Introduction
Melanin is an important factor in determining the
color of the human skin, hair and eyes (1,2). It
is produced in the melanosome through a complex process known as
melanogenesis (3–5). In addition, melanin serves an
important role in photoprotection from ultraviolet (UV) radiation
and external stress (1,3). Growth factors, cytokines, hormones
and other receptor ligands exert their function by interacting with
their receptors on the cell surface, generating a signaling cascade
and leading to distinct patterns of protein phosphorylation.
Melanocytes express several distinct receptor tyrosine kinases
(RTKs) that bind bone morphogenic protein (BMP), hepatocyte growth
factor (HGF) and c-Kit ligand. For example, BMP-2 stimulates
tyrosinase gene expression and melanogenesis in differentiated
melanocytes, and BMP signaling controls hair pigmentation via
cross-talk with the melanocortin receptor-1 pathway (6). The activation of Met in response to
HGF acts as a mitogen for melanocytes and synergistically
contributes to malignant progression with the aberrant expression
of basic fibroblast growth factor in malignant melanocytes
(7). Normal human melanocytes and
melanoma cells express the c-Kit gene and stem cell factor (SCF), a
ligand of the c-Kit receptor that upregulates the expression of
melanogenic proteins (8). In
addition, SCF/c-Kit signaling is required for cyclic regeneration
of the hair pigmentation unit (9).
Phosphorylation of these RTKs subsequently activates a series of
kinases known as mitogen-activated protein kinases (MAPKs) or other
intracellular signaling molecules such as cyclic adenosine
monophosphate (10). Then,
following the phosphorylation of proteins such as
microphthalmia-associated transcription factor (MITF), the
transcription of genes that participate in melanocyte proliferation
and melanogenesis is activated (11).
The Src kinase family (SKF) is a family of
non-receptor tyrosine kinases that is composed of nine members
including Src, Yes and Fyn. SKF interacts with many cellular
cytosolic, nuclear and membrane proteins, modifying these proteins
by phosphorylating tyrosine residues and contributing to the
progression of cellular transformation and oncogenic activity
(12). Of these, C-terminal Src
kinase (c-Src) is encoded by the SRC gene in humans; it
phosphorylates specific tyrosine residues in other proteins. c-Src
can be activated by many transmembrane proteins including RTKs,
such as platelet-derived growth factor receptor, epidermal growth
factor receptor, and c-Kit. Therefore, c-Src is closely associated
with the RTK pathways (13). As
RTKs serve a critical role in the development and progression of
many types of cancer (12), an
elevated activity level of c-Src tyrosine kinase is associated with
the progression of different types of cancers, such as pancreatic
and breast cancers (14).
Therefore, diverse Src inhibitors have been developed to prevent
cancer progression, and drugs against RTKs are widely used in
cancer therapy. However, there is little to no evidence on the
effects of SKF or its inhibitors on melanocytes. Therefore, the
present study investigated the effect of a c-Src inhibitor on
melanocytes and its associated signaling pathways.
Materials and methods
Cell cultures and chemical
treatment
Human G361 melanoma cells (cat. no. ATCC®
CRL-1424™; American Type Culture Collection, Manassas, VA, USA)
were cultured in low glucose Dulbecco's modified Eagle's medium
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and 1% penicillin/streptomycin (Gibco;
Thermo Fisher Scientific, Inc.). G361 cells were maintained at 37°C
in a humidified 5% CO2 incubator.
α-Melanocyte-stimulating hormone (α-MSH; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany), SU6656 (cat. no. S7774; Selleck Chemicals,
Houston, TX, USA), PP2 (cat. no. S7008; Selleck Chemicals), H-89
[protein kinase A (PKA) inhibitor; cat. no. B1427; Sigma-Aldrich;
Merck KGaA], SB203580 (p38 inhibitor; cat. no. S8307;
Sigma-Aldrich; Merck KGaA) dasatinib (cat. no. CDS023389;
Sigma-Aldrich; Merck KGaA) and nilotinib (cat. no. CDS023093;
Sigma-Aldrich; Merck KGaA) were utilized in the present study. For
these chemical treatments in all of the experiments,
1×105 cells were seeded in a 6-well plate for 2 days at
37°C with normal medium including 10% FBS and 1%
penicillin/streptomycin and were changed with normal media, which
included the aforementioned chemicals. All cells, except those used
for western blotting and siRNA transfection experiments, were
incubated for 9 days following chemical treatments at 37°C. For
western blotting, α-MSH treated cells were harvested at 0, 5, 15
and 30 min. SU6656 or PP2 treated cells were harvested at 0, 5, 15,
30, 60 or 120 min. For melanin content measurement, cells were
treated with α-MSH (1 µM), dasatinib and nilotinib (both at 0.1 M)
for 0, 3, 6, and 9 days, and then cells were harvested. Cells were
treated with SU6656 and PP2 at different doses (1, 10, 100 and 1000
nM) and harvested 9 days later.
Drug solution preparation
All chemical concentrations employed in the present
study were presented as stock/working concentrations as follows;
α-MSH was used at 200 µM/1 µM, and SU6656 and PP2 were used as 0.2
µM/1 nM, 2 µM/10 nM, 20 µM/100 nM and 200 µM/1,000 nM. The p38
inhibitor SB203580 was used at 400 µM/1 µM, and the final
concentration of the p38 inhibitor was 8 mM/20 µM. The PKA
inhibitor H-89 was used 1 mM/1 µM. Dasatinib and nilotinib were
used at 10 M/0.1 M.
UV irradiation
A total of 1×105 cells were seeded in
plates and incubated at 37°C for 48 h. They were washed with
phosphate-buffered saline (PBS) and covered with a thin layer of
PBS prior to UV exposure. The culture plate lid was removed and
cells were irradiated (UVB: 5 mJ/cm2) in a dark box. The
UVB irradiation apparatus (BLE-1T158) was obtained from Spectronics
Corporation (Westbury, NY, USA). The incident dose of UVB was
measured using a Waldmann UV meter (model no. 585100; Herbert
Waldmann GmbH & Co. KG, Villingen-Schwenningen, Germany).
Following UV irradiation, PBS was replaced with culture medium at
37°C. The irradiated cells were harvested at 0, 1, 5 and 10 min
following UV irradiation for western blotting. For melanin content
measurements, the irradiated cells were also harvested at 0, 3, 6
and 9 days.
siRNA transfection
G361 cells were seeded in 60-mm dishes at
1×105 cells, and Src siRNA or negative control siRNA
were transfected using Lipofectamine™ RNAiMAX (Invitrogen; Thermo
Fisher Scientific, Inc.). The sequences of Src siRNA (Bioneer
Corporation, Daejeon, Korea; cat #100545; stock concentration 100
nM/working concentration 10 or 20 nM) was as follows: Sense,
GUGUCUUAAUACUGUCCUU(dTdT) and antisense, AAGGACAGUAUUAAGACAC(dTdT).
The sequence of the negative control siRNA is commercially
unavailable (cat. no. SN-1002; Bioneer Corporation). G361 cells
were harvested following 6 days and mRNA expression levels were
analyzed via reverse transcription-quantitative polymerase chain
reaction (RT-qPCR). Transfection efficiency of siRNA was evaluated
by qPCR.
Observation of cell pellets and
measuring the melanin content
To measure the melanin content, cells were seeded in
60-mm dishes at 1×105 cells/dish, incubated at 37°C for
2 days and treated at 37°C with SU6656, PP2, α-MSH, p38 inhibitor,
PKA inhibitor, Src siRNA (cat. no. 100545), dasatinib and nilotinib
for 9 days. SU6656 and PP2 were used at 0, 1, 10, 100 and 1,000 nM,
and α-MSH was used at 1 µM. The p38 inhibitor (SB203580) was used
at 20 µM, PKA inhibitor (H-89) was used at 1 µM and Src siRNA was
used at 20 nM. Dasatinib and nilotinib were used at 0.1 M (data not
shown). Cells were trypsinized using 0.25% trypsin, harvested by
centrifugation at 13,000 × g for 1.5 min at 4°C, photographed and
solubilized in boiled 1N NaOH (80°C) for 2 h; the absorbance was
then measured at 405 nm, as described previously (15). Measurement of melanin contents
following UVB irradiation was also conducted in the same manner.
The amount of melanin measured in all experiments was normalized to
the relative value of the control group.
RNA extraction, RT-qPCR and
semi-quantitative (sq)-PCR analysis
Total RNA was extracted from G361 cells using
Favor-Prep™ Blood/Cultured cell total RNA purification mini kit
(Favorgen Biotech Corp., Ping-Tung, Taiwan) and subjected to cDNA
synthesis using oligodT and the HelixCript™ Thermo Reverse
Transcription System (NanoHelix Co., Ltd., Daejeon, Korea)
according to the manufacturer's instructions. qPCR amplification of
cDNA was performed in a total volume of 30 µl under the following
thermocycling conditions: Initial denaturation at 95°C for 5 min,
followed by 27 cycles of 95°C for 30 sec, 54°C for 20 sec and 72°C
for 30 sec, and a final extension at 72°C for 10 min. For the qPCR
reaction, BrightGreen qPCR master mix-ROX (Abcam, Cambridge, MA,
USA) was used and reaction was carried out using Applied biosystems
qPCR Machine; quantification was conducted using the
2−∆∆Cq method (16).
The GAPDH mRNA expression level was used for sample
standardization. For the quantification of sqPCR data, the
amplification conditions of all genes were 95°C for 5 min, followed
by 40 cycles of 95°C for 1 min, 55°C for 1 min and 72°C for 1 min,
and then a final extension at 72°C for 10 min. Then the samples
were loaded onto 1.5% agarose gel containing GelRed and
electrophoresed. The bands were visualized using a UV illuminator.
The band intensity of the product was calculated using ImageJ
(version 1.45; National Institutes of Health, Bethesda, MD, USA)
(17). For RT-PCR the primers used
were as follows: MITF forward, 5′-TGCCCAGGCATGAACACAC-3′, and
reverse, 5′-TGGGAAAAATACACGCTGTGAG-3′; tyrosinase-related protein 1
(TRP1) forward, 5′-TCTCTGGGCTGTATCTTCTTCC-3′ and reverse,
5′-GTCTGGGCAACACATACCACT-3′; TRP2 forward,
5′-CTTGGGCTGCAAAATCCTGC-3′ and reverse 5′-CAGCACTCCTTGTTCACTAGG-3′;
tyrosinase forward, 5′-TGCACAGAGAGACGACTCTTG-3′ and reverse
5′-GAGCTGATGGTATGCTTTGCTAA-3′; GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′.
Western blotting
For western blotting, 1×105 cells were
treated with SU6656 (1 µM), PP2 (1 µM) or α-MSH (1 µM) for 9 days
and then lysed with protein extraction solution (PRO-PREP™; Intron
Biotechnology, Inc., Seongnam, Korea). Protein quantification was
performed using a Bicinchoninic Acid (BCA) assay kit (Pierce™ BCA
Protein Assay kit; Thermo Fisher Scientific, Inc.). A total of 70
µg of protein was loaded in a 12% acrylamide gel and run for 1.5 h;
all protein was then transferred to a PVDF membrane (Immobilon-P;
Merck KGaA). The transferred membrane was blocked with 5% bovine
serum albumin (BSA; Bovogen; Bovogen Biologicals Pty Ltd., Keilor
East VIC, Australia) blocking buffer for 1 h at room temperature,
incubated with primary antibodies (1:1,000) overnight at 4°C,
washed with PBS several times and then incubated with secondary
antibodies (1:2,000) at room temperature for 1 h. The primary
antibodies used were as follows: Rabbit phosphorylated
(phospho)-Src (cat. no. #6943; Cell Signaling Technology, Inc.,
Danvers, MA, USA), rabbit Src (cat. no. 2123; Cell Signaling
Technology, Inc.), mouse phospho-p38 (cat. no. 9216; Cell Signaling
Technology, Inc.), rabbit p38 (cat. no. 9212; Cell Signaling
Technology, Inc.), rabbit phospho-cyclic adenosine monophosphate
response element binding (CREB; cat. no. 9198; Cell Signaling
Technology, Inc.), rabbit CREB (cat. no. 9197; Cell Signaling
Technology, Inc.), mouse β-actin (cat. no. sc-1615; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and mouse α-tubulin (cat. no.
sc-32293; Santa Cruz Biotechnology, Inc.). The secondary antibodies
used were as follows: Peroxidase labeled anti-mouse immunoglobulin
(Ig)-G (cat. no. PI-2000; Vector Laboratories, Inc.; Maravai
LifeSciences, San Diego, CA, USA) and peroxidase labeled
anti-rabbit IgG (cat. no. PI-1000; Vector Laboratories, Inc.;
Maravai LifeSciences). Following the addition of Enhanced
Chemiluminescence solution (Immobilon Western; Merck KGaA), western
blotting images were obtained using ImageQuant LAS 4000 (GE
Healthcare Bio-Sciences, Pittsburgh, PA, USA). Band densities were
quantified using ImageJ software (version 1.45; National Institutes
of Health, Bethesda, MD, USA). The quantification of the
phosphorylated protein was calculated as follows:
(p-protein/internal control)/(total protein/internal control).
Immunofluorescence
For immunofluorescence staining, 1×105
cells were fixed with 4% paraformaldehyde for 30 min at room
temperature, rinsed in PBS, blocked in 5% BSA-containing TBS-Tx
(supplemented with 0.2% Triton-X-100) for 1.5 h at room temperature
and incubated with the primary antibodies overnight at 4°C. The
primary antibodies used were anti-mouse phospho (p)-p38 (1:200;
cat. no. 9216; Cell Signaling Technology, Inc.) and anti-rabbit
p-CREB (1:200; cat. no. 9198; Cell Signaling Technology, Inc.).
Then, cells were incubated with the secondary antibodies, Alexa
Fluor 488 goat anti-mouse IgG (cat. no. A11001; Invitrogen; Thermo
Fisher Scientific, Inc.) and Alexa Fluor 488 goat anti-rabbit IgG
(cat. no. A11008; Invitrogen; Thermo Fisher Scientific, Inc.) with
4,6-diamidino-2-phenylindole (Sigma-Aldrich; Merck KGaA) for 1 h at
room temperature. Immunofluorescence staining was imaged using a
ZEISS LSM700 confocal microscope (magnification, ×40).
Statistical analysis
Data are presented as the mean ± standard deviation
of three independent experiments. Student's t-test was used between
two groups and one-way analysis of variance with Tukey's post hoc
test were used for comparing multiple groups. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were conducted using GraphPad Prism 5.01
(GraphPad Software, Inc., La Jolla, CA, USA).
Results
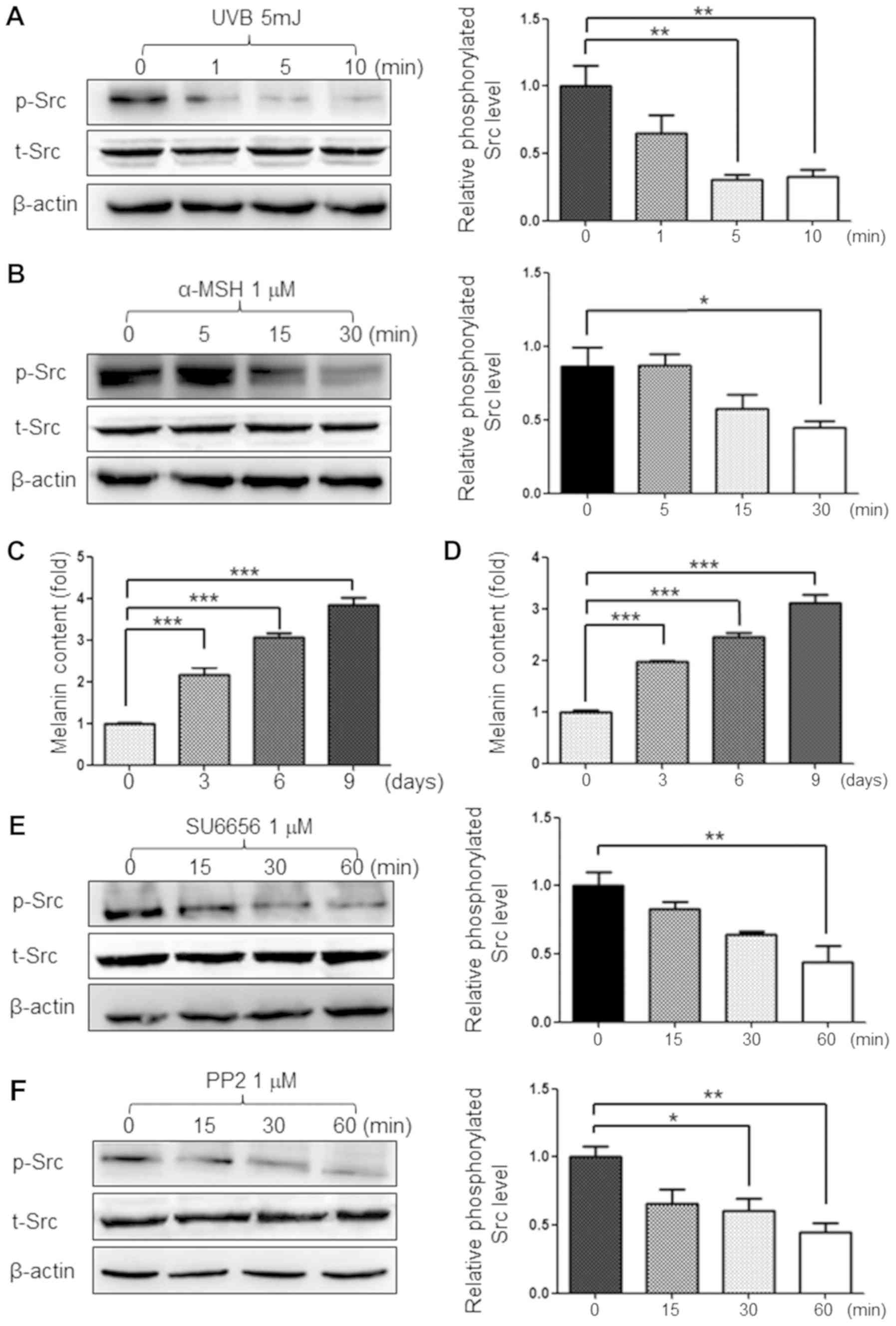
UVB and α-MSH decrease the
phosphorylation of Src protein in G361 cells
UVB radiation is a physical stimulus that increases
the amount of melanin produced in human melanoma cells. α-MSH is
also known to increase melanin production in human melanoma cells
(18). The present study treated
G361 cells with 5 mJ of UVB and 1 µM of α-MSH, which are both known
stimulants of melanogenesis. As a result, the phosphorylation of
Src protein was decreased by melanin-stimulation (Fig. 1A and B). It was also confirmed that
these melanin stimulants increased melanin production in a
time-dependent manner (Fig. 1C and
D) Therefore, it was hypothesized that Src inhibition may be
required for melanogenesis in G361 cells. The present study also
used 1 µM of the Src inhibitors SU6656 and PP2 to inhibit the
phosphorylation of Src protein (Fig.
1E and F).
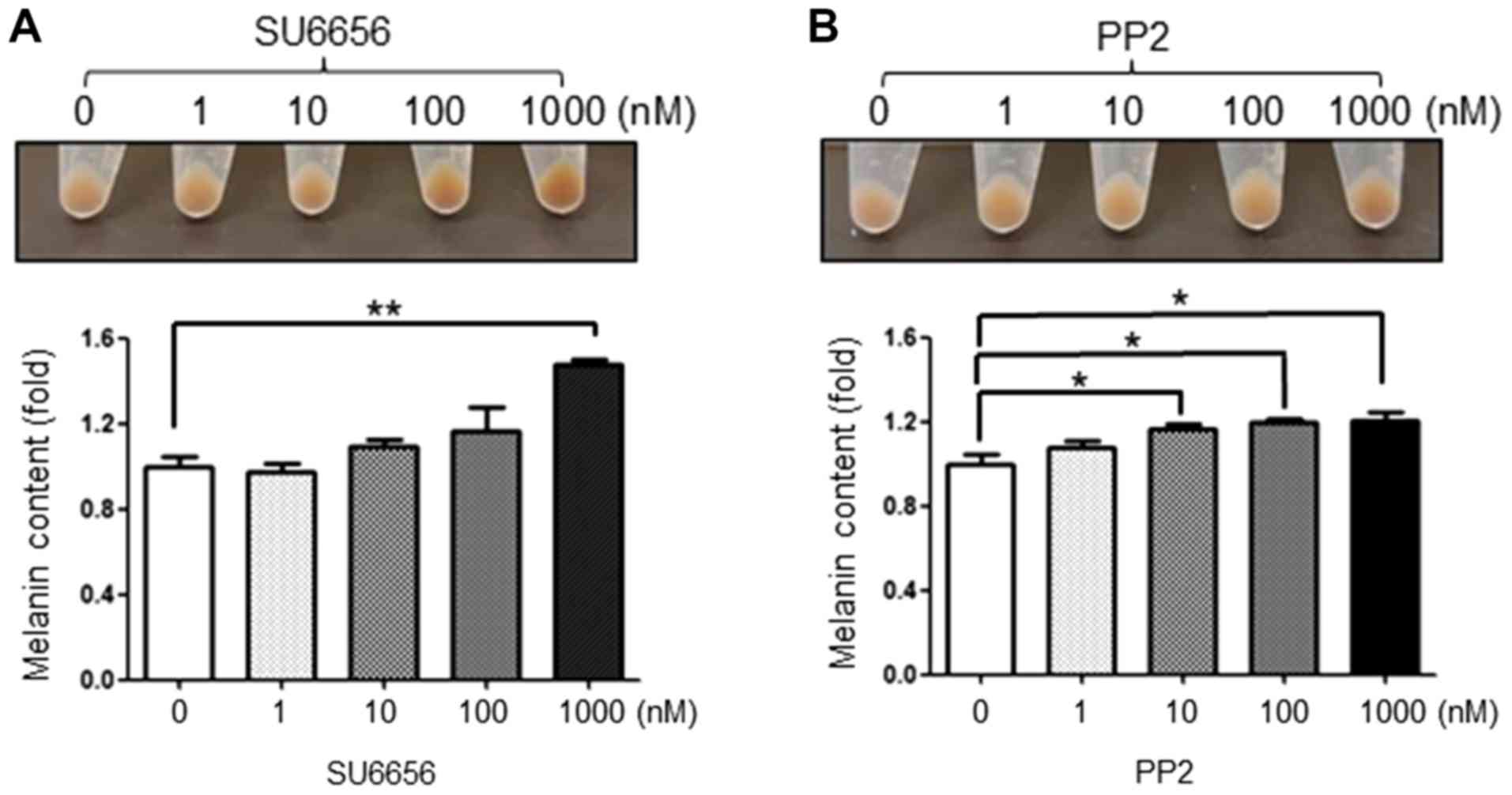
SU6656 and PP2 induce melanogenesis in
G361 cells
Up to 1,000 nM of SU6656 and PP2 were used to treat
G361 cells as these inhibitors are toxic at high concentrations.
Following SU6656 treatment, the pellet color of G361 cells became
darker in a concentration-dependent manner (Fig. 2A). In addition, PP2 also increased
the melanin content in G361 cells in a dose-dependent manner
(Fig. 2B).
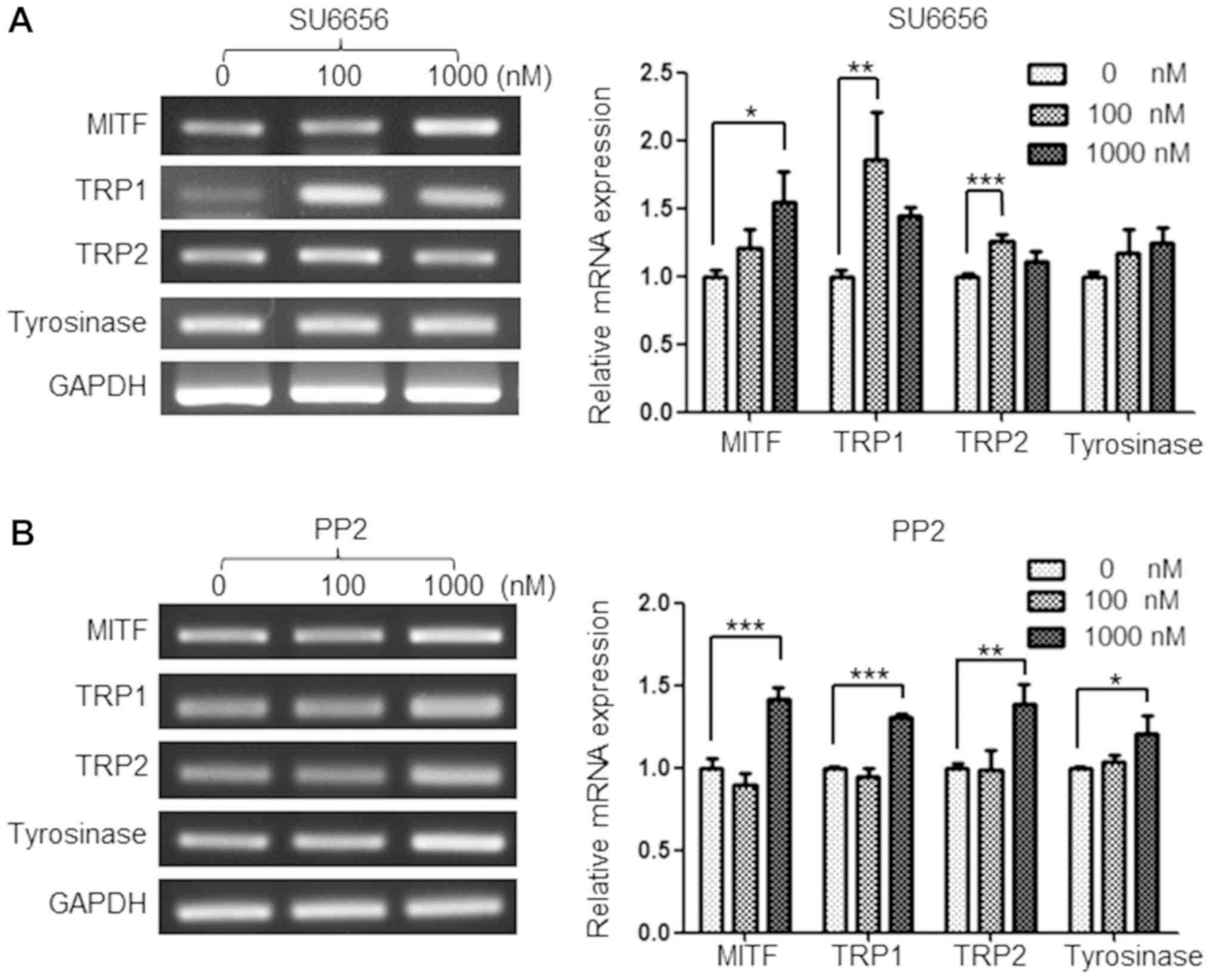
SU6656 and PP2 regulate the mRNA
expression levels of melanogenesis-associated genes
As MITF, TRP1, TRP2 and tyrosinase are key factors
in mediating melanogenesis, the present study investigated whether
Src inhibitors upregulated the mRNA expression of these genes. As
expected, treatment with 100 and 1,000 nM of SU6656 for 9 days
upregulated the mRNA expression of melanogenesis-associated genes
in G361 cells. In particular, the expression of MITF and TRP1 were
upregulated by up to 55 and 87%, respectively (Fig. 3A). In addition, PP2 upregulated the
mRNA expression of MITF, TRP1, TRP2 and tyrosinase at 1,000 nM
(Fig. 3B).
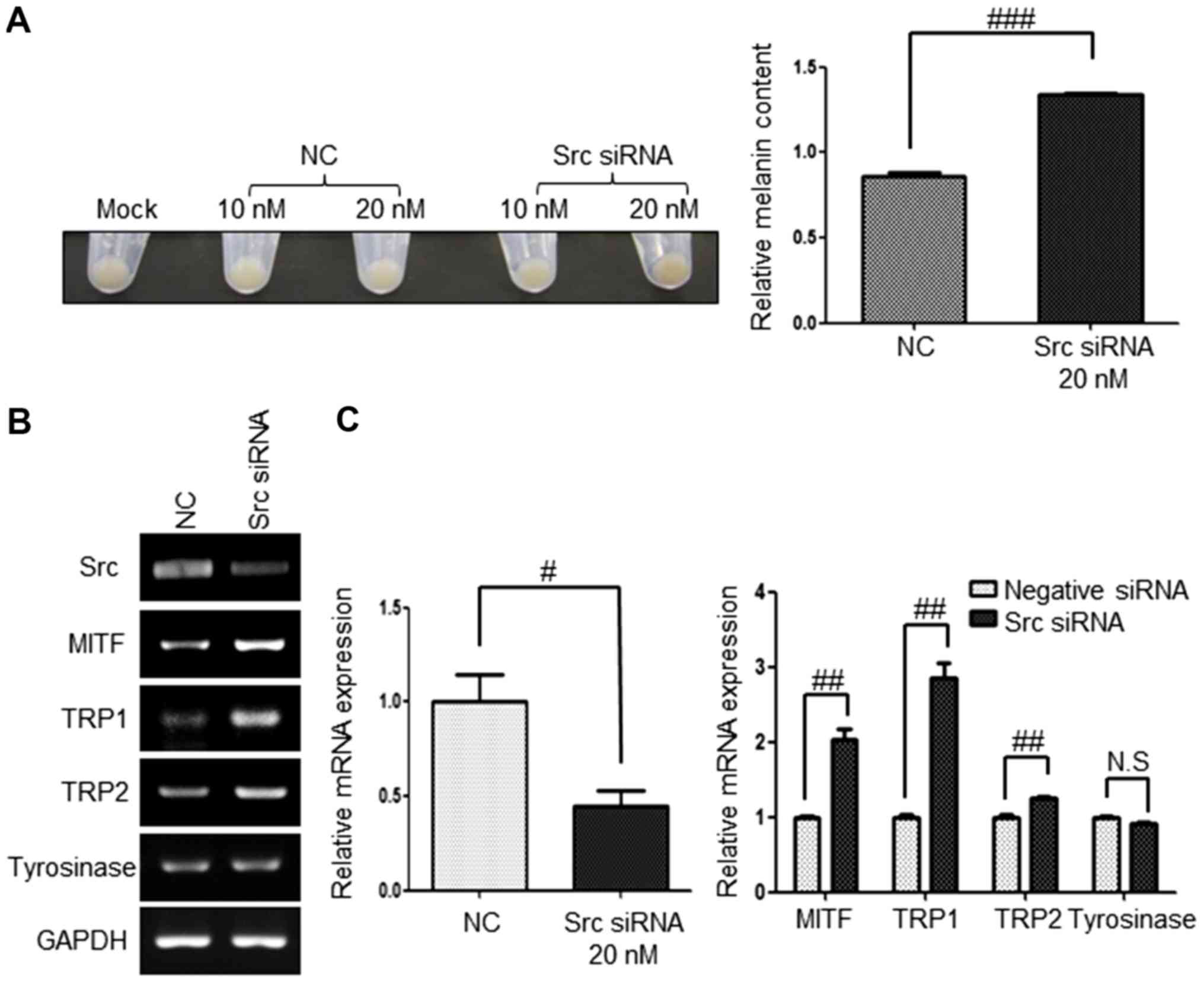
Src inhibition by siRNA upregulates
the expression of melanogenesis-associated genes
To further determine whether melanogenesis is
specifically due to Src inhibition in G361 cells, the present study
examined the pellet color, melanin contents and expression of
melanogenesis-association genes following the inhibition of Src
using Src siRNA. G361 cells were harvested 6 days post-transfection
with 20 nM of Src siRNA. Src inhibition using Src siRNA produced a
darker pellet color than the negative control (Fig. 4A) and the Src mRNA level was
downregulated effectively with ~60% efficiency (Fig. 4B and C). The mRNA expression levels
of the melanogenesis-associated genes MITF, TRP1, TRP2 and
tyrosinase were examined by RT-qPCR and sqPCR. The mRNA expression
of MITF and TRP1 in Src knockdown cells was significantly
upregulated by 2- and 2.8-fold, respectively. In addition, the
expression of TRP2 in Src knockdown cells was upregulated when
compared with that of the control (Fig. 4B and C). However, the mRNA
expression level of tyrosinase was not altered by Src siRNA.
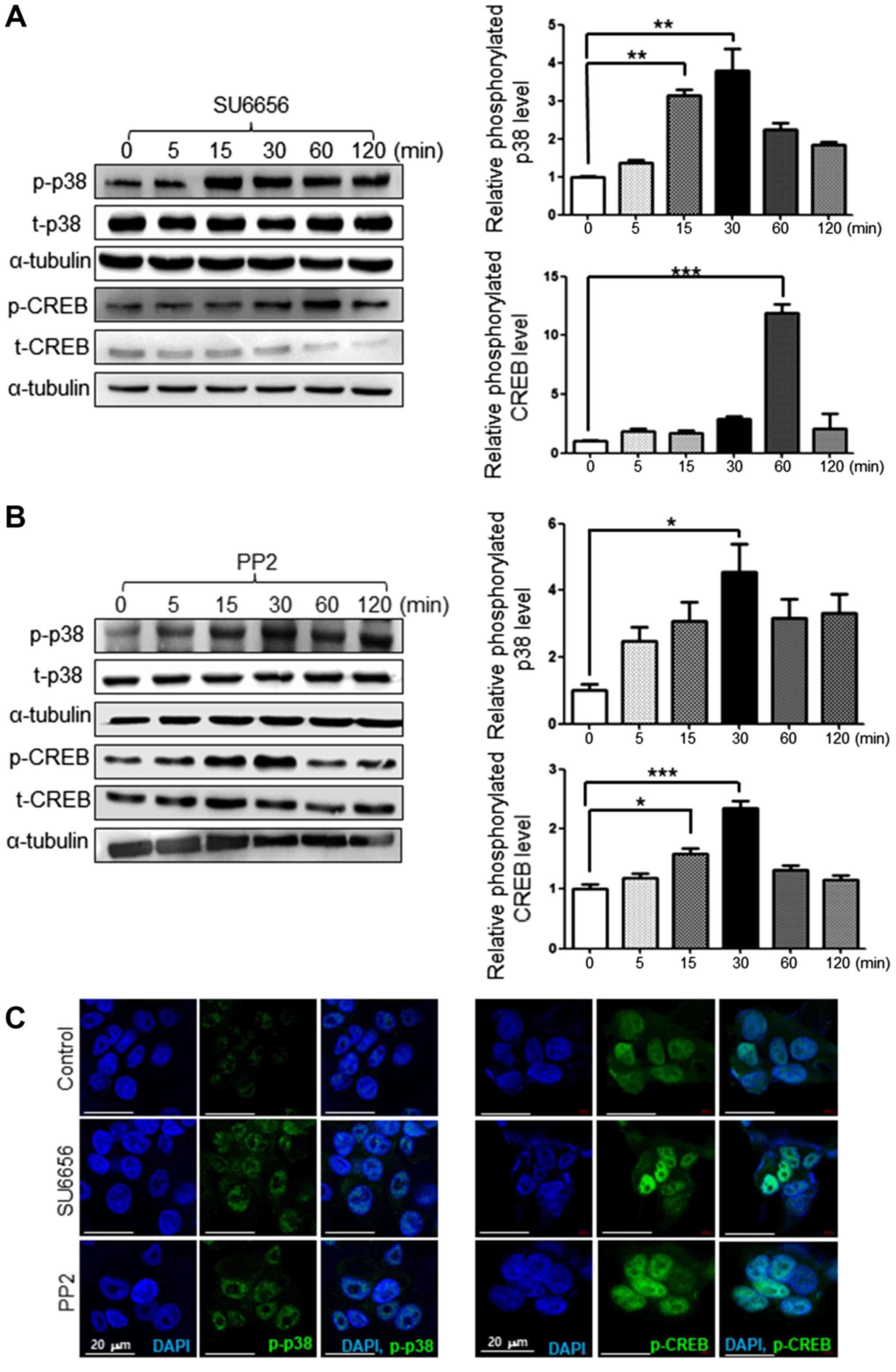
SU6656 and PP2 induce p38 and CREB
activation in G361 cells
The phosphorylation of p38 and CREB reportedly
serves a key role in melanogenesis (19). Therefore, the present study
investigated whether Src inhibitors affect the phosphorylation of
p38 and CREB by time course via western blot analysis. When G361
cells were treated with 1 µM of SU6656, the phosphorylation of p38
and CREB was increased over time as determined by western blot
analysis (Fig. 5A). Similarly, 1
µM of PP2 increased the phosphorylation of p38 and CREB over time
as determined by western blot analysis (Fig. 5B). Src inhibition by SU6656 or PP2
also increased the protein levels of phosphorylated p38 and CREB,
as presented by immunofluorescence staining (Fig. 5C).
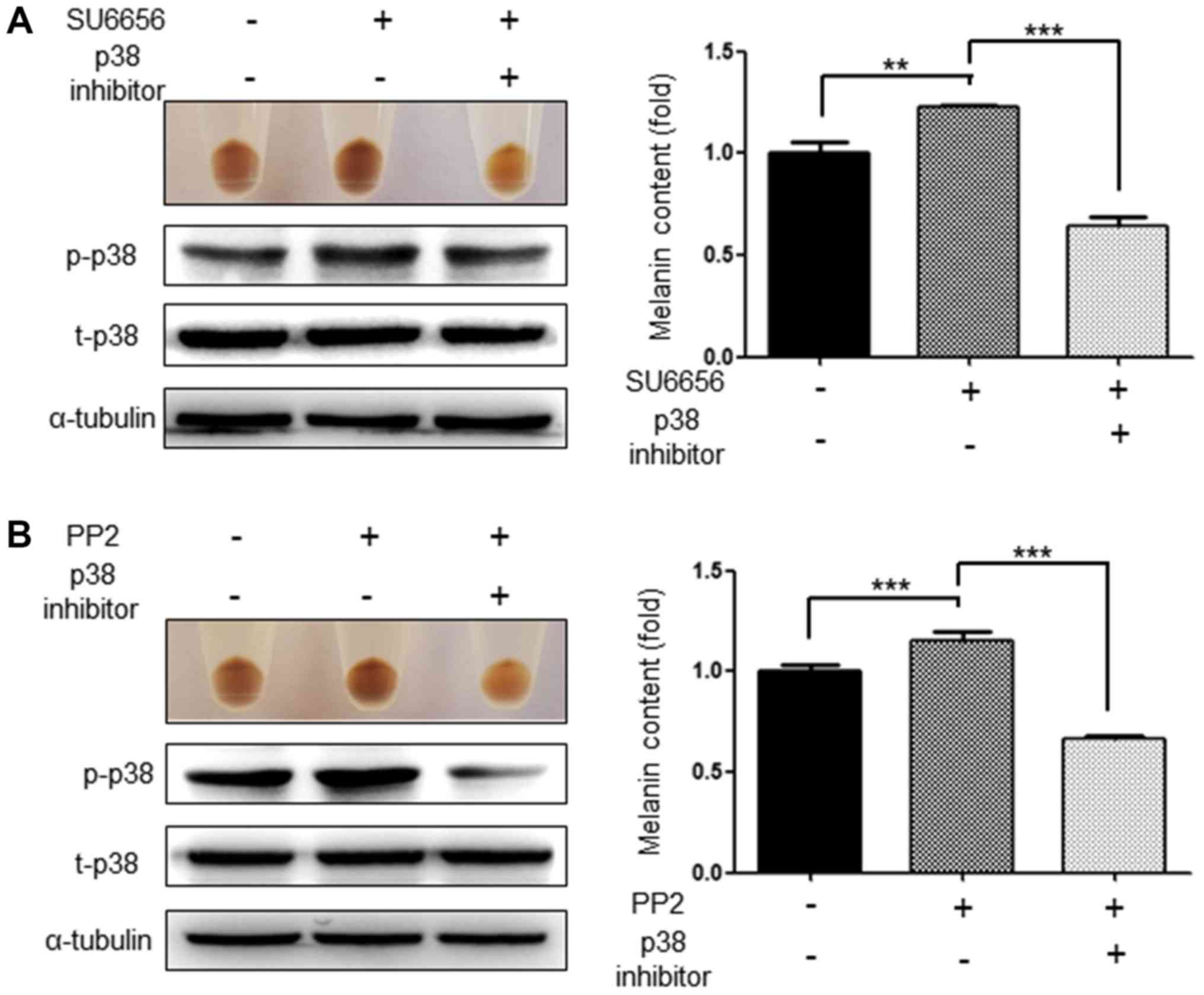
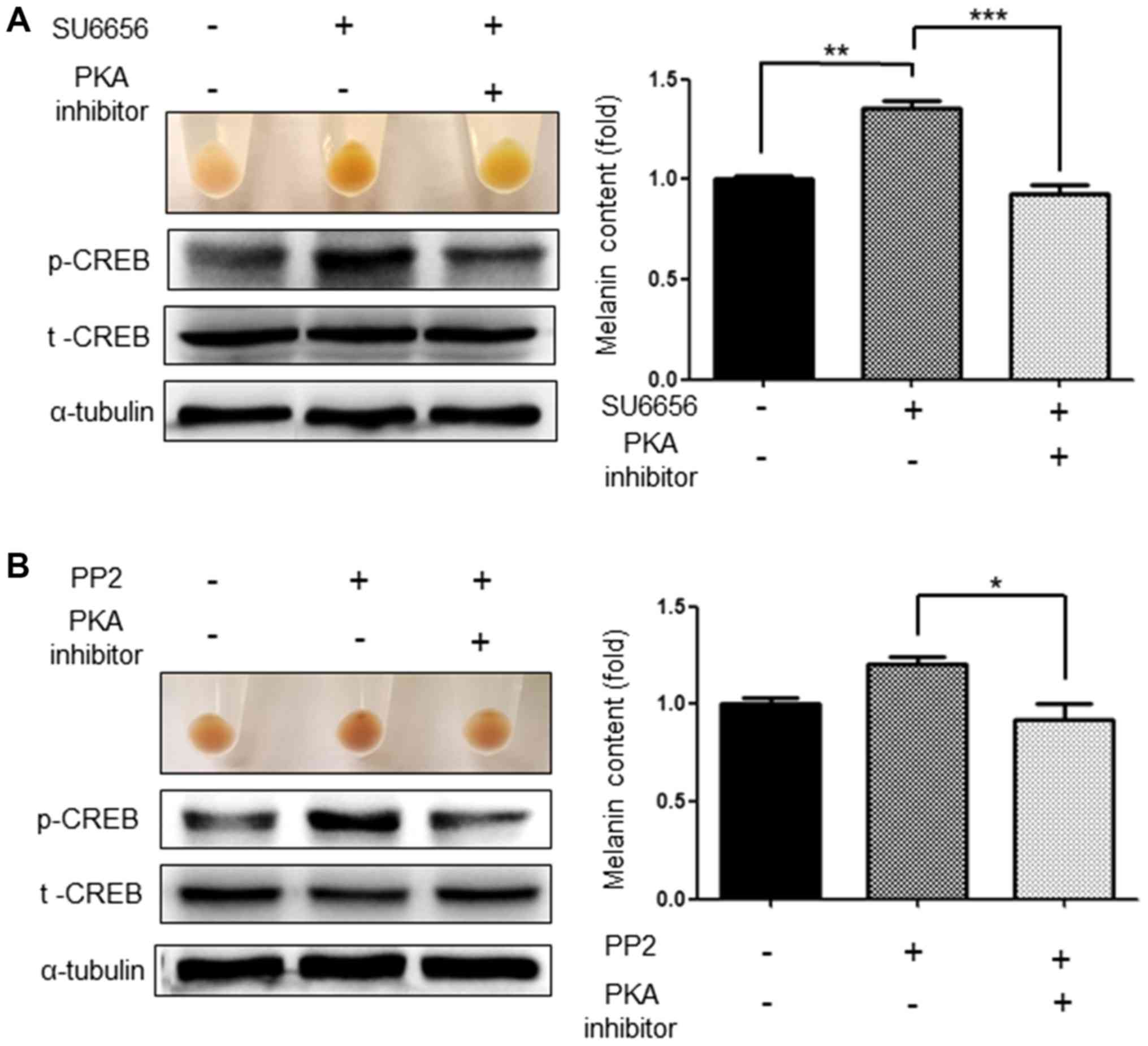
Inhibition of p38 and CREB abolishes
the increase in melanogenesis induced by SU6656 and PP2
As Src inhibitors activate the p38 MAPK signaling
pathways and the phosphorylation of CREB (19), the present study investigated
whether inhibition of these signaling pathways attenuated the
increased melanogenesis induced by SU6656 and PP2. As expected, the
p38 inhibitor, SB203580 (20 µM), significantly attenuated the
increased melanogenesis induced by the Src inhibitors SU6656 and
PP2. At the same time, it was confirmed that the activation of p38
was also markedly reduced (Fig. 6A and
B). Similarly, H-89 (1 µM), a PKA pathway inhibitor,
significantly attenuated the increased melanogenesis induced by
SU6656 and PP2. In addition, activation of CREB, which was
increased by Src inhibitors, was also markedly decreased (Fig. 7A and B).
Discussion
c-Kit is known to be expressed in melanocytes and is
associated with melanocyte proliferation, melanocyte migration and
melanogenesis in response to SCF (20,21).
c-kit is an RTK, as well as an epidermal growth factor receptor, a
fibroblast growth factor receptor and a vascular endothelial growth
factor receptor. It not only serves a role in cell survival,
proliferation and differentiation but also is closely associated
with several types of cancer, such as gastrointestinal stromal
tumors, testicular seminoma, melanoma and acute myeloid leukemia
(20,22). Therefore, RTK inhibitors, also
known as c-Kit inhibitors, including imatinib, sorafenib, sunitinib
and dasatinib, are now being used as anticancer drugs, even though
they are not specific for c-Kit only (20,22,23).
According to reports on the side effects of RTK inhibitors with
regard to pigmentation, there have been conflicting reports of the
pigmentary changes with unknown pathogenesis. Some cases have
suggested that hypopigmentation occurs as an adverse effect of
patients taking c-Kit inhibitors including imatinib (23–26).
By contrast, there have also been reports that have described
hyperpigmentation caused by chemotherapy with c-Kit inhibitors
(27–29). Therefore, to determine the effect
of c-Kit inhibitors on melanogenesis in G361 cells, c-Kit
inhibitors such as dasatinib and nilotinib (both at 0.1 M) were
evaluated in the present study. The results revealed that dasatinib
and nilotinib (both at 0.1 M) increased melanin production (data
not shown). In addition, dasatinib upregulated the mRNA expression
levels of melanogenesis-associated molecules such as MITF, TRP1,
TRP2 and tyrosinase in G361 cells (data not shown). However,
dasatinib is not as potent as the Src inhibitors SU6656 and PP2 in
melanogenesis. Dasatinib inhibited c-Kit, Src and Abl. Therefore,
the present study further examined the effect of Src inhibitors on
melanogenesis instead of the effect of c-Kit inhibitors.
First, the results demonstrated that the stimulators
of melanogenesis, UVB and α-MSH, inhibited the phosphorylation of
Src in G361 cells. This result suggests that UV- and α-MSH-induced
pigmentation could be mediated via c-Src inhibition. Src inhibition
by the chemical inhibitors SU6656 and PP2 induced melanogenesis in
G361 cells and upregulated the mRNA expression levels of the
melanogenesis-associated molecules MITF, TRP1, TRP2 and tyrosinase.
Src inhibition by siRNA knockdown in G361 cells also induced
melanogenesis and upregulated the mRNA expression levels of
melanogenesis-associated genes. The p38 MAPK and PKA signaling
pathways, which serve a key role in melanogenesis, were examined
for Src-mediated pigmentary regulation. Src inhibition by SU6656 or
PP2 induced the phosphorylation of p38 and CREB, as determined by
western blotting, and increased the expression levels of p-p38 and
p-CREB, as revealed by immunofluorescence. In addition, the
pigmentation and melanin contents of G361 cells when treated with
Src inhibitors were significantly inhibited by the p38 and CREB
inhibitors. Collectively, these results indicate that Src
inhibition induced melanogenesis via the p38 MAPK and PKA signaling
pathways in G361 cells. These data are also supported by a previous
study that indicated that Src inhibition increased p-p38 expression
levels (30). Additionally, the
suppression of c-Src activity by PP1 and SU6656 stimulated muscle
differentiation via p38 MAPK activation (31). Dasatinib, a c-Kit inhibitor, also
exerted an antileukemic effect via the activation of the p38 MAPK
signaling pathways (32). Although
there is little to no evidence on whether Src inhibitors affect
CREB phosphorylation, it is reasonable to assume that p38
activation can activate the phosphorylation of CREB, which is known
to be a downstream signal of p38 during UV-induced
melanogenesis.
The activation of c-Src increases the proliferation,
survival and invasion of cancer cells, and Src
phosphorylation/activation is involved in 50% of colon, liver,
breast and pancreatic tumors (33). Therefore, a number of tyrosine
kinase inhibitors against c-Src, including dasatinib, have been
developed for cancer therapy (34). c-Src is a non-receptor tyrosine
kinase, but c-Src can be activated by many transmembrane proteins
including adhesion receptors, RTKs, G-protein coupled receptors and
cytokine receptors (13).
Melanocytes possess a variety of receptors, such as melanocortin 1
receptor (MC1R; a type of G-protein coupled receptor) and c-Kit (a
type of RTK), and melanin production in melanocytes is regulated
through external stimulation to the receptors (35). Therefore, the MC1R and c-Kit
signaling pathways in melanocytes may be closely associated with
c-Src signaling. Understanding the role of the c-Src pathway in
melanocytes is critical for understanding melanocyte physiology,
and melanoma development and progression. The results of the
present study will also help predict pigmentation side effects when
Src inhibitors are used for anticancer therapy and develop novel
promising hypopigmentation agents for hyperpigmentary disorders,
such as melasma and aging spots. In conclusion, Src inhibition in
melanocytes may increase melanogenesis through the p38 and CREB
signaling pathways.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
National Research Foundation (grant no. NRF2018R1A6A1A03023718)
funded by the Korean government.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KEK and JHS designed the experiments. KEK developed
the methodology and performed the experiments. KEK, NC, SHO, WSK,
WS and JHS analyzed the data. KEK and JHS wrote the paper. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lin JY and Fisher DE: Melanocyte biology
and skin pigmentation. Nature. 445:843–850. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yun WJ, Kim EY, Park JE, Jo SY, Bang SH,
Chang EJ and Chang SE: Microtubule-associated protein light chain 3
is involved in melanogenesis via regulation of MITF expression in
melanocytes. Sci Rep. 6:199142016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brenner M and Hearing VJ: The protective
role of melanin against UV damage in human skin. Photochem
Photobiol. 84:539–549. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fitzpatrick TB and Breathnach AS: The
epidermal melanin unit system. Dermatol Wochenschr. 147:481–489.
1963.(In German). PubMed/NCBI
|
|
5
|
Videira IF, Moura DF and Magina S:
Mechanisms regulating melanogenesis. An Bras Dermatol. 88:76–83.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sharov AA, Fessing M, Atoyan R, Sharova
TY, Haskell-Luevano C, Weiner L, Funa K, Brissette JL, Gilchrest BA
and Botchkarev VA: Bone morphogenetic protein (BMP) signaling
controls hair pigmentation by means of cross-talk with the
melanocortin receptor-1 pathway. Proc Natl Acad Sci USA. 102:93–98.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Halaban R, Rubin JS, Funasaka Y, Cobb M,
Boulton T, Faletto D, Rosen E, Chan A, Yoko K and White W: Met and
hepatocyte growth factor/scatter factor signal transduction in
normal melanocytes and melanoma cells. Oncogene. 7:2195–2206.
1992.PubMed/NCBI
|
|
8
|
Luo D, Chen H, Searles G and Jimbow K:
Coordinated mRNA expression of c-Kit with tyrosinase and TRP-1 in
melanin pigmentation of normal and malignant human melanocytes and
transient activation of tyrosinase by Kit/SCF-R. Melanoma Res.
5:303–309. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Botchkareva NV, Khlgatian M, Longley BJ,
Botchkarev VA and Gilchrest BA: SCF/c-kit signaling is required for
cyclic regeneration of the hair pigmentation unit. FASEB J.
15:645–658. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Katz M, Amit I and Yarden Y: Regulation of
MAPKs by growth factors and receptor tyrosine kinases. Biochim
Biophys Acta. 1773:1161–1176. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wellbrock C and Arozarena I:
Microphthalmia-associated transcription factor in melanoma
development and MAP-kinase pathway targeted therapy. Pigment Cell
Melanoma Res. 28:390–406. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zwick E, Bange J and Ullrich A: Receptor
tyrosine kinase signalling as a target for cancer intervention
strategies. Endocr Relat Cancer. 8:161–173. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hubbard SR and Miller WT: Receptor
tyrosine kinases: Mechanisms of activation and signaling. Curr Opin
Cell Biol. 19:117–123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wheeler DL, Iida M and Dunn EF: The role
of Src in solid tumors. Oncologist. 4:667–678. 2009. View Article : Google Scholar
|
|
15
|
Park J, Chung H, Bang SH, Han AR, Seo EK,
Chang SE, Kang DH and Oh ES:
(E)-4-(3,4-Dimethoxyphenyl)but-3-en-1-ol enhances melanogenesis
through increasing upstream stimulating factor-1-mediated
tyrosinase expression. PLoS One. 10:e01419882015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee KE, Lee SK, Jung SE, Lee Zh and Kim
JW: Functional splicing assay of DSPP mutations in hereditary
dentin defects. Oral Dis. 17:690–695. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hennessy A, Oh C, Diffey B, Wakamatsu K,
Ito S and Rees J: Eumelanin and pheomelanin concentrations in human
epidermis before and after UVB irradiation. Pigment Cell Res.
18:220–223. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Niwano T, Terazawa S, Nakajima H and
Imokawa G: The stem cell factor-stimulated melanogenesis in human
melanocytes can be abrogated by interrupting the phosphorylation of
MSK1: Evidence for involvement of the p38/MSK1/CREB/MITF axis. Arch
Dermatol Res. 10:187–196. 2018. View Article : Google Scholar
|
|
20
|
Tatro JB, Atkins M, Mier JW, Hardarson S,
Wolfe H, Smith T, Entwistle ML and Reichlin S: Melanotropin
receptors demonstrated in situ in human melanoma. J Clin Invest.
85:1825–1832. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thody AJ, Hunt G, Donatien PD and Todd C:
Human melanocytes express functional melanocyte-stimulating hormone
receptors. Ann NY Acad Sci. 680:381–390. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hocker TL, Singh MK and Tsao H: Melanoma
genetics and therapeutic approaches in the 21st century: Moving
from the benchside to the bedside. J Invest Dermatol.
128:2575–2595. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Buscà R, Bertolotto C, Ortonne JP and
Ballotti R: Inhibition of the phosphatidylinositol
3-kinase/p70(S6)-kinase pathway induces B16 melanoma cell
differentiation. J Biol Chem. 271:31824–31830. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheli Y, Ohanna M, Ballotti R and
Bertolotto C: Fifteen-year quest for microphthalmia-associated
transcription factor target genes. Pigment Cell Melanoma Res.
23:27–40. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kobayashi T, Urabe K, Winder A,
Jiménez-Cervantes C, Imokawa G, Brewington T, Solano F,
García-Borrón JC and Hearing VJ: Tyrosinase related protein 1
(TRP1) functions as a DHICA oxidase in melanin biosynthesis. EMBO
J. 13:5818–5825. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ohguchi K, Akao Y and Nozawa Y:
Involvement of calpain in melanogenesis of mouse B16 melanoma
cells. Mol Cell Biochem. 275:103–107. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
de Melo Maia B, Lavorato-Rocha AM,
Rodrigues IS, Baiocchi G, Cestari FM, Stiepcich MM, Chinen LT,
Carvalho KC, Soares FA and Rocha RM: Prognostic significance of
c-KIT in vulvar cancer: Bringing this molecular marker from bench
to bedside. J Transl Med. 10:1502012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Marech I, Gadaleta CD and Ranieri G:
Possible prognostic and therapeutic significance of c-Kit
expression, mast cell count and microvessel density in renal cell
carcinoma. Int J Mol Sci. 15:13060–13076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yavuz AS, Lipsky PE, Yavuz S, Metcalfe DD
and Akin C: Evidence for the involvement of a hematopoietic
progenitor cell in systemic mastocytosis from single-cell analysis
of mutations in the c-kit gene. Blood. 100:661–665. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mora Vidal R, Regufe da Mota S, Hayden A,
Markham H, Douglas J, Packham G and Crabb SJ: Epidermal growth
factor receptor family inhibition identifies p38 Mitogen-activated
protein kinase as a potential therapeutic target in bladder cancer.
Urology. 112:225.e1–225.e7. 2018. View Article : Google Scholar
|
|
31
|
Lim MJ, Seo YH, Choi KJ, Cho CH, Kim BS,
Kim YH, Lee J, Lee H, Jung CY, Ha J, et al: Suppression of c-Src
activity stimulates muscle differentiation via p38 MAPK activation.
Arch Biochem Biophys. 465:197–208. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pan C, Olsen JV, Daub H and Mann M: Global
effects of kinase inhibitors on signaling networks revealed by
quantitative phosphoproteomics. Mol Cell Proteomics. 8:2796–2808.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu W, Kovacevic Z, Peng Z, Jin R, Wang P,
Yue F, Zheng M, Huang ML, Jansson PJ, Richardson V, et al: The
molecular effect of metastasis suppressors on Src signaling and
tumorigenesis: new therapeutic targets. Oncotarget. 6:35522–35541.
2015.PubMed/NCBI
|
|
34
|
Johnson FM, Saigal B, Talpaz M and Donato
NJ: Dasatinib (BMS-354825) tyrosine kinase inhibitor suppresses
invasion and induces cell cycle arrest and apoptosis of head and
neck squamous cell carcinoma and non-small cell lung cancer cells.
Clin Cancer Res. 11:6924–6932. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Carlson JA, Linette GP, Aplin A, Ng B and
Slominski A: Melanocyte receptors: Clinical implications and
therapeutic relevance. Dermatol Clin. 25:541–ix. 2007. View Article : Google Scholar : PubMed/NCBI
|