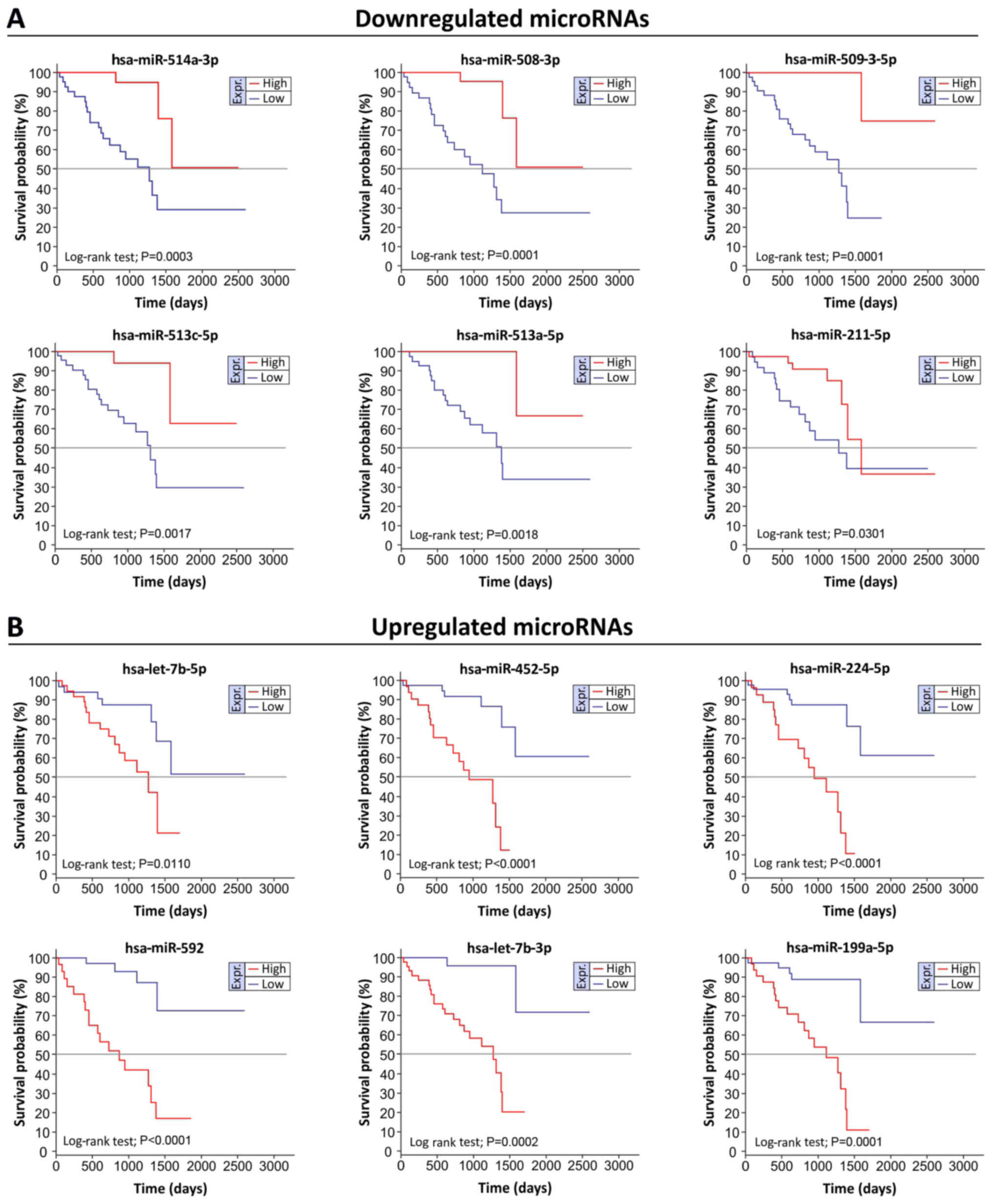
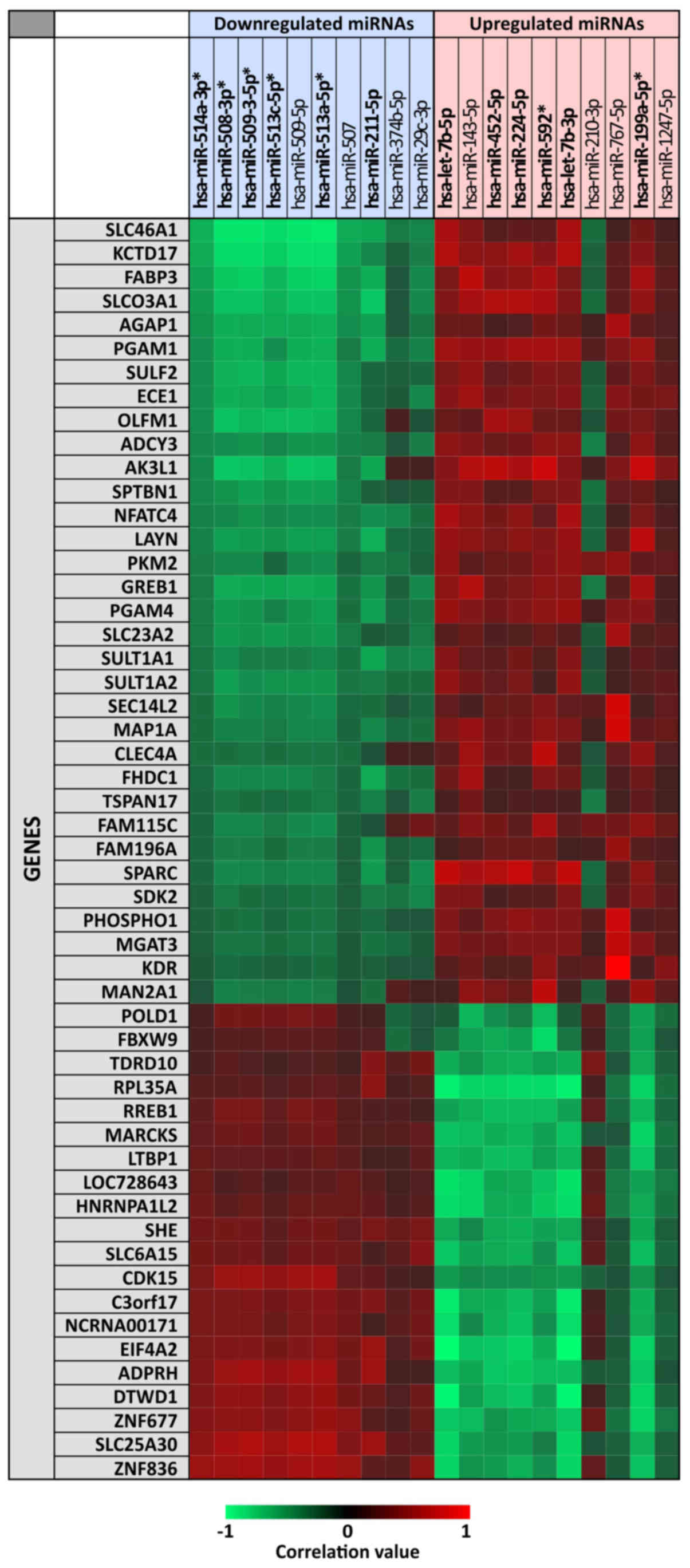
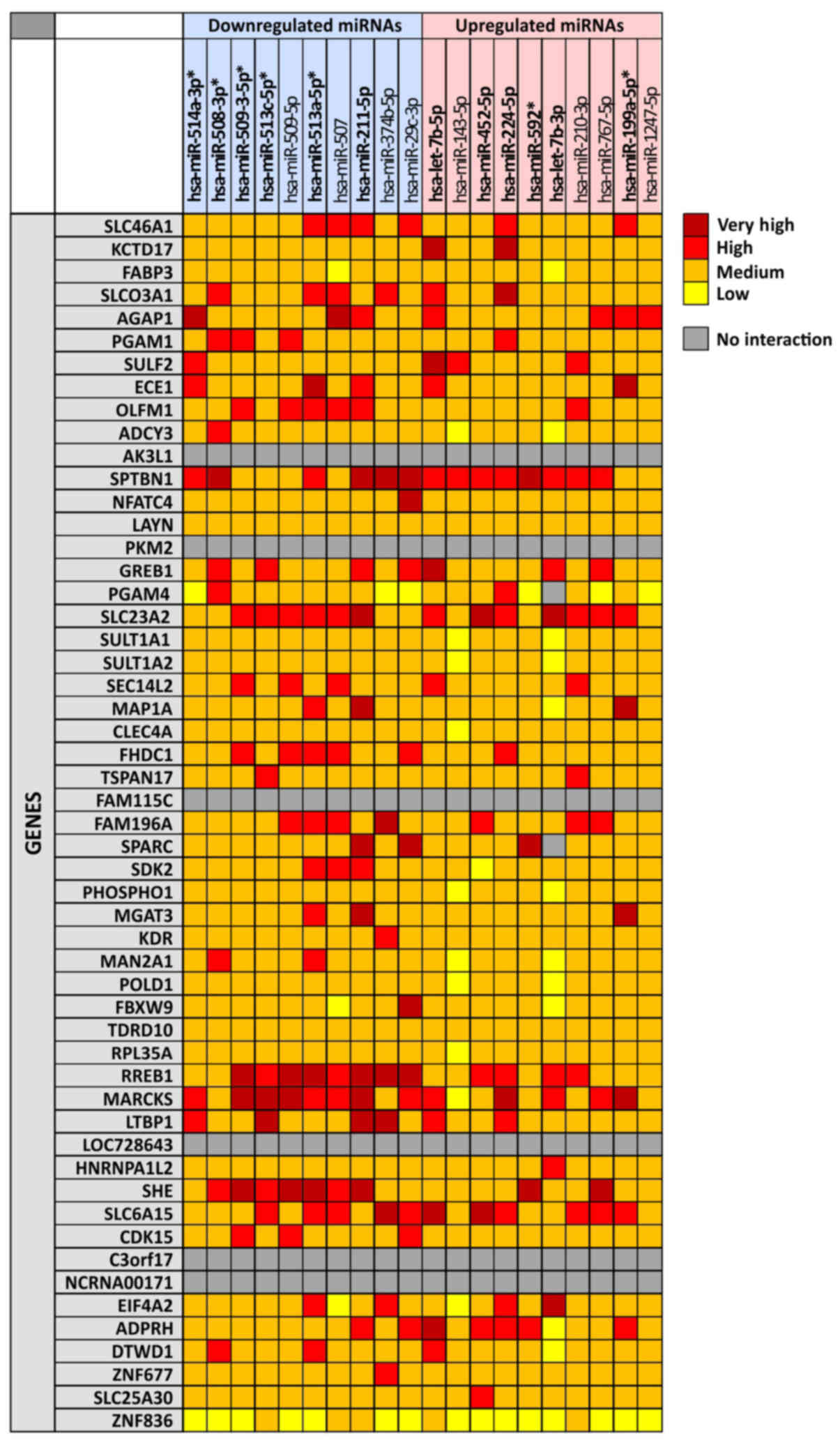
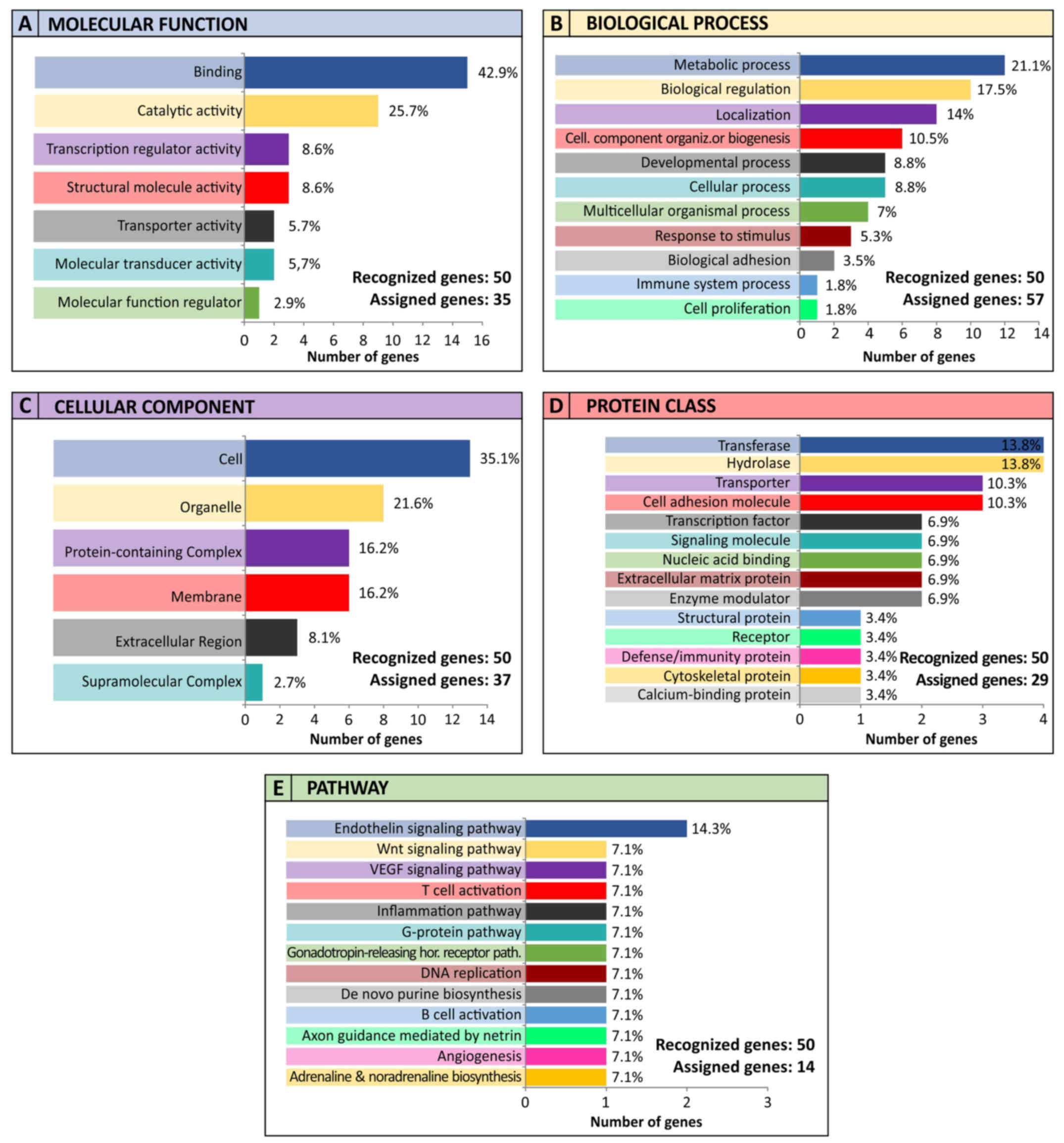
|
1
|
Shields CL, Kaliki S, Furuta M, Mashayekhi
A and Shields JA: Clinical spectrum and prognosis of uveal melanoma
based on age at presentation in 8,033 cases. Retina. 1363–1372.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nayman T, Bostan C, Logan P and Burnier MN
Jr: Uveal melanoma risk factors: A systematic review of
meta-analyses. Curr Eye Res. 42:1085–1093. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weis E, Shah CP, Lajous M, Shields JA and
Shields CL: The association between host susceptibility factors and
uveal melanoma: A meta-analysis. Arch Ophthalmol. 124:54–60. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Falzone L, Marconi A, Loreto C, Franco S,
Spandidos DA and Libra M: Occupational exposure to carcinogens:
Benzene, pesticides and fibers (Review). Mol Med Rep. 14:4467–4474.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Griewank KG, Murali R, Schilling B, Scholz
S, Sucker A, Song M, Süsskind D, Grabellus F, Zimmer L, Hillen U,
et al: TERT promoter mutations in ocular melanoma distinguish
between conjunctival and uveal tumours. Br J Cancer. 109:497–501.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Onken MD, Worley LA, Long MD, Duan S,
Council ML, Bowcock AM and Harbour JW: Oncogenic mutations in GNAQ
occur early in uveal melanoma. Invest Ophthalmol Vis Sci.
49:5230–5234. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Urtatiz O and Van Raamsdonk CD: Gnaq and
Gna11 in the Endothelin Signaling Pathway and Melanoma. Front
Genet. 7:592016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Masoomian B, Shields JA and Shields CL:
Overview of BAP1 cancer predisposition syndrome and the
relationship to uveal melanoma. J Curr Ophthalmol. 30:102–109.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ehlers JP, Worley L, Onken MD and Harbour
JW: Integrative genomic analysis of aneuploidy in uveal melanoma.
Clin Cancer Res. 14:115–122. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun Y, Tran BN, Worley LA, Delston RB and
Harbour JW: Functional analysis of the p53 pathway in response to
ionizing radiation in uveal melanoma. Invest Ophthalmol Vis Sci.
46:1561–1564. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shields CL, Kaliki S, Furuta M, Fulco E,
Alarcon C and Shields JA: American Joint Committee on Cancer
classification of posterior uveal melanoma (tumor size category)
predicts prognosis in 7731 patients. Ophthalmology. 120:2066–2071.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vivarelli S, Salemi R, Candido S, Falzone
L, Santagati M, Stefani S, Torino F, Banna GL, Tonini G and Libra
M: Gut microbiota and cancer: From pathogenesis to therapy. Cancers
(Basel). 11:E382019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan W, Liu B, Qu S, Liang G, Luo W and
Gong C: MicroRNAs and cancer: Key paradigms in molecular therapy.
Oncol Lett. 15:2735–2742. 2018.PubMed/NCBI
|
|
15
|
Banna GL, Torino F, Marletta F, Santagati
M, Salemi R, Cannarozzo E, Falzone L, Ferraù F and Libra M:
Lactobacillus rhamnosus GG: An Overview to Explore the
Rationale of Its Use in Cancer. Front Pharmacol. 8:6032017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baxter E, Windloch K, Gannon F and Lee JS:
Epigenetic regulation in cancer progression. Cell Biosci. 4:452014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
18
|
Yang Y, Dong X, Xie B, Ding N, Chen J, Li
Y, Zhang Q, Qu H and Fang X: Databases and web tools for cancer
genomics study. Genomics Proteomics Bioinformatics. 13:46–50. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Falzone L, Scola L, Zanghì A, Biondi A, Di
Cataldo A, Libra M and Candido S: Integrated analysis of colorectal
cancer microRNA datasets: Identification of microRNAs associated
with tumor development. Aging (Albany NY). 10:1000–1014. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Falzone L, Candido S, Salemi R, Basile MS,
Scalisi A, McCubrey JA, Torino F, Signorelli SS, Montella M and
Libra M: Computational identification of microRNAs associated to
both epithelial to mesenchymal transition and NGAL/MMP-9 pathways
in bladder cancer. Oncotarget. 7:72758–72766. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hafsi S, Candido S, Maestro R, Falzone L,
Soua Z, Bonavida B, Spandidos DA and Libra M: Correlation between
the overexpression of Yin Yang 1 and the expression levels of
miRNAs in Burkitt's lymphoma: A computational study. Oncol Lett.
11:1021–1025. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ahmed M, Nguyen H, Lai T and Kim DR:
miRCancerdb: A database for correlation analysis between microRNA
and gene expression in cancer. BMC Res Notes. 11:1032018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tokar T, Pastrello C, Rossos AEM, Abovsky
M, Hauschild AC, Tsay M, Lu R and Jurisica I: mirDIP
4.1-integrative database of human microRNA target predictions.
Nucleic Acids Res. 46:D360–D370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vlachos IS, Zagganas K, Paraskevopoulou
MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T and
Hatzigeorgiou AG: DIANA-miRPath v3.0: Deciphering microRNA function
with experimental support. Nucleic Acids Res. 43:W460–6. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Leonardi GC, Falzone L, Salemi R, Zanghì
A, Spandidos DA, Mccubrey JA, Candido S and Libra M: Cutaneous
melanoma: From pathogenesis to therapy (Review). Int J Oncol.
52:1071–1080. 2018.PubMed/NCBI
|
|
26
|
Falzone L, Salomone S and Libra M:
Evolution of cancer pharmacological treatments at the turn of the
third millennium. Front Pharmacol. 9:13002018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tandler N, Mosch B and Pietzsch J: Protein
and non-protein biomarkers in melanoma: A critical update. Amino
Acids. 43:2203–2230. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Salemi R, Falzone L, Madonna G, Polesel J,
Cinà D, Mallardo D, Ascierto PA, Libra M and Candido S: MMP-9 as a
Candidate Marker of Response to BRAF Inhibitors in Melanoma
Patients With BRAFV600E Mutation Detected in Circulating-Free DNA.
Front Pharmacol. 9:8562018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Field MG, Decatur CL, Kurtenbach S, Gezgin
G, van der Velden PA, Jager MJ, Kozak KN and Harbour JW: PRAME as
an independent biomarker for metastasis in uveal melanoma. Clin
Cancer Res. 22:1234–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Falzone L, Salemi R, Travali S, Scalisi A,
McCubrey JA, Candido S and Libra M: MMP-9 overexpression is
associated with intragenic hypermethylation of MMP9 gene in
melanoma. Aging (Albany NY). 8:933–944. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rezayi M, Farjami Z, Hosseini ZS, Ebrahimi
N and Abouzari-Lotf E: MicroRNA-based biosensors for early
detection of cancers. Curr Pharm. Jan 11–2019.(Epub ahead of
print). doi: 10.2174/1381612825666190111144525. View Article : Google Scholar
|
|
32
|
Battaglia R, Palini S, Vento ME, La
Ferlita A, Lo Faro MJ, Caroppo E, Borzì P, Falzone L, Barbagallo D,
Ragusa M, et al: Identification of extracellular vesicles and
characterization of miRNA expression profiles in human blastocoel
fluid. Sci Rep. 9:842019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Armand-Labit V and Pradines A: Circulating
cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts.
8:61–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hasin Y, Seldin M and Lusis A: Multi-omics
approaches to disease. Genome Biol. 18:832017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xin X, Zhang Y, Ling F, Wang L, Sheng X,
Qin L and Zhao X: Identification of a nine-miRNA signature for the
prognosis of Uveal Melanoma. Exp Eye Res. 180:242–249. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wan Q, Tang J, Han Y and Wang D:
Co-expression modules construction by WGCNA and identify potential
prognostic markers of uveal melanoma. Exp Eye Res. 166:13–20. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stark MS, Bonazzi VF, Boyle GM, Palmer JM,
Symmons J, Lanagan CM, Schmidt CW, Herington AC, Ballotti R,
Pollock PM, et al: miR-514a regulates the tumour suppressor NF1 and
modulates BRAFi sensitivity in melanoma. Oncotarget. 6:17753–17763.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Streicher KL, Zhu W, Lehmann KP,
Georgantas RW, Morehouse CA, Brohawn P, Carrasco RA, Xiao Z, Tice
DA, Higgs BW, et al: A novel oncogenic role for the miRNA-506-514
cluster in initiating melanocyte transformation and promoting
melanoma growth. Oncogene. 31:1558–1570. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hanniford D, Zhong J, Koetz L,
Gaziel-Sovran A, Lackaye DJ, Shang S, Pavlick A, Shapiro R, Berman
R, Darvishian F, et al: A miRNA-Based Signature Detected in Primary
Melanoma Tissue Predicts Development of Brain Metastasis. Clin
Cancer Res. 21:4903–4912. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nguyen T, Kuo C, Nicholl MB, Sim MS,
Turner RR, Morton DL and Hoon DS: Downregulation of microRNA-29c is
associated with hypermethylation of tumor-related genes and disease
outcome in cutaneous melanoma. Epigenetics. 6:388–394. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang K and Guo L: MiR-767 promoted cell
proliferation in human melanoma by suppressing CYLD expression.
Gene. 641:272–278. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hwang HW, Baxter LL, Loftus SK, Cronin JC,
Trivedi NS, Borate B and Pavan WJ: Distinct microRNA expression
signatures are associated with melanoma subtypes and are regulated
by HIF1A. Pigment Cell Melanoma Res. 27:777–787. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Luke JJ, Triozzi PL, McKenna KC, Van Meir
EG, Gershenwald JE, Bastian BC, Gutkind JS, Bowcock AM, Streicher
HZ, Patel PM, et al: Biology of advanced uveal melanoma and next
steps for clinical therapeutics. Pigment Cell Melanoma Res.
28:135–147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Y, Yang Y, Chen L and Zhang J:
Expression analysis of genes and pathways associated with liver
metastases of the uveal melanoma. BMC Med Genet. 15:292014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Brantley MA Jr and Harbour JW:
Deregulation of the Rb and p53 pathways in uveal melanoma. Am J
Pathol. 157:1795–1801. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Polo A, Crispo A, Cerino P, Falzone L,
Candido S, Giudice A, De Petro G, Ciliberto G, Montella M, Budillon
A, et al: Environment and bladder cancer: Molecular analysis by
interaction networks. Oncotarget. 8:65240–65252. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Guarneri C, Bevelacqua V, Polesel J,
Falzone L, Cannavò PS, Spandidos DA, Malaponte G and Libra M: NF κB
inhibition is associated with OPN/MMP 9 downregulation in cutaneous
melanoma. Oncol Rep. 37:737–746. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
McCubrey JA, Fitzgerald TL, Yang LV,
Lertpiriyapong K, Steelman LS, Abrams SL, Montalto G, Cervello M,
Neri LM, Cocco L, et al: Roles of GSK-3 and microRNAs on epithelial
mesenchymal transition and cancer stem cells. Oncotarget.
8:14221–14250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Helgadottir H and Höiom V: The genetics of
uveal melanoma: Current insights. Appl Clin Genet. 9:147–155. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yoshida M, Selvan S, McCue PA, DeAngelis
T, Baserga R, Fujii A, Rui H, Mastrangelo MJ and Sato T: Expression
of insulin-like growth factor-1 receptor in metastatic uveal
melanoma and implications for potential autocrine and paracrine
tumor cell growth. Pigment Cell Melanoma Res. 27:297–308. 2014.
View Article : Google Scholar : PubMed/NCBI
|