Introduction
Staphylococcus aureus (S. aureus) is
the most common bacterial pathogen, and due to the increase of
drug-resistant S. aureus strains, S. aureus infection
remains a public health threat worldwide. Methicillin-resistant
S. aureus (MRSA) strains exhibit a multidrug-resistant
phenotype and the available antimicrobial treatments are
ineffective (1). MRSA infections
account for ~50% of nosocomial and community-associated
staphylococcal infections (2).
MRSA infections are characterized by high mortality rates and
prolonged hospitalization, thus increasing the costs of health
care. At present, to the best of the authors' knowledge, there is
no effective treatment for MRSA infection, thus it is necessary to
identify novel molecules targeting MRSA strains. Antimicrobial
peptides (AMPs) are considered among the most promising drugs
against resistant strains (3).
AMPs are synthesized by multicellular organisms as a
defense mechanism against pathogenic microbes (4,5).
Previous studies demonstrated that AMPs exhibit antimicrobial and
anticancer activities, and serve as regulators of the innate immune
system (6–8). The majority of natural cationic
peptides are characterized by low biological activity or by high
toxicity, and these peptides require modification in order to
achieve high and broad-spectrum activity without toxicity (9). In addition, the potency of synthetic
peptides is identical to that of natural peptides, and it is
possible to produce large quantities of highly pure AMPs ready to
be used in clinical applications (10). A 66-amino acid peptide was designed
in the laboratory at the Department of Nanlou Pulmonology &
National Clinical Research Center of Geriatrics Disease, Chinese
People's Liberation Army General Hospital and derived from
LCT-EF128 enterocin (11). Based
on the 66-amino-acid peptide, three AMPs composed of 9–12 amino
acids were designed and their antimicrobial activities against
gram-positive and gram-negative strains were examined in
vitro. The present results suggested that L12 is able to target
gram-positive strains, particularly S. aureus. In addition,
the synergistic action of L12 combined with various antibacterial
drugs was tested, and its antibacterial mechanism was
investigated.
Materials and methods
Bacterial strains and antibiotics
The bacteria selected in the present study were
gram-positive and gram-negative isolates, and common clinical and
standard strains (Table I). The
gram positive clinical isolates (S. aureus, Staphylococcus
epidermidis, Enterococcus faecalis and Enterococcus
faecium) and gram negative clinical isolates (Pseudomonas
aeruginosa, Escherichia coli, Klebsiella pneumoniae and
Acinetobacter baumannii) were collected from The Southwest
Hospital (Chongqing, China) and are not commercially available. The
standard strains (Vichita, N315, FDA Strain PCI 1200, RP62A, Boston
41501 and FDA strain Seattle 1946) were purchased from China Center
for Type Culture Collection (Wuhan, China) and stored in the
laboratory of the Pulmonology Department of Southwest Hospital,
Third Military Medical University (Army Medical University),
Chongqing, China. Oxacillin and linezolid were purchased from
Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). The following
antibiotics were purchased from The National Institute for the
Control of Pharmaceutical and Biological Products (Beijing, China):
erythromycin, tetracycline, ceftazidime, levofloxacin, gentamycin
and vancomycin. The antibiotics were diluted in water or the
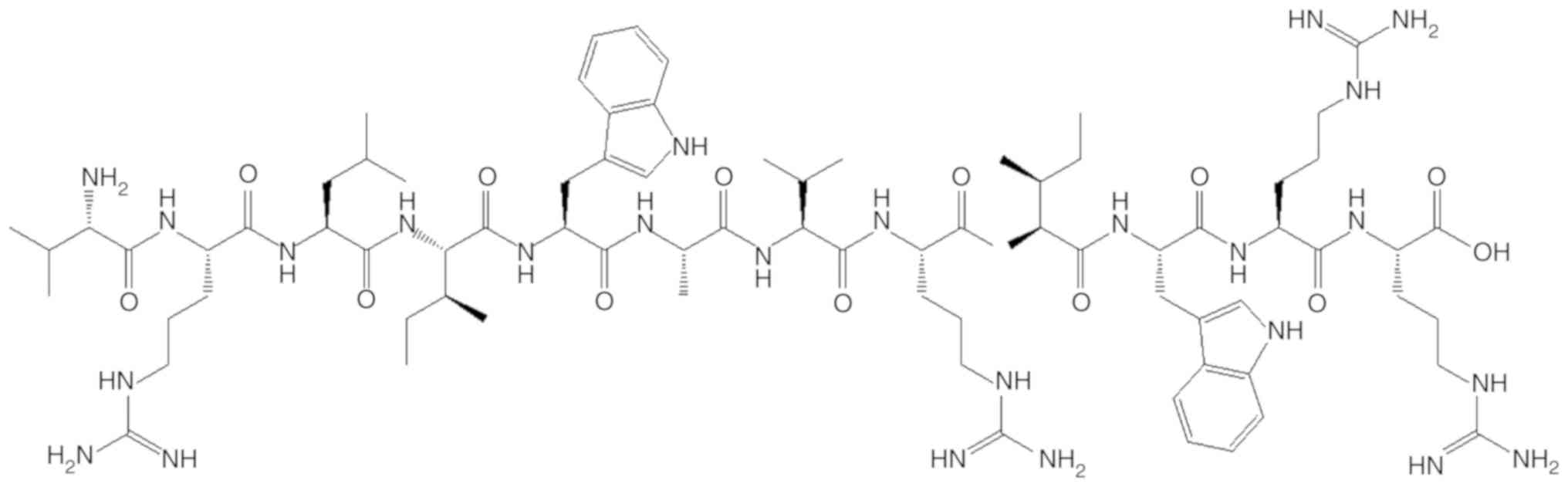
recommended solvent to obtain a working concentration. L12, whose
structure is presented in Fig. 1,
was purchased from Sangon Biotech Co., Ltd. (Shanghai, China).
 | Table I.MICs of L12 against gram-positive and
gram-negative bacteria. |
Table I.
MICs of L12 against gram-positive and
gram-negative bacteria.
| A, Gram-positive
bacteria |
|---|
|
|---|
| Species | Strain | MIC of L12,
µg/ml |
|---|
| Staphylococcus
aureus |
|
| Wichita | 8 |
|
| N315 | 8 |
|
| S26 | 8 |
|
| S28 | 16 |
|
| S29 | 4 |
| Staphylococcus
epidermidis |
|
| FDA strain | 16 |
|
| PCI 1200 |
|
|
| RP62A | 16 |
|
| 48 | 16 |
|
| 49 | 16 |
|
| 50 | 32 |
| Enterococcus
faecalis |
|
| 167 | 32 |
|
| 169 | 16 |
|
| 170 | 32 |
|
| 171 | 32 |
|
| 172 | 16 |
| Enterococcus
faecium |
|
| 175 | 32 |
|
| 176 | 4 |
|
| 177 | 4 |
|
| 178 | 16 |
|
| 179 | 32 |
|
| B, Gram-negative
bacteria |
|
| Species | Strain | MIC of L12,
µg/ml |
|
| Pseudomonas
aeruginosa |
|
| Boston 41501 | >256 |
|
| P496 | >256 |
|
| P501 | >256 |
|
| P544 | >256 |
|
| P630 | >256 |
| Escherichia
coli |
|
| FDA strain | 64 |
|
| Seattle 1946 |
|
| E259 | >256 |
|
| E260 | >256 |
|
| E270 | >256 |
|
| E282 | >256 |
| Klebsiella
pneumoniae |
|
| 1206 | >256 |
|
| 1220 | >256 |
|
| 1240 | >256 |
|
| 1248 | >256 |
|
| 1314 | >256 |
| Acinetobacter
baumannii |
|
| aba658 | >256 |
|
| aba659 | >256 |
|
| aba660 | >256 |
|
| aba661 | >256 |
|
| aba662 | >256 |
Analysis of antibacterial
activity
The minimal inhibitory concentrations (MICs) of L12
and various antibiotics were calculated using the broth
microdilution method (12) for
each type of strain, according to the guidelines of The Clinical
and Laboratory Standards Institute (2015) (13). The MIC for each drug was set as the
lowest concentration required to inhibit bacterial growth. S.
aureus (American Type Culture Collection, Manassas, VA, USA;
cat. no. 29213) strain Wichita was used as a control strain.
Checkerboard assays
The checkerboard assay method was used to measure
the combinatorial effects of L12 with multiple antibiotics
(14). Solutions containing two
drugs were prepared in 96-well plates. Serial two-fold dilutions
were prepared for each column, relative to drug ‘A’, and for each
row, relative to drug ‘B’. The starting concentration was two times
the MIC for each drug. Bacterial suspensions at the mid-log phase
of growth (1-5×105 Cfu/ml) were added to the plates, and
the plates were subsequently incubated at 37°C for 24 h. The
effects of the combinations were analyzed by calculating the
fractional inhibitory concentration index (FICI) of each
combination as follows: (MICdrug‘A’ in
combination)/(MICdrug‘A’ alone)+(MICdrug‘B’
in combination)/(MICdrug‘B’ alone). The effect was
considered synergistic if the FICI was ≤0.5, additive if the FICI
was >0.5 and ≤4.0, and antagonistic if the FICI was >4.0
(15).
Time-kill curves of various
antibiotics alone and in combination with L12
Freshly prepared colonies of S. aureus were
suspended in tryptic soy broth (TSB) medium (Solarbio Science and
Technology Co., Ltd., Beijing, China) and incubated at 37°C in a
shaker at 180 rpm for 18 h. The cultures were diluted to
5×105 Cfu/ml with TSB to obtain a final volume of 10 ml.
Vancomycin and L12 alone or in combination were added to the
prepared bacterial suspensions to meet the MIC. The negative
control was not treated with any drugs. The treated samples were
incubated at 37°C in a shaker at 180 rpm, and bacterial counts were
measured at 0, 15, 30, 60, 120, 240, 360, 480, 720 and 1,440 min.
The results were expressed in Cfu/ml on a logarithmic scale. The
limit of detection was defined as 100 Cfu/ml and lower bacterial
numbers were considered not detectable.
Transmission electron microscope (TEM)
examination
The standard control S. aureus strain Wichita
was treated with or without L12 at the MIC in TSB medium and
subsequently incubated at 37°C in a shaker at 180 rpm for 30 min.
The cultures were centrifuged at 7,620 × g for 15 min prior to
harvesting. The bacteria were subsequently fixed in 2.5%
glutaraldehyde overnight at 4°C. The samples were washed three
times with PBS and incubated in 1% osmium tetraoxide for 2 h at
4°C. Following washing with water, the samples were dehydrated with
an acetone series (50, 70, 90 and 100%) for 15 min at each
concentration. The samples were subsequently embedded in epoxy
resin at 70°C for ~9 h, cut into ultrathin (60 nm) sections, and
stained with uranyl acetate and lead citrate for 15 min
respectively at room temperature prior to examination. Each
specimen was examined using a TEM (H-600; Hitachi, Ltd., Tokyo,
Japan; magnification, ×50,000).
Scanning electron microscope (SEM)
examination
The standard control S. aureus strain Wichita
was treated with or without L12 at the MIC in TSB medium and
incubated at 37°C in a shaker at 180 rpm for 30 min. The solutions
were centrifuged at 7,620 × g for 2 min and washed twice with PBS.
The bacteria were resuspended in PBS and subsequently added to
polylysine-treated 8-mm cover glasses. The samples were dried at
room temperature and fixed with 2.5% glutaraldehyde overnight at
4°C. Following fixation, the samples were washed with 0.9% NaCl for
5 min. The samples were subsequently dehydrated at room temperature
with increasing concentrations of ethanol (30, 50, 70, 95 and
100%), and the solvent was replaced with a tert-butanol series for
6 min following incubation with each concentration. The samples
were point dried with CO2, mounted on aluminum stubs and
sputter-coated with gold. The samples were examined using an SEM
(S-3400N II; Hitachi, Ltd.; magnification, ×10,000).
Biofilm assay
Biofilm formation and removal were tested using the
96-well crystal violet staining method. An overnight culture of
S. aureus was diluted 1:1,000 in TSB. The effect of L12 on
the biofilm formation of S. aureus was tested as follows:
100 µl bacterial suspensions were mixed with 100 µl L12 at
concentrations ranging between the MIC and MIC/32, resulting in
final concentrations ranging between MIC/2 and MIC/64, or with 100
µl TSB. The resulting solutions were added to the 96-well plate.
The effect of L12 on biofilm removal was evaluated as follows: 200
µl bacterial suspension was added to the 96-well plate, and the
plate was incubated at 37°C for 24 h. The medium was removed, and
the wells were gently rinsed three times with sterile distilled
water. Subsequently, 200 µl TSB was mixed with L12 at a
concentration ranging between 2× MIC and 10× MIC, or without L12,
and the resulting solution was added to the 96-well plate.
Following incubation at 37°C for 24 h, the media from the two
experiments were removed, and the wells were gently rinsed three
times with sterile distilled water. The plates were air-dried,
stained with 1% crystal violet for 10 min at room temperature,
rinsed three times with distilled water, and air-dried. Following
drying, 100 µl acetic acid at 30% were added to each well. The
biofilms were examined at 590 nm using a MicroELISA reader
(Dynatech Laboratories, Inc., Alexandria, VA, USA). Each assay was
performed in triplicate, and wells without biofilm were used as
blank controls.
Statistical analysis
All the experiments were repeated independently
three times. The bacterial biofilm data are presented as the mean ±
standard deviation and were analyzed by one-way analysis of
variance with the Least Significant Difference post hoc test, using
SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). The correlation
between the MICs of L12 and antibiotic resistance was calculated by
Spearman's correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Antibacterial activity analysis
To investigate the antimicrobial effect of L12, five
strains of each species of bacteria were selected. The MICs of L12
against gram-positive and gram-negative bacteria are presented in
Table I. The MICs of L12 against
gram-positive bacteria ranged between 4 and 32 µg/ml. However, the
majority of MICs of L12 against gram-negative bacteria were >256
µg/ml. In the present study, L12 exhibited an increased effect on
S. aureus compared with the other three types of
gram-positive bacteria. Therefore, only S. aureus strains
were analyzed in the following experiments. Since the present study
aimed to investigate the potential of L12 in treating infections
caused by resistant strains, 30 MRSA isolates (which are not
commercially available) were selected. The MICs of L12 against MRSA
strains ranged between 4 and 32 µg/ml. Specifically, the MICs
relative to MRSA strains 11, 10, 5 and 4 were 4, 8, 16 and 32
µg/ml, respectively (data not shown). The antibiotic resistance of
MRSA strains and the correlation between antibiotic resistance and
the MICs of L12 for all the 30 MRSA strains are presented in
Table II. The MRSA strains were
highly resistant to the majority of the antibiotics tested, the
resistance rates of oxacillin, erythromycin, tetracycline,
levofloxacin and gentamycin were 100.0% (30 strains), 83.3% (25
strains), 76.7% (23 strains), 76.7% (23 strains), 73.3% (22
strains) and 66.7% (20 strains). However, they were wholly
sensitive to vancomycin and linezolid. A correlation analysis
suggested that no correlation was present between the MICs of L12
and antibiotic resistance, the strains resistant to more
antibiotics were not more resistant to L12.
 | Table II.Resistance of methicillin-resistant
Staphylococcus aureus strains to antibiotics, and the
correlations between antibiotic resistance and L12 minimal
inhibitory concentration. |
Table II.
Resistance of methicillin-resistant
Staphylococcus aureus strains to antibiotics, and the
correlations between antibiotic resistance and L12 minimal
inhibitory concentration.
| Antibiotic | Resistance rate,
% | Resistant strains,
n | Spearman's
correlation coefficient r | P-value |
|---|
| Oxacillin | 100.0 | 30 | 0.060 | 0.751 |
| Erythromycin |
83.3 | 25 | 0.110 | 0.563 |
| Tetracycline |
76.7 | 23 | −0.045 | 0.813 |
| Ceftazidime |
76.7 | 23 | 0.238 | 0.205 |
| Levofloxacin |
73.3 | 22 | 0.006 | 0.976 |
| Gentamycin |
66.7 | 20 | −0.147 | 0.437 |
| Vancomycin |
0.0 | 0 | 0.342 | 0.064 |
| Linezolid |
0.0 | 0 | 0.173 | 0.362 |
Checkerboard assays
The synergistic effect of L12 with various
antibiotics was examined by calculating the FICI. The MICs of the
antibiotics against the majority of the MRSA strains were not
obtained since the tested concentrations were not sufficient to
inhibit the growth of the strains, thus five MRSA strains were
selected for this experiment. The MICs of L12 against five MRSA
strains ranged between 4 and 32 µg/ml, the MICs of vancomycin,
levofloxacin, tetracycline, gentamycin and ceftazidime were 1–2,
16–64, 16–64, 64–128 and 32–128 µg/ml, respectively. As presented
in Table III, L12 manifested a
synergistic effect when used in combination with vancomycin and
levofloxacin. However, L12 exhibited an additive effect when used
in combination with gentamicin, tetracycline and ceftazidime.
 | Table III.Fractional inhibitory concentration
indexes of L12 in combination with multiple antibiotics. |
Table III.
Fractional inhibitory concentration
indexes of L12 in combination with multiple antibiotics.
|
| Treatment |
|---|
|
|
|
|---|
| Strain | Vancomycin+L12 | Gentamycin+L12 |
Levofloxacin+L12 |
Tetracycline+L12 |
Ceftazidime+L12 |
|---|
| S26 | 0.375 | 0.750 | 0.313 | 0.750 | 1.000 |
| S29 | 0.531 | 0.531 | 0.531 | 0.750 | 2.000 |
| S37 | 0.531 | 0.531 | 0.531 | 0.531 | 0.625 |
| S47 | 0.281 | 0.750 | 0.500 | 1.000 | 0.531 |
| S49 | 0.188 | 0.625 | 0.125 | 0.750 | 2.000 |
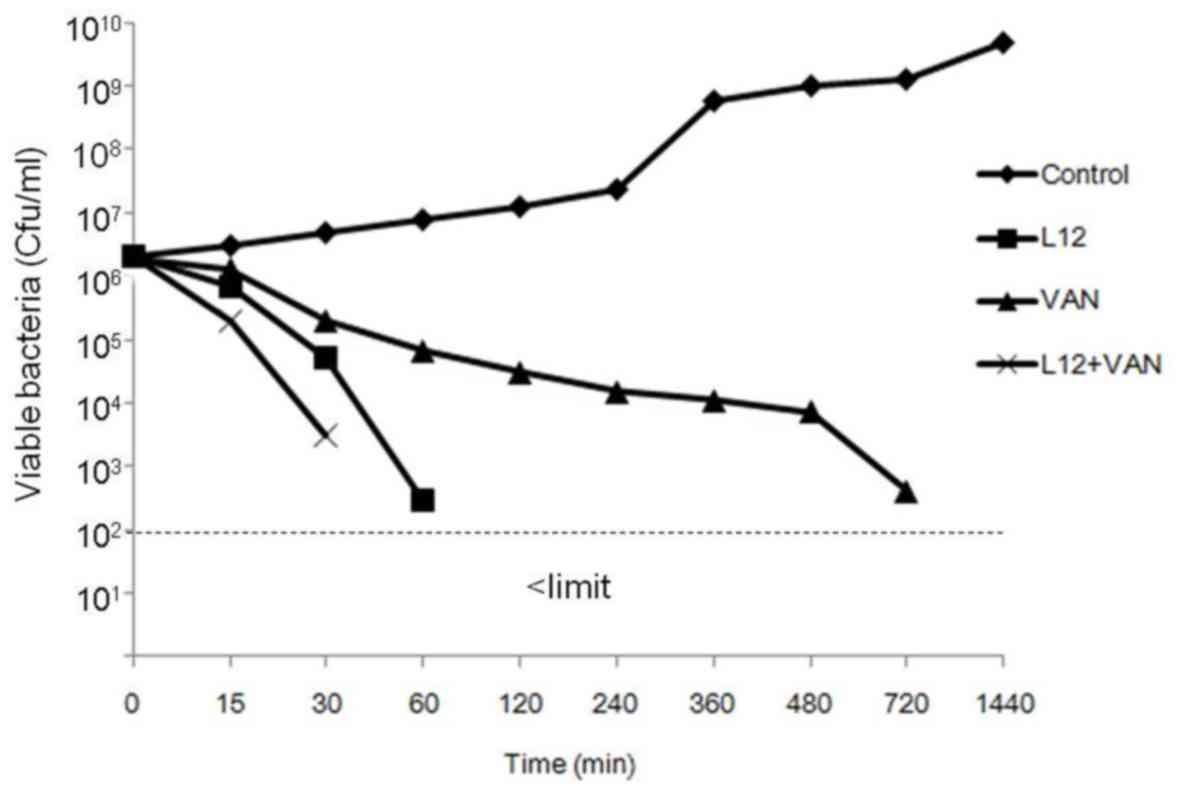
Time-kill curves
The time-killing curves suggested that L12 induced
bacterial death of S. aureus in 60 min, and the number of
bacteria decreased continuously when treated with vancomycin.
Furthermore, samples cotreated with L12 and vancomycin induced
bacterial death in 30 min (Fig.
2).
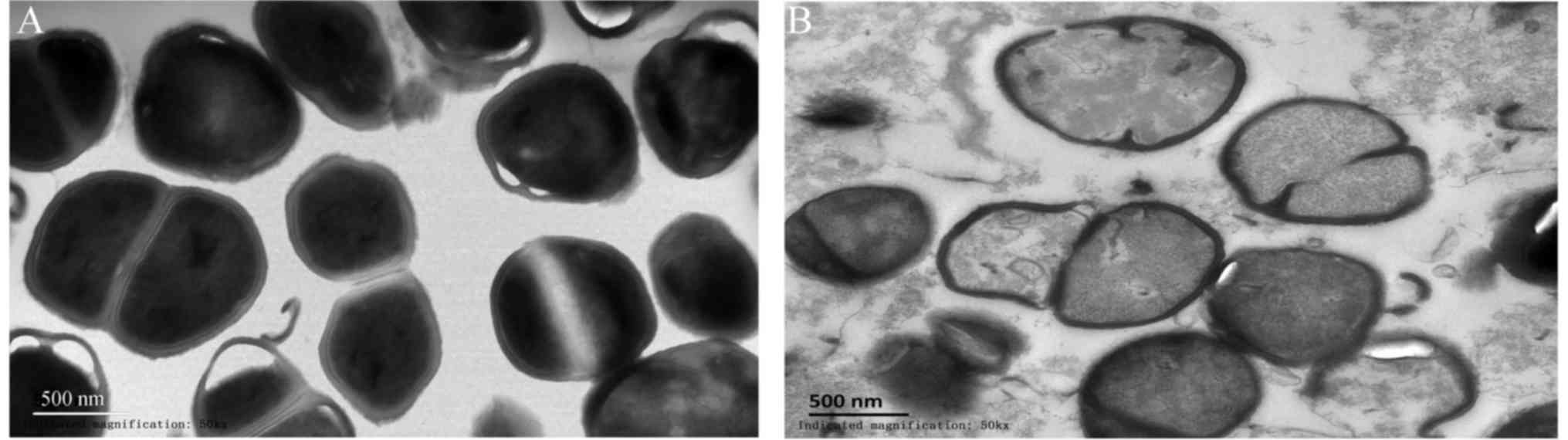
TEM observations
TEM images of S. aureus cells treated with
L12 are presented in Fig. 3.
Untreated S. aureus cells exhibited an intact cellular
architecture with a uniform cytoplasmic density, whereas, the cells
treated with L12 presented severe damage, disrupted cell walls,
leakage of the cytoplasmic contents and misshapen or fragmented
cells.
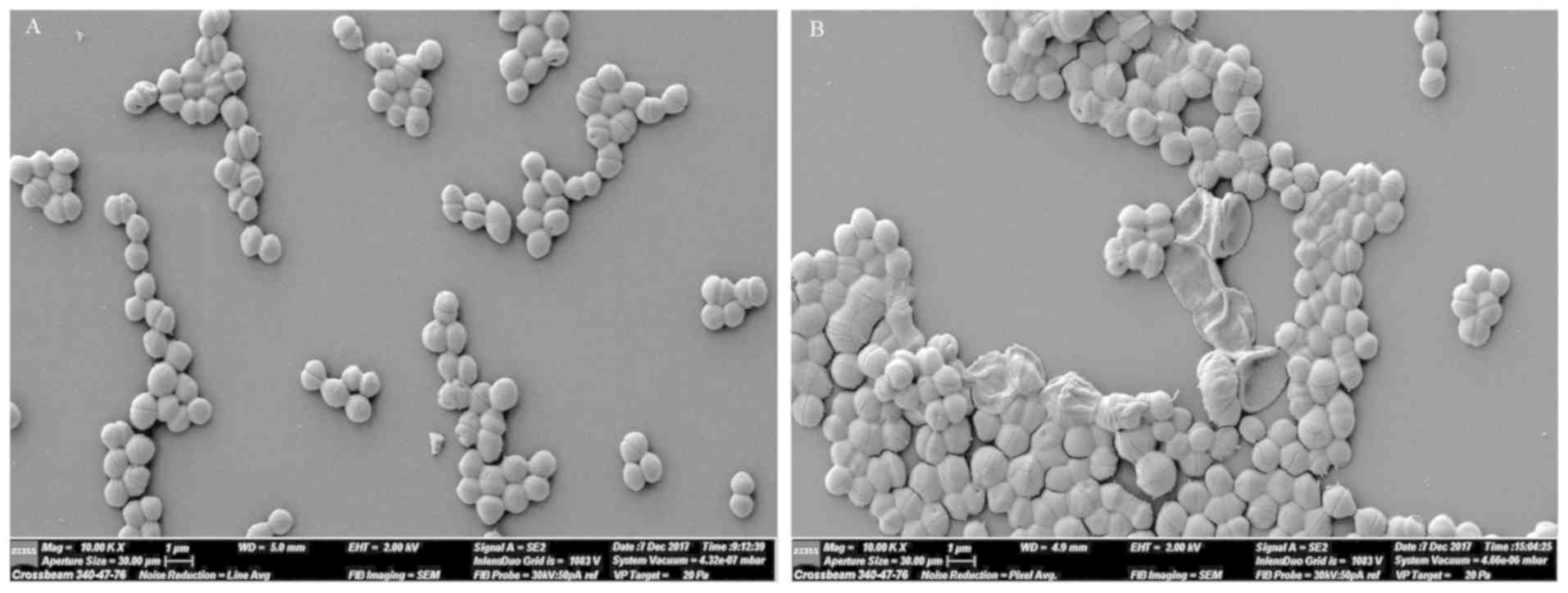
SEM observations
SEM images of S. aureus cells that were
treated with L12 are presented in Fig.
4. The untreated cells were round and plump, whereas, the
majority of the cells treated with L12 were shriveled, and
exhibited a disrupted cell wall and cell membrane.
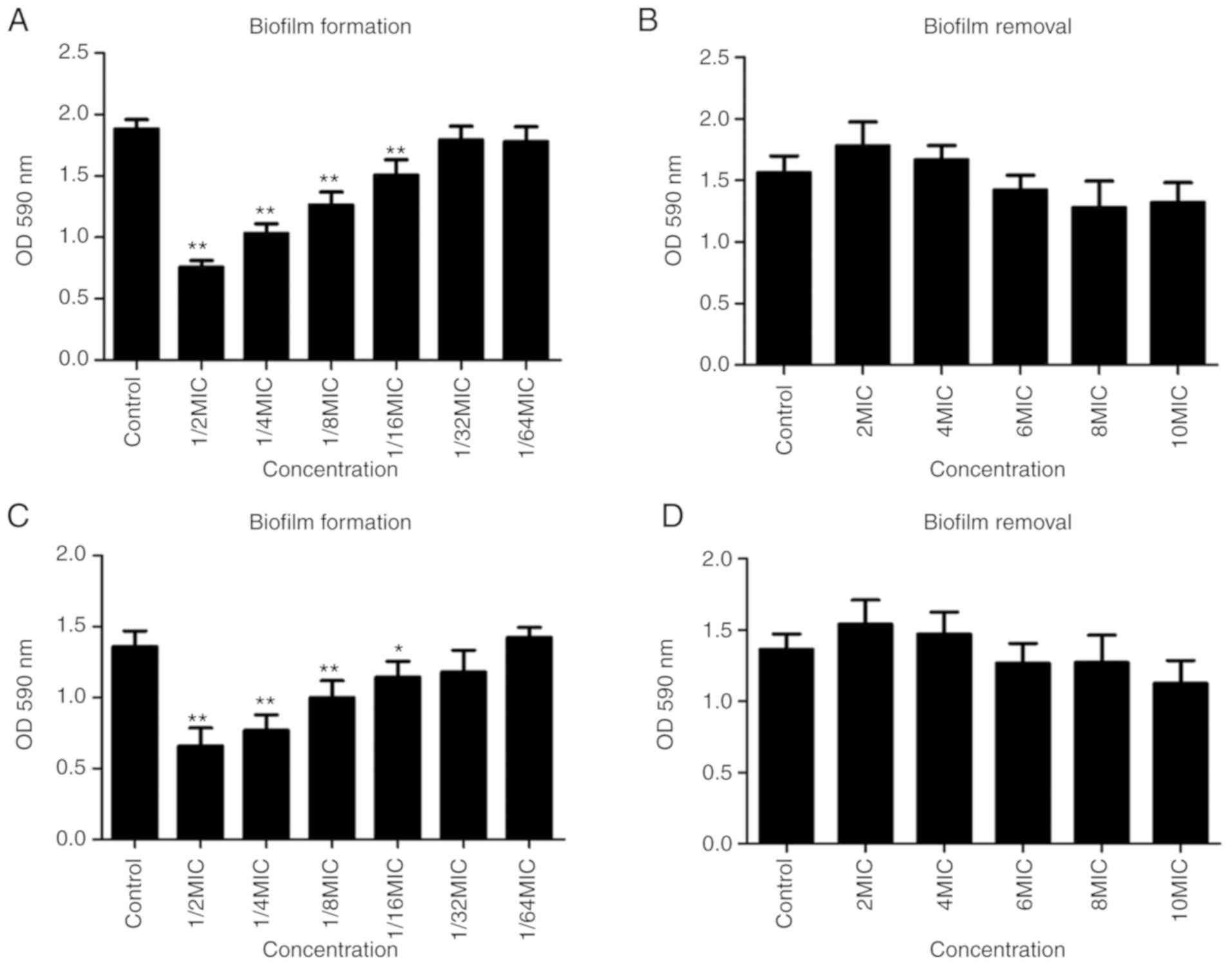
Biofilm assay
The effect of L12 on biofilms of S. aureus is
presented in Fig. 5. The MRSA
isolate S37, which exhibits the strongest biofilm formation, and
the standard strain N315, which is the biofilm-positive strain were
selected. Compared with the control group, concentrations ranging
between MIC/2 and MIC/16 of L12 significantly inhibited the biofilm
formation of S. aureus strains N315 and S37 (Fig. 5A and C). However, concentrations
ranging between 2× MIC and 10× MIC L12 did not degrade the
previously formed biofilms of S. aureus strains N315 and S37
(Fig. 5B and D).
Discussion
To resolve the problem of antimicrobial resistance,
multiple AMPs have been developed in recent years as alternative
antibiotics (16–19), and a number of AMPs exhibited high
antimicrobial efficacy and broad-spectrum activity (17,20).
The present study suggested that L12 was effective in
susceptibility tests against gram-positive bacteria, particularly
S. aureus. The decreased effectiveness of L12 against other
gram-positive bacteria may be due to differences in the cell wall
composition. In particular, Enterococcus has a thick cell wall and
grows at a slow rate (21), and
these two features may be responsible for the observed poor
antibacterial activity of L12. However, the molecular mechanism
underlying Enterococcus resistance to L12 requires further
investigation in future studies. The MIC of L12 against S.
aureus was similar to traditional antibacterial drugs, and the
antibacterial activity of L12 was comparable with other AMPs
(22–25). Furthermore, the MICs of L12
exhibited no correlation with resistance to the tested antibiotics,
the strains resistant to more antibiotics were not more resistant
to L12, which suggested that L12 may be used to treat infections
caused by MRSA strains. However, L12 exhibited little effect on
gram-negative bacteria. The variable susceptibility between
gram-negative and gram-positive bacteria was likely due to
structural variations in the cell wall and the cell membrane. The
cell membrane is the target of the majority of AMPs (26,27).
Notably, according to SEM and TEM analysis, L12 may additionally
target the cell membrane.
In the present study, L12 exerted a synergistic
effect with vancomycin and levofloxacin. The majority of AMPs
exhibit synergistic effects with traditional antibacterial drugs
(24,28), which may broaden the scope of their
use. The additional antibacterial molecules tested presented
additive effects. A previous study demonstrated that antibacterial
drugs with similar molecular mechanisms or that target the same
molecular components, exert improved synergistic effects compared
with combinations of drugs with dissimilar function (17). Additionally, vancomycin and
ceftazidime inhibit the cell wall synthesis of gram-positive
bacteria by multiple mechanisms (29), which is hypothesized to be
responsible for their synergistic effects with L12. According to
the present study, ceftazidime was not among the most effective
drugs against S. aureus and exhibited a decreased
antibacterial effect compared with vancomycin. Since synergistic
effects are due to similar antibacterial mechanism, further studies
are necessary to investigate the antibacterial mechanisms of the
tested compounds, thus improving the clinical use of these
antimicrobial drugs.
S. aureus is considered one of the most
important pathogens involved in biomaterial-associated infections.
S. aureus adheres to the surfaces of medical devices to form
biofilms resistant to the action of the immune system (30). The results of the present study
suggested that sub-MIC of L12 may inhibit biofilm formation in a
dose-dependent manner; however, concentrations ≤10× MIC were not
sufficient to degrade previously formed biofilms. Biofilms are
colonies of bacteria embedded by a self-secreted extracellular
polymeric substance, and this extracellular polymeric substance
serves as a selective barrier that allows the exchange of nutrients
with the surroundings, preventing xenobiotics from entering the
biofilm. L12 is a large molecule that is not able to penetrate the
biofilm (31). The present results
suggested that L12 may only serve as a preventive drug and not as a
therapeutic drug for the treatment of biofilm infection.
A number of peptides have been developed as novel
pharmaceuticals and tested in clinical trials (5,32)
and these antimicrobial peptides possess promising potential.
Further technological developments may allow researchers to
precisely modify AMP sequences in order to modulate their
antibacterial potency and cytotoxicity. Furthermore, it is crucial
to identify novel strategies to shorten the peptide sequences to
improve their clinical application (33). In the present study, it was
demonstrated that L12, derived from enterocin, a molecule isolated
from E. faecium (14),
exerts antibacterial effects against S. aureus and biofilm
production. The present study suggested that L12 may represent a
potential therapeutic drug for the treatment of S. aureus
infection.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Nature
Science Foundation of Beijing, China (grant no. 7152132).
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
FX designed the present study and wrote the
manuscript. XD and YL performed the experiments. RW, LA and YW
analyzed the data. ZC was involved in the design of the present
study and provided financial support for the present study. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Palavecino EL: Clinical, epidemiologic,
and laboratory aspects of methicillin-resistant Staphylococcus
aureus infections. Methods Mol Biol. 1085:1–24. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bassetti M and Righi E:
Multidrug-resistant bacteria: What is the threat? Hematology Am Soc
Hematol Educ Program. 2013:428–432. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kang HK, Kim C, Seo CH and Park Y: The
therapeutic applications of antimicrobial peptides (AMPs): A patent
review. J Microbiol. 55:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zasloff M: Antimicrobial peptides of
multicellular organisms. Nature. 415:389–395. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fjell CD, Hiss JA, Hancock RE and
Schneider G: Designing antimicrobial peptides: Form follows
function. Nat Rev Drug Discov. 11:37–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zasloff M: Antibiotic peptides as
mediators of innate immunity. Curr Opin Immunol. 4:3–7. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tripathi AK, Kumari T, Tandon A, Sayeed M,
Afshan T, Kathuria M, Shukla PK, Mitra K and Ghosh JK: Selective
phenylalanine to proline substitution for improved antimicrobial
and anticancer activities of peptides designed on phenylalanine
heptad repeat. Acta Biomater. 57:170–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kazmirchuk T, Dick K, Burnside DJ, Barnes
B, Moteshareie H, Hajikarimlou M, Omidi K, Ahmed D, Low A, Lettl C,
et al: Designing anti-Zika virus peptides derived from predicted
human-Zika virus protein-protein interactions. Comput Biol Chem.
71:180–187. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xia X, Cheng L, Zhang S, Wang L and Hu J:
The role of natural antimicrobial peptides during infection and
chronic inflammation. Antonie Van Leeuwenhoek. 111:5–26. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ciociola T, Giovati L, Conti S, Magliani
W, Santinoli C and Polonelli L: Natural and synthetic peptides with
antifungal activity. Future Med Chem. 8:1413–1433. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Z, Chang D, Zou Y, Su L, Zhu Y, Fang
X, Wang J, Guo Y, Zhao J, Li D, et al: Genome sequence of
enterococcus faecium clinical isolate LCT-EF128. J Bacteriol.
194:47652012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ouyang J, Sun F, Feng W, Sun Y, Qiu X,
Xiong L, Liu Y and Chen Y: Quercetin is an effective inhibitor of
quorum sensing, biofilm formation and virulence factors in
Pseudomonas aeruginosa. J Appl Microbiol. 120:966–974. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Clinical Laboratory Standards Institute, .
Performance Standards for Antimicrobial Susceptibility Testing;
25th Informational Supplement. CLSI document M100-S24. Clinical
Laboratory Standards Institute. (Wayne, PA). 2015.
|
|
14
|
Pankey GA and Ashcraft DS: In vitro
synergy of ciprofloxacin and gatifloxacin against
ciprofloxacin-resistant Pseudomonas aeruginosa. Antimicrob
Agents Chemother. 49:2959–2964. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Menegucci TC, Albiero J, Migliorini LB,
Alves JL, Viana GF, Mazucheli J, Carrara-Marroni FE, Cardoso CL and
Tognim MC: Strategies for the treatment of polymyxin B-resistant
acinetobacter baumannii infections. Int J Antimicrob Agents.
47:380–385. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wuerth KC, Falsafi R and Hancock REW:
Synthetic host defense peptide IDR-1002 reduces inflammation in
Pseudomonas aeruginosa lung infection. PloS One.
12:e01875652017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu X, Li Z, Li X, Tian Y, Fan Y, Yu C,
Zhou B, Liu Y, Xiang R and Yang L: Synergistic effects of
antimicrobial peptide DP7 combined with antibiotics against
multidrug-resistant bacteria. Drug Des Devel Ther. 11:939–946.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bechinger B and Gorr SU: Antimicrobial
peptides: Mechanisms of action and resistance. J Dent Res.
96:254–260. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mangoni ML, McDermott AM and Zasloff M:
Antimicrobial peptides and wound healing: Biological and
therapeutic considerations. Exp Dermatol. 25:167–173. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ge Y, MacDonald DL, Holroyd KJ,
Thornsberry C, Wexler H and Zasloff M: In vitro antibacterial
properties of pexiganan, an analog of magainin. Antimicrob Agents
Chemother. 43:782–788. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wellinghausen N, Chatterjee I, Berger A,
Niederfuehr A, Proctor RA and Kahl BC: Characterization of clinical
Enterococcus faecalis small-colony variants. J Clin Microbiol.
47:2802–2811. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schillaci D, Cusimano MG, Spinello A,
Barone G, Russo D, Vitale M, Parrinello D and Arizza V: Paracentrin
1, a synthetic antimicrobial peptide from the sea-urchin
paracentrotus lividus, interferes with Staphylococcal and
Pseudomonas aeruginosa biofilm formation. AMB Express.
4:782014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gottschalk S, Gottlieb CT, Vestergaard M,
Hansen PR, Gram L, Ingmer H and Thomsen LE: Amphibian antimicrobial
peptide fallaxin analogue FL9 affects virulence gene expression and
DNA replication in Staphylococcus aureus. J Med Microbiol.
64:1504–1513. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu Y, Liu A, Vaudrey J, Vaiciunaite B,
Moigboi C, McTavish SM, Kearns A and Coates A: Combinations of
beta-lactam or aminoglycoside antibiotics with plectasin are
synergistic against methicillin-sensitive and methicillin-resistant
Staphylococcus aureus. PloS One. 10:e01176642015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Zhang L, Wang J, Wei D, Shi B and
Shan A: Synergistic interaction of PMAP-36 and PRW4 with
aminoglycoside antibiotics and their antibacterial mechanism. World
J Microbiol Biotechnol. 30:3121–3128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sani MA and Separovic F: Antimicrobial
peptide structures: From model membranes to live cells. Chemistry.
24:286–291. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marin-Medina N, Ramirez DA, Trier S and
Leidy C: Mechanical properties that influence antimicrobial peptide
activity in lipid membranes. Appl Microbiol Biotechnol.
100:10251–10263. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Amani J, Barjini KA, Moghaddam MM and
Asadi A: In vitro synergistic effect of the CM11 antimicrobial
peptide in combination with common antibiotics against clinical
isolates of six species of multidrug-resistant pathogenic bacteria.
Protein Pept Lett. 22:940–951. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Karampatakis V, Papanikolaou T, Giannousis
M, Goulas A, Mandraveli K, Kilmpasani M, Alexiou-Daniel S and
Mirtsou-Fidani V: Stability and antibacterial potency of
ceftazidime and vancomycin eyedrops reconstituted in BSS against
Pseudomonas aeruginosa and Staphylococcus aureus.
Acta Ophthalmol. 87:555–558. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Montanaro L, Speziale P, Campoccia D,
Ravaioli S, Cangini I, Pietrocola G, Giannini S and Arciola CR:
Scenery of staphylococcus implant infections in orthopedics. Future
Microbiol. 6:1329–1349. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stewart EJ, Ganesan M, Younger JG and
Solomon MJ: Artificial biofilms establish the role of matrix
interactions in staphylococcal biofilm assembly and disassembly.
Sci Rep. 5:130812015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Domingues MM, Inacio RG, Raimundo JM,
Martins M, Castanho MA and Santos NC: Biophysical characterization
of polymyxin B interaction with LPS aggregates and membrane model
systems. Biopolymers. 98:338–344. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Haney EF, Mansour SC and Hancock RE:
Antimicrobial peptides: An introduction. Methods Mol Biol.
1548:3–22. 2017. View Article : Google Scholar : PubMed/NCBI
|