Introduction
Glaucoma is the leading cause of irreversible
blindness and is characterized by the progressive degeneration of
the optic nerve and loss of the visual field (1). A total of 76 million and 111.8
million individuals worldwide are estimated to have glaucoma by
2020 and 2040, respectively (2,3).
Increased intraocular pressure (IOP) has been identified as the
main risk factor for glaucoma (4).
IOP is maintained by the aqueous humor, which is produced by the
ciliary epithelium; the aqueous humor enters into the posterior
chamber around the lens and iris, and then goes through the
trabecular meshwork (TM) to the Schlemm's canal, eventually
entering the episcleral venous circulation via the conventional
route (5). The TM presents as a
porous, sponge-like structure, and is composed of strands or beams
of connective tissues that contain a core of collagenous and
elastic fibers, and is covered by flat cells (6). Since the TM is an important structure
in the aqueous humor outflow tract, it plays a major role in
maintaining IOP balance. Variations in IOP are usually a result of
changes in the physiological status of the TM (6).
Reactive oxygen species (ROS) is a collective term
used to define oxygen radicals such as superoxide and non-radicals
such as hydrogen peroxide (H2O2) (7). Excessive ROS can not only cause
direct damage to the DNA, protein and lipids, but it can also lead
to oxidative stress via the activation of various signaling
pathways, such as the mitogen-activated protein kinase (MAPK),
PI3K/AKT, NF-κB and p53 signaling pathways (8). These pathways play important roles in
determining cell death and survival (8). Although there are multiple
hypotheses, oxidative stress has been widely accepted as the
pathological mechanism of glaucoma (9). Exposure of eyes to sunlight can
result in the acceleration of the generation of ROS due to
ultraviolet radiation (7), while
also resulting in oxidative stress to the ocular structures
(10). Previous research has also
demonstrated that oxidative stress can result in pathological
alteration of the anterior chamber and TM (10). The TM has shown greater sensitivity
to oxidative damage compared to other ocular tissues, possibly due
to the lack of an effective antioxidant mechanism in the TM
(9). Furthermore, oxidative DNA
damage has been shown to be significantly higher in the TM of
glaucoma patients (11). It has
been hypothesized that repetitive oxidative stress due to in
vivo H2O2 can impair TM cell adhesion and
cause cell loss, resulting in elevated resistance to aqueous humor
outflow (12). Thus, oxidative
stress is an important mechanism that regulates TM pathology.
Long non-coding RNAs (lncRNAs) are a group of
non-coding transcripts of >200 nucleotides in length (13). lncRNAs form the largest percentage
of mammalian non-coding transcriptomes (13). lncRNAs play a significant role in
regulating gene expression, protein modification, cell
differentiation, immune response and other critical biochemical
pathways (14,15). Studies have identified roles for
lncRNAs in several neurodegenerative diseases, such as Alzheimer's,
Parkinson's and Huntington's disease (16–19).
In 2011, a hypothesis was proposed, which suggested that lncRNAs
were microRNA (miRNA) sponges that suppressed miRNA interactions
with mRNAs and affected translation of protein-coding genes
(20). The study referred to the
miRNA sponges as competing endogenous RNAs (ceRNAs). Given this
lncRNA-miRNA-mRNA regulation network, in the present study a ceRNA
network that further describes the interactions of these RNAs was
elucidated.
Over the years, there have been extensive studies
into the effects of lncRNAs on various types of diseases (19,21).
Nevertheless, the effects of the lncRNA-associated ceRNA network on
human trabecular meshwork cells (HTMCs) under oxidative stress have
not yet been completely described. In this study, the aim was to
analyze the effects of a lncRNA-associated ceRNA network on HTMCs
under oxidative stress.
Materials and methods
Tissue samples from Gene Expression
Omnibus (GEO) database and bioinformatics analysis
The RNA expression data were downloaded from the
NCBI GEO database (GSE126170). The series contained three HTMC
samples that were treated with 300 µm H2O2 in
serum-free medium for 2 h and three control samples that were
treated with vehicle. Agilent-078298 human ceRNA array V1.0 4X180K
(Agilent Technologies, Inc.) was utilized to perform all the
treatments. Approval from an ethics committee was not required as
the data were downloaded from the GEO database.
Analysis of differentially expressed
genes
The limma package in the R software (version 3.5.2;
www.R-project.org) (22) was used to identify differentially
expressed lncRNAs and mRNAs with |log2 Fold Change (FC)| thresholds
>1.0 and adjusted P<0.05 in the treatment and control groups.
The volcano map was obtained using the pheatmap package in the R
software (version 3.5.2; www.R-project.org). The Encyclopedia of DNA Elements
(version 38; http://www.encodeproject.org/) and Ensembl (version
96; htps://www.ensembl.org/) were used in
order to define and annotate the differentially expressed RNAs
(mRNAs and lncRNAs).
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) functional enrichment
analysis
In order to explore the underlying biological
functions and processes of these differentially expressed genes,
the GO database (http://www.geneontology.org), KEGG (http://www.kegg.jp/), and the clusterProfilerGOpackage
and clusterProfilerKEGG packages in R software (version 3.5.2;
www.R-project.org) (23) were used to perform functional
enrichment analysis. Records with P<0.05 and enrichment >2.0
were defined as ‘conserved’.
Construction of the lncRNA-miRNA-mRNA
ceRNA networks
The miRNA targets of lncRNAs were predicted using
the mircode database (version 11; http://www.mircode.org/) (24) and mRNA targets of miRNAs using
miRTarBase (version 7.0; http://mirtarbase.mbc.nctu.edu.tw/) (25), miRDB (version 5.0, http://mirdb.org/) (26) and TargetScan (version 7.2;
http://www.targetscan.org/) (27). Targeted mRNAs were cross-matched
with differentially expressed mRNAs. The lncRNAs, miRNAs and mRNAs
with |log2FC| >1.0 and P<0.05 were recorded. Finally, a
lncRNA-associated ceRNA network was constructed and visualized
using Cytoscape v3.6.1 (28).
Results
Differential expression of lncRNAs
induced by oxidative stress in HTMCs
A total of 70 differentially expressed lncRNAs were
discovered from HTMCs treated with 300 µm
H2O2 in serum-free medium for 2 h and HTMCs
treated with vehicle and having |log2FC| >1 and adjusted
P<0.05 (Table SI). Of these
differentially expressed lncRNAs, 24 were part of the ceRNA network
that was constructed using the following steps (Table I). The volcano map of
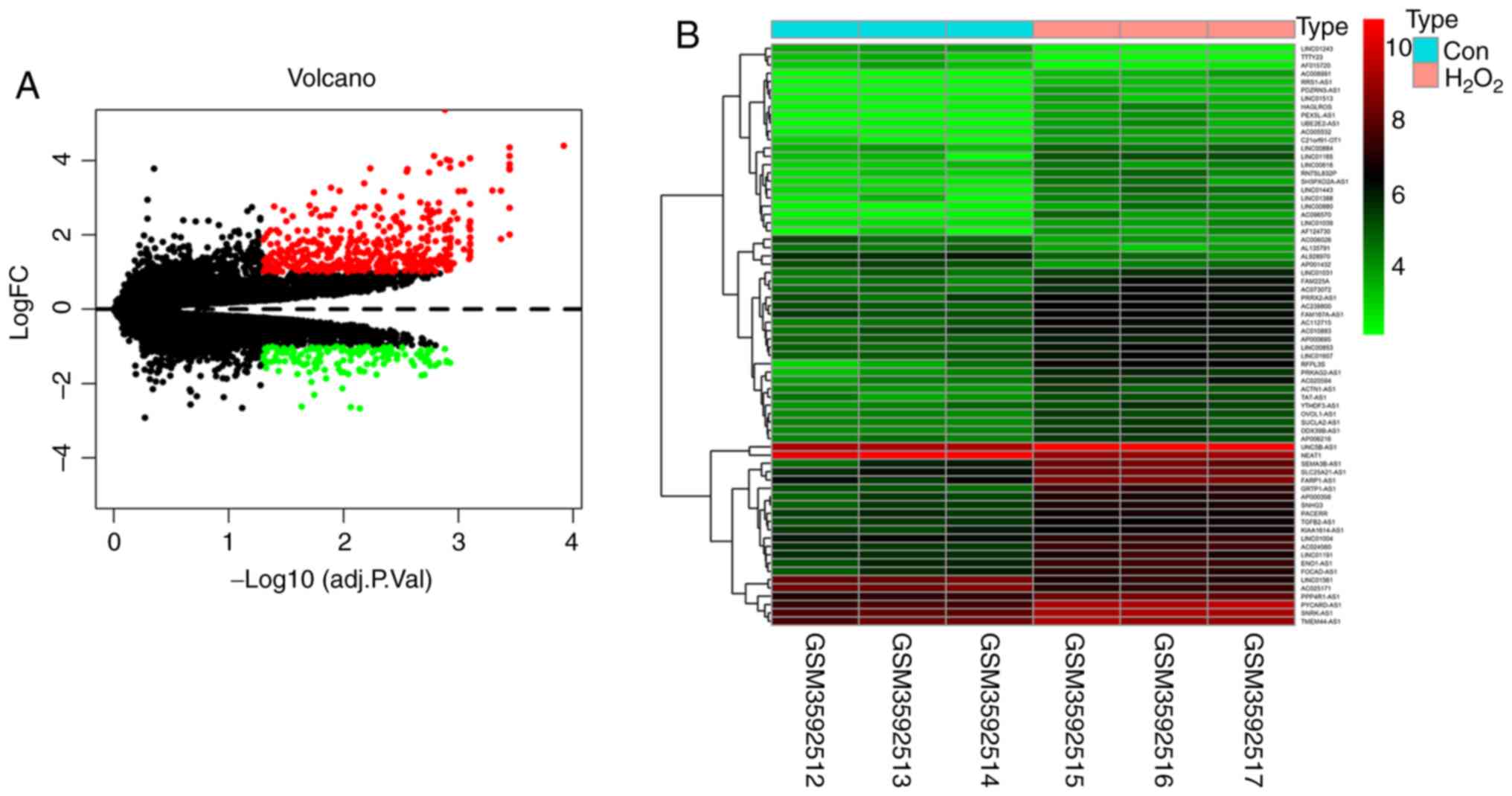
differentially expressed lncRNAs and mRNAs was obtained from the
limma package and pheatmap package in the R software (Fig. 1A). The heatmap of differentially
expressed lncRNAs showed clear differences in expression between
the two groups (Fig. 1B).
 | Table I.Differentially expressed lncRNAs in
the ceRNA network. |
Table I.
Differentially expressed lncRNAs in
the ceRNA network.
| lncRNA | Gene ID | Regulation | LogFC (T/N) | adj.P.Val |
|---|
| ACTN1-AS1 | 161159 | Up | 1.129092 | 0.016997 |
| AC005532 | – | Up | 1.406952 | 0.002765 |
| AC020594 | – | Up | 1.518925 | 0.026473 |
| AC024560 | – | Up | 1.922623 | 0.001163 |
| AC073072 | – | Up | 1.532428 | 0.013798 |
| AC096570 | – | Up | 1.747278 | 0.025237 |
| AF124730 | – | Up | 1.346528 | 0.042546 |
| AP000356 | – | Up | 1.720098 | 0.005243 |
| AP000695 | – | Up | 1.10531 | 0.004826 |
| AP006216 | – | Up | 1.004154 | 0.009124 |
| C21orf91-OT1 | 246312 | Up | 1.157345 | 0.013636 |
| DDX39B-AS1 | 106478957 | Up | 1.315506 | 0.001423 |
| ENO1-AS1 | 100505975 | Up | 1.829472 | 0.006297 |
| FARP1-AS1 | 100874080 | Up | 2.605723 | 0.003072 |
| GRTP1-AS1 | 100874068 | Up | 2.368667 | 0.003501 |
| PDZRN3-AS1 | 101927249 | Up | 1.152825 | 0.010978 |
| PEX5L-AS1 | 100874040 | Up | 1.606603 | 0.001311 |
| SNHG3 | 8420 | Up | 1.669148 | 0.00915 |
| SNRK-AS1 | 100873954 | Up | 1.375921 | 0.002028 |
| AC006026 | – | Down | −1.1049 | 0.031015 |
| AC025171 | – | Down | −1.13395 | 0.009238 |
| AF015720 | – | Down | −1.01988 | 0.041587 |
| NEAT1 | 283131 | Down | −1.47837 | 0.006227 |
| TTTY23 | 252955 | Down | −1.10446 | 0.021033 |
GO enrichment and KEGG pathway
analysis of differentially expressed mRNAs
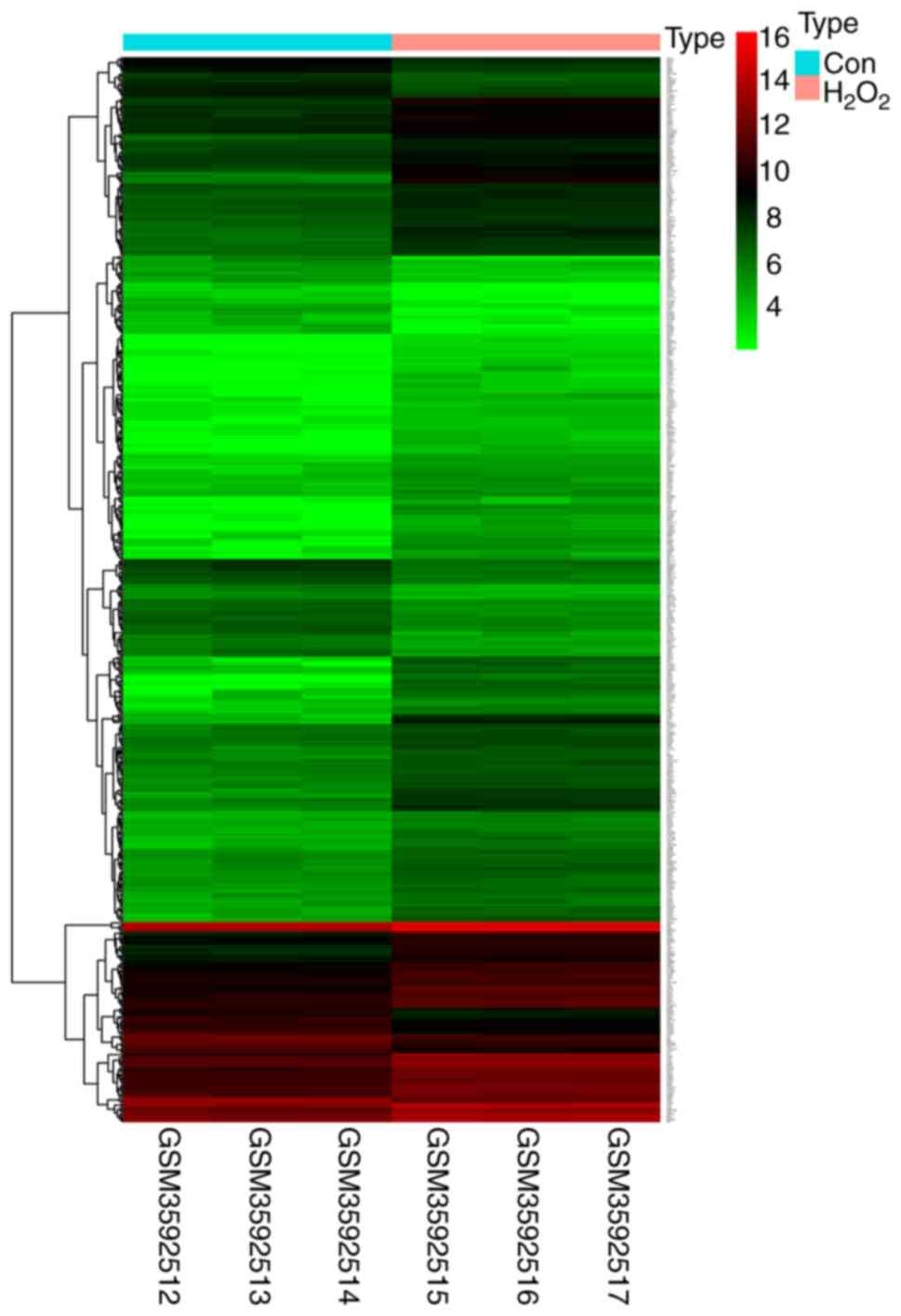
As shown in the heatmap in Fig. 2, a total of 518 differentially
expressed mRNAs were identified. In order to explore the functions
and signaling pathways of these genes GO and KEGG analyses were
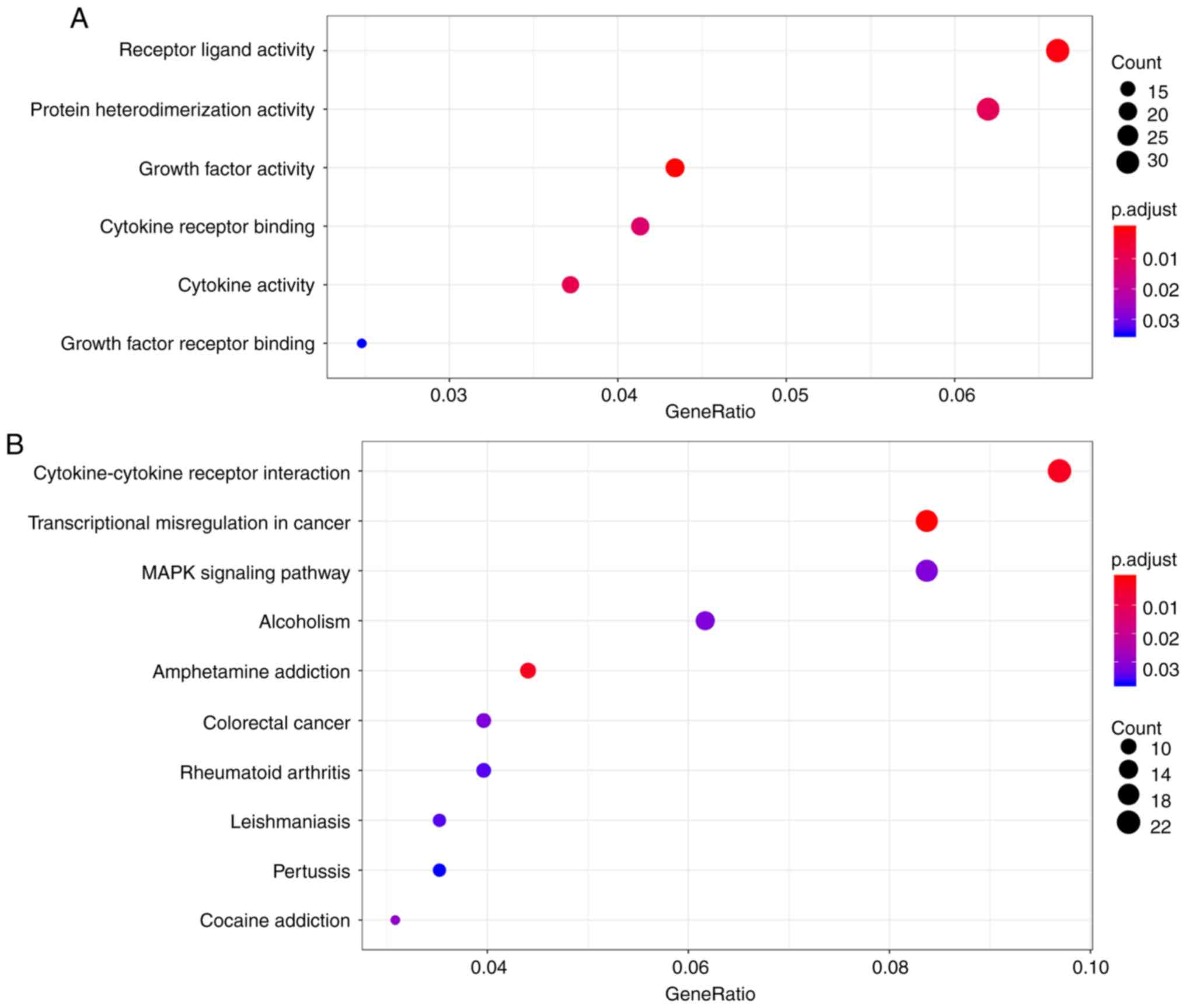
carried out. A total of 6 GO enrichment terms were listed (Fig. 3A) and the functions that were the
most enriched were receptor ligand activity (GO: 0048018), protein
heterodimerization activity (GO: 0046982) and growth factor
activity (GO: 0008083) in 32, 21 and 20 genes, respectively. As
shown in the bubble chart in Fig.
3B, 10 KEGG pathways were also identified. Among the KEGG
pathways, cytokine-cytokine receptor interaction, transcriptional
misregulation in cancer and the MAPK signaling pathway ranked top
3, and were implicated in 22, 19 and 19 genes, respectively.
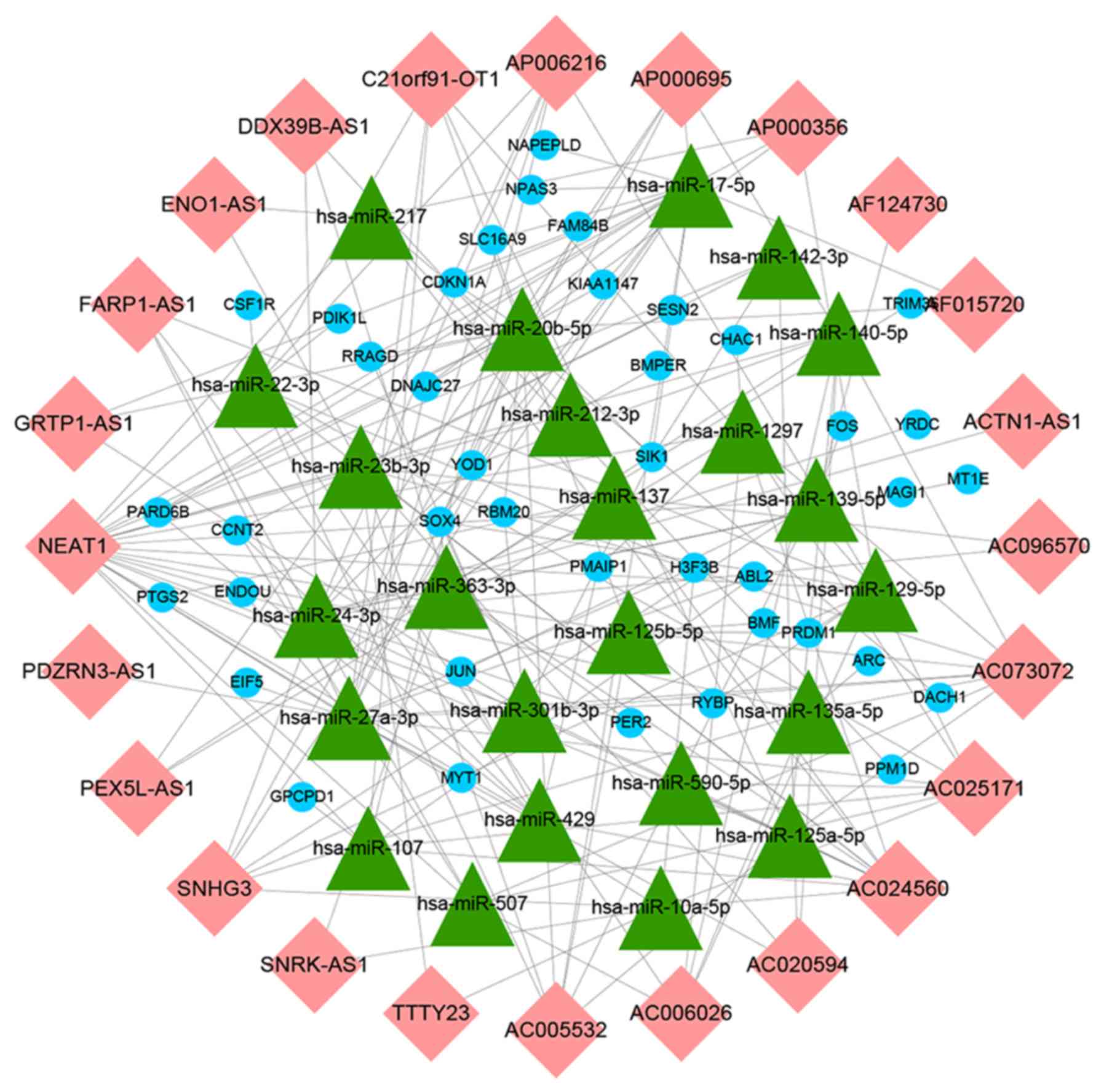
Construction of ceRNA network
To further understand how lncRNAs and miRNAs
regulate mRNA in HTMCs under oxidative stress, an lncRNA-miRNA-mRNA
network was designed. Using mircode, 24 miRNAs that interacted with
24 lncRNAs were predicted (Table
II). It was found that the lncRNA, nuclear enriched abundant
transcript 1 (NEAT1), was downregulated the most and was predicted
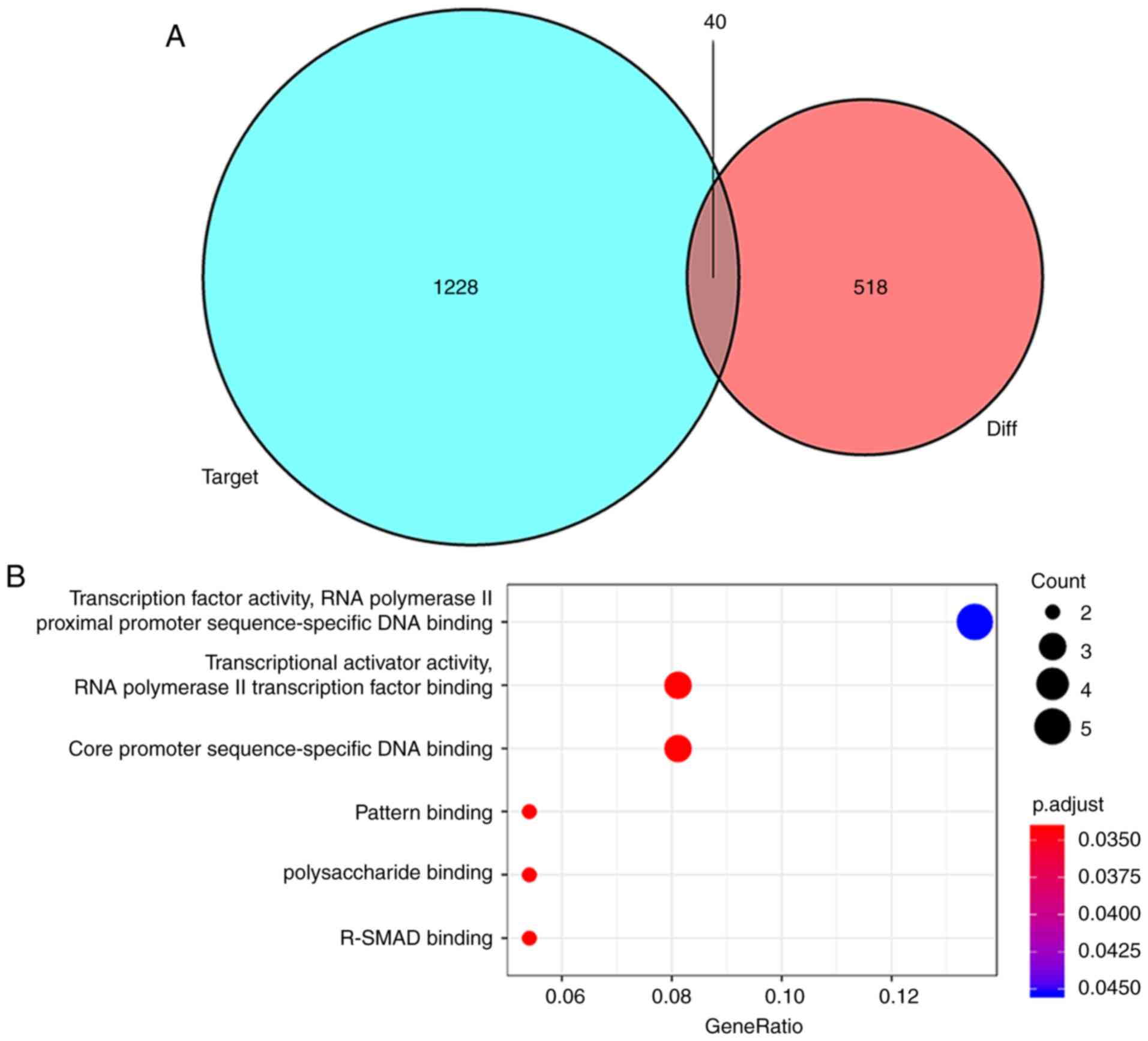
to interact with 21 miRNAs. A total of 1,228 mRNA targets of these
24 miRNAs were predicted using miRDB, miRTarBase and TargetScan
databases. miRNA targets of mRNAs that were not included in the 518
differentially expressed mRNAs were discarded and 40 mutual mRNAs
were preserved (Fig. 4A; Table III). Among the mRNAs that were
preserved, the expressions of 27 mRNAs, including ABL2, activity
regulated cytoskeleton associated protein, fos proto-oncogene, AP-1
transcription factor subunit and
phorbol-12-myristate-13-acetate-induced protein 1 were upregulated,
and that of 13 mRNAs, including dachshund family transcription
factor 1, KIAA1147, membrane associated guanylate kinase, WW and
PDZ domain containing 1, and SOX4, were downregulated (Table SII). Moreover, the GO enrichment
analysis of mRNAs that were a part of the ceRNA network
demonstrated the role of transcription factor activity, RNA
polymerase II transcription factor binding, core promoter
sequence-specific DNA binding and pattern binding (Fig. 4B). Finally, based on these
lncRNA-miRNA and miRNA-mRNA interactions, a ceRNA network
containing 24 lncRNAs, 24 miRNAs and 40 mRNAs was constructed and
visualized using Cytoscape v3.7.0 (Fig. 5).
 | Table II.lncRNAs and target miRNAs in the
competing endogenous RNA network. |
Table II.
lncRNAs and target miRNAs in the
competing endogenous RNA network.
| Key lncRNAs | miRNAs |
|---|
| AC005532 | miR-125a-5p,
miR-125b-5p, miR-139-5p, miR-17-5p, miR-20b-5p, miR-22-3p,
miR-23b-3p |
| AC006026 | miR-129-5p,
miR-1297, miR-135a-5p, miR-140-5p, miR-22-3p, miR-507 |
| AC020594 | miR-140-5p,
miR-142-3p, miR-23b-3p, miR-27a-3p, miR-107 |
| AC024560 | miR-10a-5p,
miR-1297, miR-137, miR-217, miR-22-3p, miR-23b-3p, miR-24-3p,
miR-301b-3p, miR-363-3p, miR-429, miR-590-5p |
| AC025171 | miR-107,
miR-10a-5p, miR-129-5p, miR-1297, miR-22-3p, miR-27a-3p, miR-507,
miR-590-5p |
| AC073072 | miR-125a-5p,
miR-125b-5p, miR-1297, miR-135a-5p, miR-140-5p, miR-23b-3p,
miR-27a-3p, miR-301b-3p, miR-507 |
| AC096570 | miR-23b-3p,
miR-24-3p |
| ACTN1-AS1 | miR-24-3p |
| AF015720 | miR-17-5p,
miR-20b-5p |
| AF124730 | miR-139-5p |
| AP000356 | miR-129-5p,
miR-17-5p, miR-20b-5p, miR-217 |
| AP000695 | miR-140-5p,
miR-17-5p, miR-20b-5p, miR-24-3p, miR-27a-3p |
| AP006216 | miR-107,
miR-129-5p, miR-22-3p, miR-23b-3p, miR-24-3p, miR-27a-3p |
| C21orf91-OT1 | miR-107,
miR-125a-5p, miR-125b-5p, miR-129-5p, miR-22-3p, miR-27a-3p |
| DDX39B-AS1 | miR-212-3p,
miR-24-3p, miR-363-3p |
| ENO1-AS1 | miR-217,
miR-23b-3p |
| FARP1-AS1 | miR-129-5p,
miR-27a-3p, miR-429, miR-507 |
| GRTP1-AS1 | miR-17-5p,
miR-20b-5p, miR-301b-3p |
| NEAT1 | miR-107,
miR-10a-5p, miR-125a-5p, miR-125b-5p, miR-129-5p, miR-1297,
miR-135a-5p, miR-139-5p, miR-140-5p, miR-142-3p, miR-17-5p,
miR-20b-5p, miR-212-3p, miR-217, miR-22-3p, miR-23b-3p, miR-24-3p,
miR-27a-3p, miR-301b-3p, miR-429, miR-507 |
| PDZRN3-AS1 | miR-27a-3p |
| PEX5L-AS1 | miR-1297,
miR-140-5p, miR-23b-3p |
| SNHG3 | miR-10a-5p,
miR-129-5p, miR-135a-5p, miR-139-5p, miR-17-5p, miR-20b-5p,
miR-24-3p |
| SNRK-AS1 | miR-10a-5p,
miR-363-3p |
| TTTY23 | miR-10a-5p,
miR-23b-3p |
 | Table III.miRNAs and target mRNAs in the
competing endogenous RNA network. |
Table III.
miRNAs and target mRNAs in the
competing endogenous RNA network.
| miRNA | mRNA |
|---|
| miR-107 | ABL2, GPCPD1 |
| miR-10a-5p | H3F3B |
| miR-125a-5p | PRDM1, BMF, PRDM1,
RYBP |
| miR-129-5p | PRDM1, RYBP,
SOX4 |
| miR-1297 | CHAC1, PMAIP1 |
| miR-135a-5p | ARC |
| miR-137 | PTGS2 |
| miR-139-5p | FOS, JUN |
| miR-140-5p | PRDM1, YOD1 |
| miR-142-3p | CCNT2, SIK1 |
| miR-17-5p | CDKN1A, DNAJC27,
KIAA1147, NAPEPLD, NPAS3, PARD6B, RRAGD, SIK1, SLC16A9, SOX4,
YOD1 |
| miR-20b-5p | CDKN1A, DNAJC27,
KIAA1147, PARD6B, RBM20, RRAGD, SIK1, SLC16A9, SOX4, YOD1 |
| miR-212-3p | BMPER |
| miR-217 | DACH1, PPM1D |
| miR-22-3p | CSF1R, H3F3B,
PDIK1L |
| miR-23b-3p | SESN2 |
| miR-24-3p | MAGI1, MT1E, PER2,
SESN1, YOD1, YRDC |
| miR-27a-3p | ABL2, DNAJC27,
EIF5, ENDOU, FAM84B, H3F3B, MYT1, RYBP |
| miR-301b-3p | RBM20, RRAGD, SIK1,
SOX4 |
| miR-363-3p | PER2, SOX4,
TRIM36 |
| miR-429 | CCNT2, JUN, PARD6B,
PMAIP1 |
| miR-507 | CCNT2 |
| miR-590-5p | PER2 |
Discussion
lncRNAs can act as ceRNA and suppress miRNA
function, thus preventing miRNAs from binding to and interacting
with target mRNAs (20). In the
present analysis, 24 dysregulated lncRNAs that have miRNA targets
were identified. Among these, NEAT1 was downregulated the most, and
was predicted to interact with 21 miRNAs. NEAT1 plays an important
role in the formation of nuclear structure (29). A previous study suggested that the
downregulation of NEAT1 lowers cell viability in hepatocellular
carcinoma and esophageal squamous cell carcinoma by serving as a
ceRNA and influencing the expression of miR-129-5p (30,31).
Another study showed that NEAT1 regulates the miR-107-mediated
expression of cyclin-dependent kinase 6 in laryngeal squamous cell
cancer (32). A number of studies
have demonstrated that the expression of NEAT1 can be activated by
NF-κB and p53, and activate PI3K/AKT pathways (33–35).
However, the function of NEAT1 in the pathogenesis of glaucoma has
not yet been investigated. Apart from directly participating in
biological processes, lncRNAs play an important role in various
signal pathways, such as NF-κB, PI3K/AKT, Notch and Wnt/β-catenin
(36–40). Multiple types of NF-κB-associated
lncRNAs, including NEAT1, have been reported to interact with
different sites in the NF-κB/IκB complex in order to orchestrate
opposite effects. For example, the interaction of NF-κB with NF-κB
interacting lncRNA (NKILA) can result in the binding and inhibition
of the phosphorylation site of IκB, thus suppressing NF-κB
activation (37). NKILA can, in
turn, be upregulated by NF-κB (37). p50-associated cyclooxygenase-2
extragenic RNA enhances NF-κB signaling by combining with free p50
to decrease the concentration of p50-p50 heterodimers and increase
the concentration of p65-p50 heterodimers (40). lncRNA AK023948 was found to
stabilize p85 by interacting with p85 and DExH-box helicase 9, as
well as elevating AKT activity in the PI3K/AKT pathway (38).
miRNAs, such as miR-107, miR-137, miR-22 and
miR-590, have been reported to regulate several biochemical
pathways in glaucoma (41). HTMCs
injured by oxidative stress can release miR-107, miR-149, miR-21
and miR-450 into the aqueous humor, which then travels via the
uveoscleral pathway to the peripapillary retina, thus resulting in
the transmission of damaging signals to the optic nerve (42). miR-27a was demonstrated to play a
critical role in cell proliferation, differentiation and apoptosis
by regulating f-box and WD repeat domain containing 7 (43). The upregulation of miR-24 can be a
limitation in the activation of transforming growth factor β
(TGFβ)1 under mechanical stress, which can in turn contribute to
the pathogenesis of glaucoma (44). Additionally, miR-23-27-24 gene
clusters can enhance angiogenesis (45) and inhibit neuronal apoptosis by
suppressing the expression of apoptotic peptidase activating factor
1 (46). These miRNAs were
detected in the present analysis, suggesting that miRNAs in the
ceRNA network play an important role in the pathological changes
associated with glaucoma.
The functional enrichment of differentially
expressed mRNAs was mainly associated with the activities of
cytokines and cytokine receptors. Previously, it was demonstrated
that the TGFβ pathway plays an important role in the regulation of
aqueous humor by promoting the expression of genes related to cell
contractility and senescence of TM (47). These processes are coordinated via
numerous signaling molecules and crosstalk communication between
various pathways, including MAPK, JNKs and Rho GTPase signaling
pathways (47). Other pathways,
such as the brain-derived neurotrophic factor, JNK, PI3K/AKT, PTEN,
Bcl-2, caspase and calcium-calpain pathways have been shown to
participate in the pathological process of glaucoma (48). Consistent with previous studies,
the pathway analysis that was performed also demonstrated an
enrichment of these cytokine-cytokine receptor activities and the
MAPK pathway, thus indicating the effect of these signaling
pathways in the progression of glaucoma.
The majority of lncRNAs target more than one miRNA,
and miRNAs target a number of mRNAs as well (Tables II and III). For example, lncRNAs such as
NEAT1, AC005532, AC006026, AC024560, AC025171, AC073072, AP006216,
C21orf91-OT1 and small nucleolar RNA host gene 3 all interact with
more than 6 miRNAs (Table II),
implying the complex interaction between these genes in biological
processes. There is a similar relationship between miRNAs and
mRNAs, such as miR-125a-5p, miR-17-5p, miR-20b-5p, miR-27a-3p,
miR-301b-3p and miR-429, thus creating a comprehensive network of
lncRNA-miRNA-mRNA. By establishing the ceRNA network, associated
genes were linked together.
The most crucial metabolic process in the body is
the utilization of molecular oxygen and the formation of adenosine
triphosphate. ROS are generated from the metabolic pathways, such
as the tricarboxylic acid cycle and the respiratory chain.
Oxidative stress occurs once ROS overloads in the tissue (7). H2O2 was used to
mimic the oxidative stress condition in glaucoma (12). The interplay of lncRNAs and
signaling pathways has been partly demonstrated in previous
studies. When oxidative stress occurs, PI3K/Akt, NF-κB and p53
signaling pathways are directly activated by ROS, thus causing
damage to ocular structures (7,9). The
ROS regulate lncRNAs, and some lncRNAs influence these pathways
(33–40). A total of 70 differentially
expressed lncRNAs were obtained, of which 24 participate in the
lncRNA-miRNA-mRNA interaction network. As a ‘sponge’ of miRNA,
lncRNAs play an important role in this network.
A total of 24 lncRNAs, 24 miRNAs and 40 mRNAs made
up the ceRNA network. As stated before, the functions and
associations of some remarkable RNAs have been discussed. lncRNA
could not only have effects on various signaling pathways, but also
combine with miRNAs to regulate the expression of mRNA, which might
lead to morphotype or property changes of TM under oxidative
stress. By constructing the ceRNA network, it was possible to
clearly demonstrate the complex association between these RNAs and
different signaling pathways. Thus, this contributed to the
understanding of their interactions and the pathological processes
of TM under oxidative stress. Once a typical association to explain
pathogenesis is discovered, a new treatment theory for glaucoma
could be found.
There were also limitations in the present analysis.
Due to the value of HTMCs, the sample size was not large. This also
limits the ability to carry out cell experiments to confirm lncRNA
function and lncRNA-miRNA-mRNA interactions. There are plans to
conduct further experiments, providing that HTMCs are available, in
order to explore the role of lncRNA-associated ceRNA networks in
glaucoma.
To conclude, the differential expression profiles of
lncRNAs were demonstrated, and a lncRNA-associated ceRNA network
was constructed in HTMCs under oxidative stress. This could
contribute to finding a new pathological mechanism or a potential
therapeutic target for glaucoma.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YY, KY and HZ conceived and designed the study. KY,
YY, PJ, CD and FL contributed analysis tools and carried out the
data analysis. YY, KY and CD wrote the paper. KY, YY and FL
reviewed and edited the manuscript. All authors read and approved
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
miRNA
|
microRNA
|
|
ceRNA
|
competing endogenous RNA
|
|
HTMC
|
human trabecular meshwork cell
|
|
IOP
|
intraocular pressure
|
|
TM
|
trabecular meshwork
|
|
ROS
|
reactive oxygen species
|
|
MAPK
|
mitogen-activated protein kinase
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Quigley HA: Glaucoma. Lancet.
377:1367–1377. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quigley HA and Broman AT: The number of
people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol.
90:262–267. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tham YC, Li X, Wong TY, Quigley HA, Aung T
and Cheng CY: Global prevalence of glaucoma and projections of
glaucoma burden through 2040: A systematic review and
meta-analysis. Ophthalmology. 121:2081–2090. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weinreb RN, Aung T and Medeiros FA: The
pathophysiology and treatment of glaucoma: A review. JAMA.
311:1901–1911. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McDougal DH and Gamlin PD: Autonomic
control of the eye. Compr Physiol. 5:439–473. 2015.PubMed/NCBI
|
|
6
|
Braunger BM, Fuchshofer R and Tamm ER: The
aqueous humor outflow pathways in glaucoma: A unifying concept of
disease mechanisms and causative treatment. Eur J Pharm Biopharm.
95:173–181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bayir H: Reactive oxygen species. Crit
Care Med. 33 (12 Suppl):S498–S501. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martindale JL and Holbrook NJ: Cellular
response to oxidative stress: Signaling for suicide and survival. J
Cell Physiol. 192:1–15. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Izzotti A, Sacca SC, Longobardi M and
Cartiglia C: Sensitivity of ocular anterior chamber tissues to
oxidative damage and its relevance to the pathogenesis of glaucoma.
Invest Ophthalmol Vis Sci. 50:5251–5258. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sacca SC, Gandolfi S, Bagnis A, Manni G,
Damonte G, Traverso CE and Izzotti A: From DNA damage to functional
changes of the trabecular meshwork in aging and glaucoma. Ageing
Res Rev. 29:26–41. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Izzotti A, Sacca SC, Cartiglia C and De
Flora S: Oxidative deoxyribonucleic acid damage in the eyes of
glaucoma patients. Am J Med. 114:638–646. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou L, Li Y and Yue BY: Oxidative stress
affects cytoskeletal structure and cell-matrix interactions in
cells from an ocular tissue: The trabecular meshwork. J Cell
Physiol. 180:182–189. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nature reviews. Genetics.
10:155–159. 2009.PubMed/NCBI
|
|
14
|
Caley DP, Pink RC, Trujillano D and Carter
DR: Long noncoding RNAs, chromatin, and development.
ScientificWorldJournal. 10:90–102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Akhade VS, Pal D and Kanduri C: Long
noncoding RNA: Genome organization and mechanism of action. Adv Exp
Med Biol. 1008:47–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou X and Xu J: Identification of
Alzheimer's disease-associated long noncoding RNAs. Neurobiol
Aging. 36:2925–2931. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gstir R, Schafferer S, Scheideler M,
Misslinger M, Griehl M, Daschil N, Humpel C, Obermair GJ,
Schmuckermair C, Striessnig J, et al: Generation of a
neuro-specific microarray reveals novel differentially expressed
noncoding RNAs in mouse models for neurodegenerative diseases. RNA.
20:1929–1943. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie Y, Hayden MR and Xu B: BDNF
overexpression in the forebrain rescues Huntington's disease
phenotypes in YAC128 mice. J Neurosci. 30:14708–14718. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wan P, Su W and Zhuo Y: The role of long
noncoding RNAs in neurodegenerative diseases. Mol Neurobiol.
54:2012–2021. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43((Database Issue)): D146–D152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar :
|
|
28
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sunwoo H, Dinger ME, Wilusz JE, Amaral PP,
Mattick JS and Spector DL: MEN epsilon/beta nuclear-retained
non-coding RNAs are up-regulated upon muscle differentiation and
are essential components of paraspeckles. Genome Res. 19:347–359.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fang L, Sun J, Pan Z, Song Y, Zhong L,
Zhang Y, Liu Y, Zheng X and Huang P: Long non-coding RNA NEAT1
promotes hepatocellular carcinoma cell proliferation through the
regulation of miR-129-5p-VCP-IκB. Am J Physiol Gastrointest Liver
Physiol. 313:G150–G156. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Chen D, Gao X, Li X and Shi G:
LncRNA NEAT1 regulates cell viability and invasion in esophageal
squamous cell carcinoma through the miR-129/CTBP2 axis. Dis
Markers. 2017:53146492017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H,
Xuan L, Wang X, Tian L, Sun Y, et al: Long noncoding RNA NEAT1
promotes laryngeal squamous cell cancer through regulating
miR-107/CDK6 pathway. J Exp Clin Cancer Res. 35:222016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cheng N and Guo Y: Long noncoding RNA
NEAT1 promotes nasopharyngeal carcinoma progression through
regulation of miR-124/NF-κB pathway. Onco Targets Ther.
10:5843–5853. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fuschi P, Carrara M, Voellenkle C,
Garcia-Manteiga JM, Righini P, Maimone B, Sangalli E, Villa F,
Specchia C, Picozza M, et al: Central role of the p53 pathway in
the noncoding-RNA response to oxidative stress. Aging (Albany NY).
9:2559–2586. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu H, Li J and Zhou ZG: NEAT1 promotes
cell proliferation in multiple myeloma by activating PI3K/AKT
pathway. Eur Rev Med Pharmacol Sci. 22:6403–6411. 2018.PubMed/NCBI
|
|
36
|
Zhang A, Xu M and Mo YY: Role of the
lncRNA-p53 regulatory network in cancer. J Mol Cell Biol.
6:181–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X,
Lin L, Yao H, Su F, Li D, et al: A cytoplasmic NF-κB interacting
long noncoding RNA blocks IκB phosphorylation and suppresses breast
cancer metastasis. Cancer Cell. 27:370–381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Koirala P, Huang J, Ho TT, Wu F, Ding X
and Mo YY: LncRNA AK023948 is a positive regulator of AKT. Nat
Commun. 8:144222017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Trimarchi T, Bilal E, Ntziachristos P,
Fabbri G, Dalla-Favera R, Tsirigos A and Aifantis I: Genome-wide
mapping and characterization of Notch-regulated long noncoding RNAs
in acute leukemia. Cell. 158:593–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Romano GL, Platania CB, Forte S, Salomone
S, Drago F and Bucolo C: MicroRNA target prediction in glaucoma.
Prog Brain Res. 220:217–240. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Izzotti A, Ceccaroli C, Longobardi MG,
Micale RT, Pulliero A, La Maestra S and Saccà SC: Molecular damage
in glaucoma: From anterior to posterior eye segment. The MicroRNA
role. Microrna. 4:3–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang L, Ye X, Liu Y, Wei W and Wang Z:
Aberrant regulation of FBW7 in cancer. Oncotarget. 5:2000–2015.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Luna C, Li G, Qiu J, Epstein DL and
Gonzalez P: MicroRNA-24 regulates the processing of latent TGFβ1
during cyclic mechanical stress in human trabecular meshwork cells
through direct targeting of FURIN. J Cell Physiol. 226:1407–1414.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou Q, Gallagher R, Ufret-Vincenty R, Li
X, Olson EN and Wang S: Regulation of angiogenesis and choroidal
neovascularization by members of microRNA-23~27~24 clusters. Proc
Natl Acad Sci USA. 108:8287–8292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen Q, Xu J, Li L, Li H, Mao S, Zhang F,
Zen K, Zhang CY and Zhang Q: MicroRNA-23a/b and microRNA-27a/b
suppress Apaf-1 protein and alleviate hypoxia-induced neuronal
apoptosis. Cell Death Dis. 5:e11322014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pervan CL: Smad-independent TGF-β2
signaling pathways in human trabecular meshwork cells. Exp Eye Res.
158:137–145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gauthier AC and Liu J: Epigenetics and
signaling pathways in glaucoma. Biomed Res Int. 2017:57123412017.
View Article : Google Scholar : PubMed/NCBI
|