Introduction
Head and neck cancer (HNC) is the sixth most common
malignant tumor in the world, with the main pathological type being
head and neck squamous cell carcinoma (HNSCC) (1). HNSCC involves numerous organs, such
as the oral cavity, nasopharynx, oropharynx, larynx and salivary
glands. The pathogenesis of HNC has not been fully elucidated,
however, a number of studies have shown that smoking, drinking and
papillomavirus infection are important environmental factors for
HNC initiation (2,3). Patients with HNC have poor quality of
life, and the majority have difficulty in communication, breathing
and swallowing (4). Although the
effectiveness of treatment for patients with HNC has improved in
recent years, the prognosis is still poor (5). HNC remains one of the most important
diseases threatening life and health, and is a major health problem
that needs to be solved.
With in-depth research on non-coding RNAs and
increasing research on microRNAs, the relationship between
microRNAs and HNC prognosis has received unprecedented attention.
Numerous studies have reported that the abnormal expression of
microRNAs is closely related to the initiation, progression and
prognosis of various tumors (6–10),
including HNC (11,12). The screening and identification of
microRNAs related to the prognosis of HNC will provide important
clues for the prevention, intervention and treatment of the
high-risk population. Moreover, it is helpful to find new targets
for HNC-targeted therapy. The identification of potential
prognostic biomarkers in HNC, based on the analysis of microRNA
expression profiles and the exploration of appropriate
interventions, may improve the prognosis of patients with HNC.
The Cancer Genome Atlas (TCGA) database covers
abundant genetic information and detailed clinical information of
33 types of tumors. The database includes six genome-wide platforms
that assayed tumor DNA (exome sequencing, DNA methylation, and copy
number), RNA (mRNA and microRNA sequencing) and a cancer-relevant
set of proteins and phosphoproteins (13). This data can be directly downloaded
through the TCGA Data Portal (https://portal.gdc.cancer.gov/) and provide convenient
multidimensional omics data resources for tumor researchers
(14,15). The present study aimed to identify
microRNAs that were closely related to HNSCC prognosis by
integrating the genomic, transcriptomic and clinicopathological
information in the TCGA database. It is expected that the results
from the present study will provide clues for the diagnosis and
treatment of patients with HNC.
Materials and methods
Collection and analysis of microRNA
expression profiles of HNC tissues in the TCGA database to identify
prognostic microRNAs
The collection and analysis of differential
expression profiles of microRNAs between HNC tissues (525 cases)
and adjacent cancer tissues (44 cases) in the TCGA database
(http://cancergenome.nih.gov) were
performed through the TCGA Data Portal and R software (https://www.r-project.org, v3.5.1, logFC >2;
P<0.01). Univariate and multivariate Cox regression analyses
were further performed on microRNAs with differential expression to
determine the independent risk factors significantly associated
with patient survival using SPSS 22.0 (IBM Corp.). The identified
risk factors were then verified by survival and receiver operating
characteristic (ROC) curve analyses. To better predict prognosis, a
prognostic model (risk equation) was established: Risk
score=(−0.1597 × hsa-miR-99a) + (0.1871 × hsa-miR-499a) + (0.1033 ×
hsa-miR-1911), according to the risk coefficient of each microRNA
calculated by the multivariate Cox regression analysis.
Analysis of the effect of
clinicopathological characteristics on HNC prognosis
Univariate and multivariate Cox regression analyses
were performed on clinicopathological characteristics to analyze
their roles in HNC survival. These findings were then verified by
Kaplan-Meier survival analysis and calculated using the log-rank
test with GraphPad Prism7 (GraphPad Software Inc.).
Clinicopathological characteristics analyzed included age, gender,
tumor-node-metastasis (TNM) stage, Fuhrman grade, ethnicity,
radiotherapy, human papillomavirus (HPV) status and mutation
number.
Kyoto Encyclopedia of Genes and
Genomes (KEGG) and Gene Ontology (GO) analyses to explore the
signaling pathways related to HNC prognosis
HNC cases in TCGA database were divided into
high-risk and low-risk groups according the median risk score
calculated by the risk equation (cut-off value=1.03), and
subsequently a differential gene expression analysis was performed.
KEGG (https://www.genome.jp/kegg/) and GO
(http://geneontology.org/) analyses were performed
on the upregulated and downregulated genes to explore the signaling
pathways related to the microRNAs aforementioned using R cluster
Profiler package (16).
Results
Identification of three microRNAs as
independent risk factors in HNC prognosis
The differential expression profiles of microRNAs in
HNC tissues (525 cases) and adjacent tumor tissues (44 cases) in
the TCGA database (logFC >2; P<0.01) were analyzed. A total
of 89 differentially expressed microRNAs were found, of which 38
were upregulated and 51 were downregulated in HNC tissues compared
with those in adjacent tumor tissues. After excluding two HNC cases
that lacked clinicopathological data, 523 patients with HNC were
randomly divided into the test group (n=300) and the validation
group (n=223). The general clinicopathological data are shown in
Table I.
 | Table I.General clinicopathological data of
head and neck cancer cases in The Cancer Genome Atlas database. |
Table I.
General clinicopathological data of
head and neck cancer cases in The Cancer Genome Atlas database.
| Characteristic | Test series n=300
(%) | Validation series
n=223 (%) | Entire series n=523
(%) |
|---|
| Sex |
|
|
|
| Male | 209/297 (70.4) | 170/222 (76.6) | 379/519 (73.0) |
|
Female | 88/297 (29.6) | 52/222 (23.4) | 140/519 (27.0) |
| Age (years) |
|
|
|
| ≥60 | 171/297 (57.6) | 115/222 (51.8) | 286/519 (55.1) |
|
<60 | 126/297 (42.4) | 107/222 (48.2) | 233/519 (44.9) |
| TNM stage |
|
|
|
| I | 20/252 (7.9) | 7/198 (3.5) | 27/450 (6.00) |
| II | 50/252 (19.8) | 26/198 (13.1) | 76/450 (16.9) |
| III | 35/252 (13.9) | 43/198 (21.7) | 78/450 (17.3) |
| IV | 147/252 (58.3) | 122/198 (61.6) | 269/450 (59.8) |
| Fuhrman grade |
|
|
|
| I | 30/282 (10.6) | 33/215 (15.3) | 63/498 (12.7) |
| II | 172/283 (60.8) | 133/215 (61.9) | 305/498 (61.2) |
|
III | 76/283 (26.9) | 47/215 (21.9) | 123/498 (24.7) |
| IV | 5/283 (1.8) | 2/215 (0.9) | 7/498 (1.4) |
| Ethnicity |
|
|
|
|
Non-white | 32/288 (11.1) | 28/216 (13.0) | 60/504 (11.9) |
|
White | 256/288 (88.9) | 188/216 (87.0) | 444/504 (88.1) |
| Radiation |
|
|
|
| No | 94/255 (36.9) | 62/197 (31.5) | 156/452 (34.5) |
|
Yes | 161/255 (63.1) | 135/197 (68.5) | 296/452 (65.5) |
In the test group, 11 of the 89 differentially
expressed microRNAs were determined as risk factors through the
univariate Cox regression analysis, as shown in Table II. To establish a more accurate
prediction model, these 11 microRNAs were further analyzed by a
multivariate Cox regression analysis. A total of three microRNAs,
namely, hsa-miR-99a (P=0.0078), hsa-miR-499a (P=0.0023) and
hsa-miR-1911 (P=0.0304), were identified as independent risk
factors significantly associated with patient survival (Table III). These three microRNAs
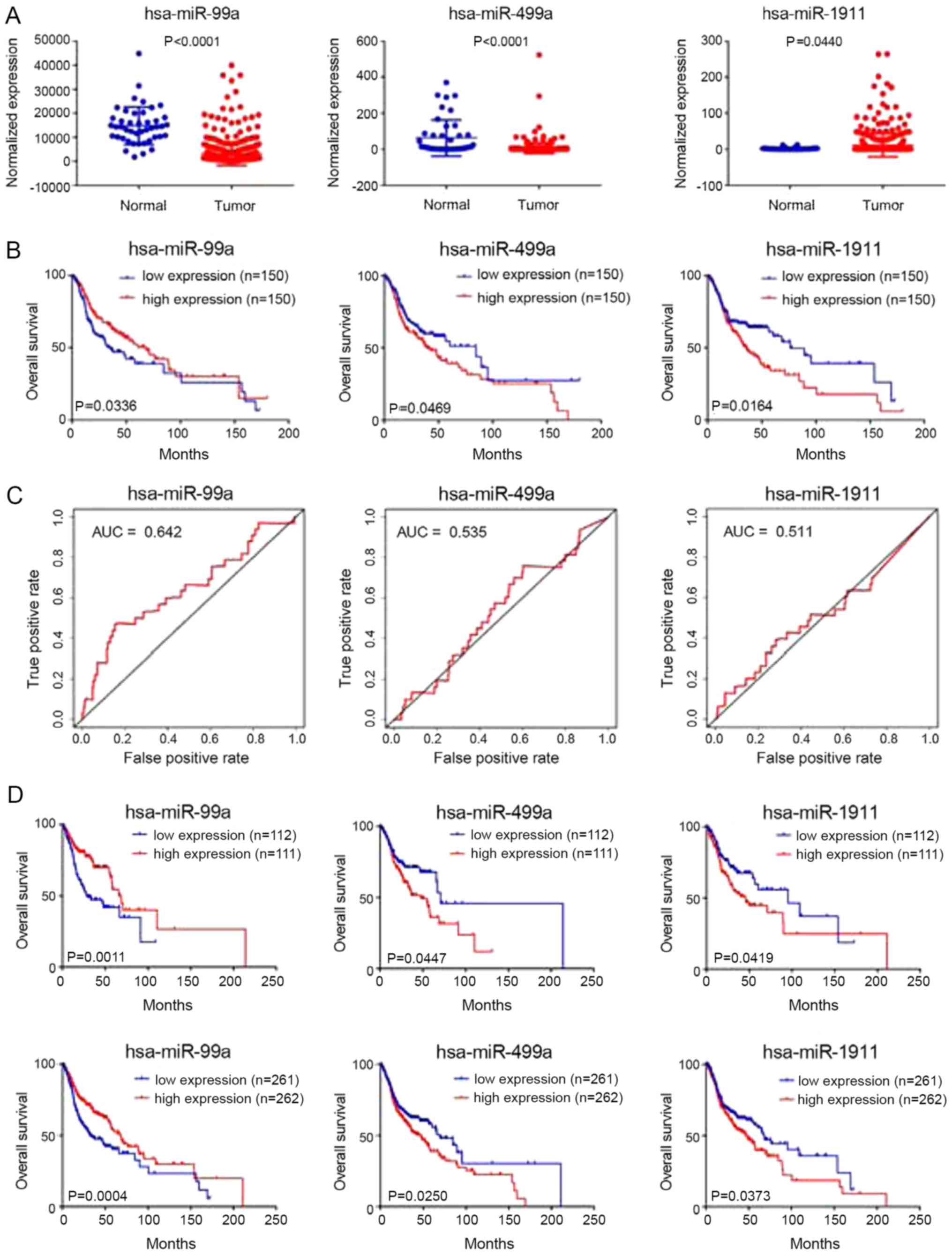
exhibited differential expression in HNC tissues and adjacent tumor
tissues (Fig. 1A). Survival and
ROC curve analyses were performed on the three microRNAs in the
test group, and the results showed that they are closely related to
prognosis (Fig. 1B and C).
 | Table II.Eleven microRNAs as risk factors of
head and neck cancer prognosis. |
Table II.
Eleven microRNAs as risk factors of
head and neck cancer prognosis.
| MicroRNA | Hazard ratio | z-score | P-value |
|---|
| hsa-miR-1911 | 1.134803307 | 3.034161031 | 0.002412056 |
| hsa-miR-4510 | 0.693911755 | −2.814364084 | 0.004887384 |
| hsa-miR-99a | 0.868724953 | −2.712379627 | 0.006680204 |
| hsa-miR-410 | 1.172183233 | 2.486637243 | 0.012895682 |
| hsa-miR-381 | 1.182780455 | 2.405357775 | 0.016156640 |
| hsa-miR-411 | 1.177476467 | 2.329398742 | 0.019837952 |
| hsa-miR-499a | 1.131263045 | 2.209846565 | 0.027115813 |
| hsa-miR-548f-1 | 1.117557677 | 2.176162956 | 0.029543078 |
| hsa-miR-1-1 | 1.056771872 | 2.151940931 | 0.031402007 |
| hsa-miR-1-2 | 1.053267713 | 2.000451927 | 0.045451486 |
| hsa-miR-4652 | 1.114090000 | 1.981273393 | 0.047560623 |
 | Table III.Three microRNAs as independent risk
factors in head and neck cancer prognosis. |
Table III.
Three microRNAs as independent risk
factors in head and neck cancer prognosis.
| MicroRNA | Regression
coefficient | Hazard ratio | P-value |
|---|
| hsa-miR-99a | −0.1597 | 0.8524 | 0.0078 |
| hsa-miR-499a | 0.1871 | 1.2057 | 0.0023 |
| hsa-miR-1911 | 0.1033 | 1.1088 | 0.0304 |
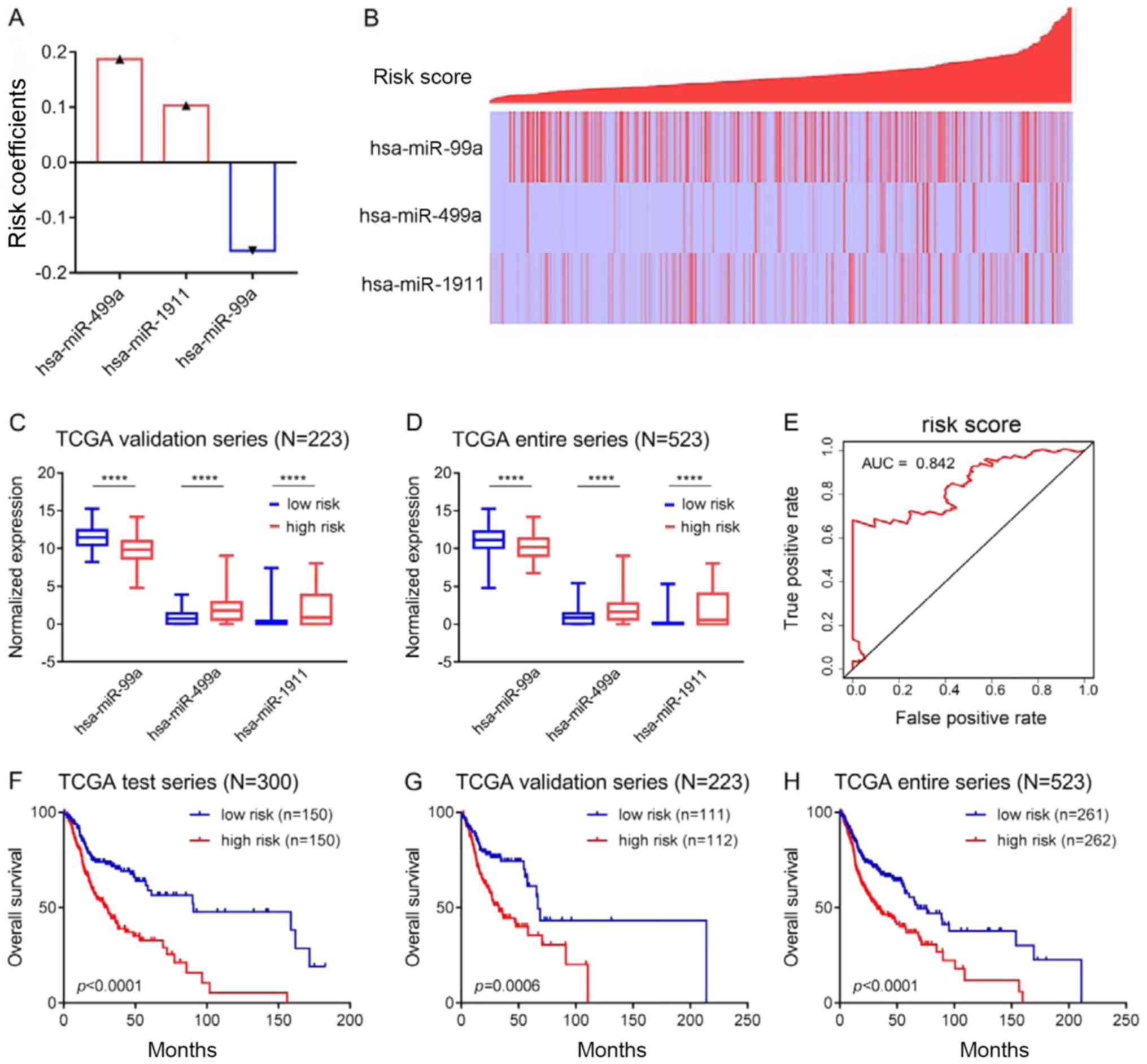
Establishing a combined prognostic
model/risk equation
To better predict prognosis, the three
aforementioned microRNAs were combined to establish a prognostic
model. The risk equation was as follows: Risk score=(−0.1597 ×
hsa-miR-99a) + (0.1871 × hsa-miR-499a) + (0.1033 × hsa-miR-1911),
according to the risk coefficient of each microRNA calculated by
the multivariate Cox regression analysis. A positive coefficient
indicates that high expression is associated with a poor prognosis
(for example, hsa-miR-499a and hsa-miR-1911), while a negative
coefficient indicates that high expression is associated with a
good prognosis (for example, hsa-miR-99a; Fig. 2A). Risk scores for each patient
were calculated and ranked. The expression of hsa-miR-499a,
hsa-miR-1911 and hsa-miR-99a and the distribution of the risk
scores are shown in Fig. 2B. As a
protective factor, hsa-miR-99a was highly expressed in the low-risk
group, while as risk factors, hsa-miR-499a and hsa-miR-1911 were
highly expressed in the high-risk group (Fig. 2B). Similar results were obtained in
the validation group and in the entire series, as shown in Fig. 2C and D. ROC curve (AUC=0.842;
Fig. 2E) and survival analyses
(Fig. 2F) were then carried out in
the test group. Patients in the high-risk group had significantly
shorter overall survival than those in the low-risk group
(P<0.0001, Fig. 2F). Moreover,
survival analysis was also carried out in the validation group and
in the entire series, and the results were similar to those of the
test group (Fig. 2G and H).
Risk score used to predict prognosis
is not affected by clinicopathological characteristics
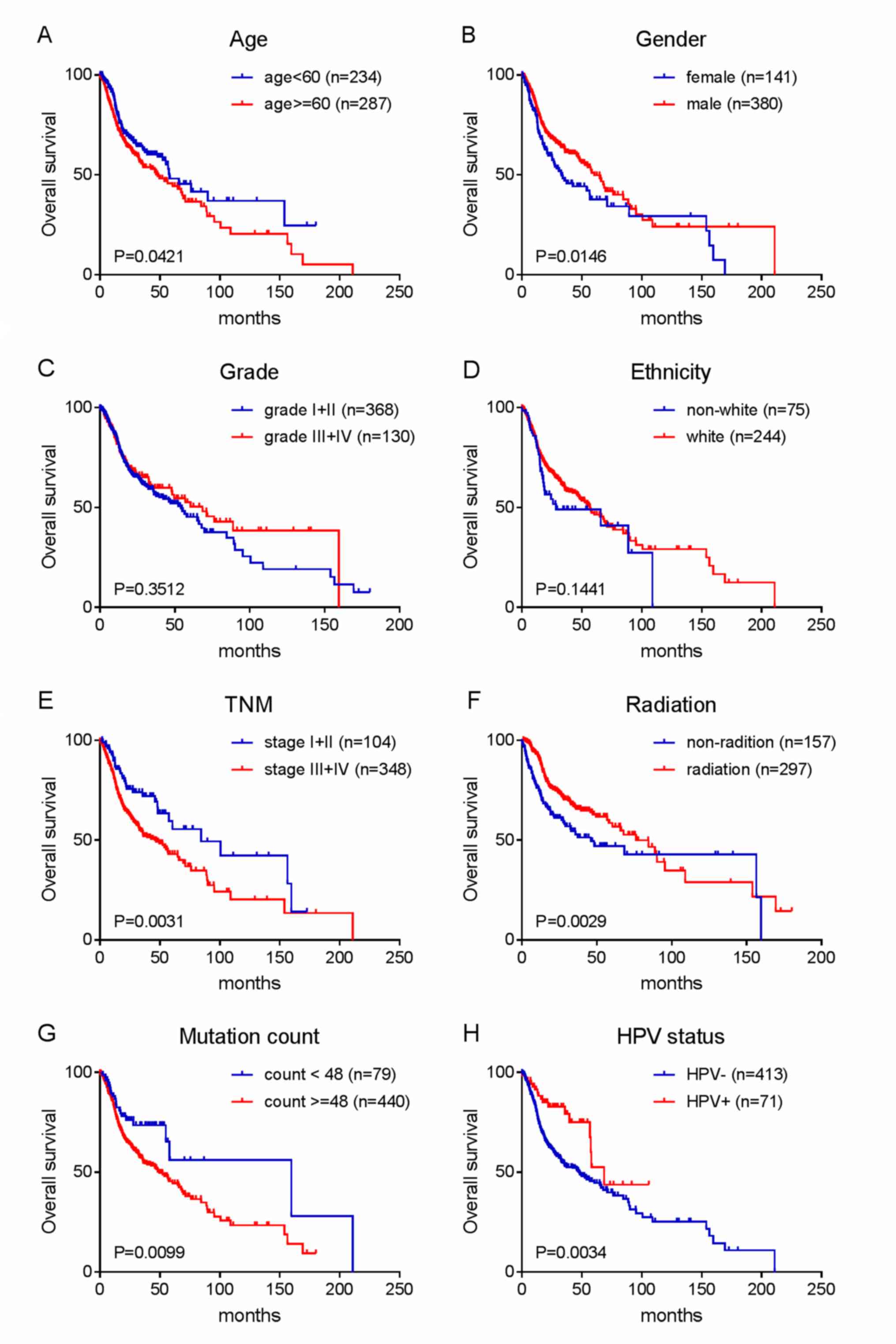
Considering that the prognosis of patients with HNC
is associated with clinicopathological characteristics, univariate
and multivariate Cox regression analyses were conducted in the test
group, the verification group and in the entire series based on the
risk score and clinicopathological data. The results showed that
age (entire series), gender (validation and entire series), TNM
stage (test, validation and entire series), mutation number (entire
series) and risk score (test, validation and entire series) were
risk factors in the univariate Cox regression analysis.
Contrastingly, radiotherapy (test and entire series) and HPV
infection (entire series) were protective factors. The multivariate
Cox regression analysis of these factors showed that the risk score
(test, validation and entire series) and TNM stage (test,
validation and entire series) were independent risk factors, while
radiotherapy (test, validation and entire series) was an
independent protective factor (Table
IV). Kaplan-Meier survival analysis was then performed on age,
gender, TNM stage, grade, ethnicity, radiotherapy, HPV status and
mutation number in the entire series to verify the above
conclusions. The results are shown in Fig. 3A-H.
 | Table IV.Risk score and clinicopathological
characteristics affect head and neck cancer prognosis. |
Table IV.
Risk score and clinicopathological
characteristics affect head and neck cancer prognosis.
| A, Test series
(n=300) |
|---|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Risk score (High
vs. low) | 2.435 | 1.691–3.508 | <0.0001 | 2.020 | 1.262–3.233 | 0.0030 |
| Age (≥60 vs.
<60) | 1.432 | 0.997–2.057 | 0.0520 | – | – | – |
| Gender (Male vs.
female) | 0.762 | 0.531–1.094 | 0.1410 | – | – | – |
| TNM (I, II, III,
IV) | 1.384 | 1.121–1.709 | 0.0020 | 1.797 | 1.329–2.431 | <0.0001 |
| Grade | 0.735 | 0.507–1.065 | 0.1040 | – | – | – |
| Ethnicity (White
vs. non-white) | 0.759 | 0.459–1.253 | 0.2810 | – | – | – |
| Radiation (Yes vs.
no) | 0.602 | 0.403–0.901 | 0.0140 | 0.395 | 0.242–0.644 | <0.0001 |
|
| B, Validation
series (n=223) |
|
|
| Univariate
analysis | Multivariate
analysis |
|
|
|
|
|
Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|
| Risk score (High
vs. low) | 2.006 | 1.298–3.101 | 0.0020 | 2.045 | 1.204–3.476 | 0.0080 |
| Grade | 0.960 | 0.597–1.544 | 0.8680 | – | – | – |
| Age (≥60 vs.
<60) | 1.188 | 0.776–1.816 | 0.4280 | – | – | – |
| Gender (Male vs.
female) | 0.616 | 0.389–0.977 | 0.0400 | – | – | – |
| TNM (I, II, III,
IV) | 1.530 | 1.113–2.104 | 0.0090 | 2.654 | 1.713–4.112 | <0.0001 |
| Ethnicity (White
vs. non-white) | 1.009 | 0.659–1.545 | 0.9670 | – | – | – |
| Radiation (Yes vs.
no) | 0.664 | 0.409–1.078 | 0.0980 | 0.333 | 0.188–0.591 | <0.0001 |
| Entire series
(n=523) |
|
|
|
|
|
|
|
|
| Univariate
analysis | Multivariate
analysis |
|
|
|
|
|
Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|
| Risk score (High
vs. low) | 1.869 | 1.422–2.457 | <0.0001 | 1.799 | 1.252–2.586 | 0.002 |
| Age (≥60 vs.
<60) | 1.327 | 1.009–1.745 | 0.043 | – | – | – |
| Gender (Male vs.
female) | 0.704 | 0.530–0.934 | 0.015 | 0.652 | 0.450–0.944 | 0.024 |
| Radiation (Yes vs.
no) | 0.628 | 0.462–0.855 | 0.003 | 0.368 | 0.246–0.549 | <0.0001 |
| HPV status
(Positive vs. negative) | 0.484 | 0.294–0.795 | 0.004 | – | – | – |
| Mutation count (≥48
vs. <48) | 1.797 | 1.144–2.821 | 0.011 | – | – | – |
| Ethnicity (White
vs. non-white) | 0.759 | 0.523–1.100 | 0.145 | – | – | – |
| Grade | 0.817 | 0.610–1.093 | 0.174 | – | – | – |
| TNM (I, II, III,
IV) | 1.421 | 1.192–1.694 | <0.0001 | 2.108 | 1.615–2.752 | <0.0001 |
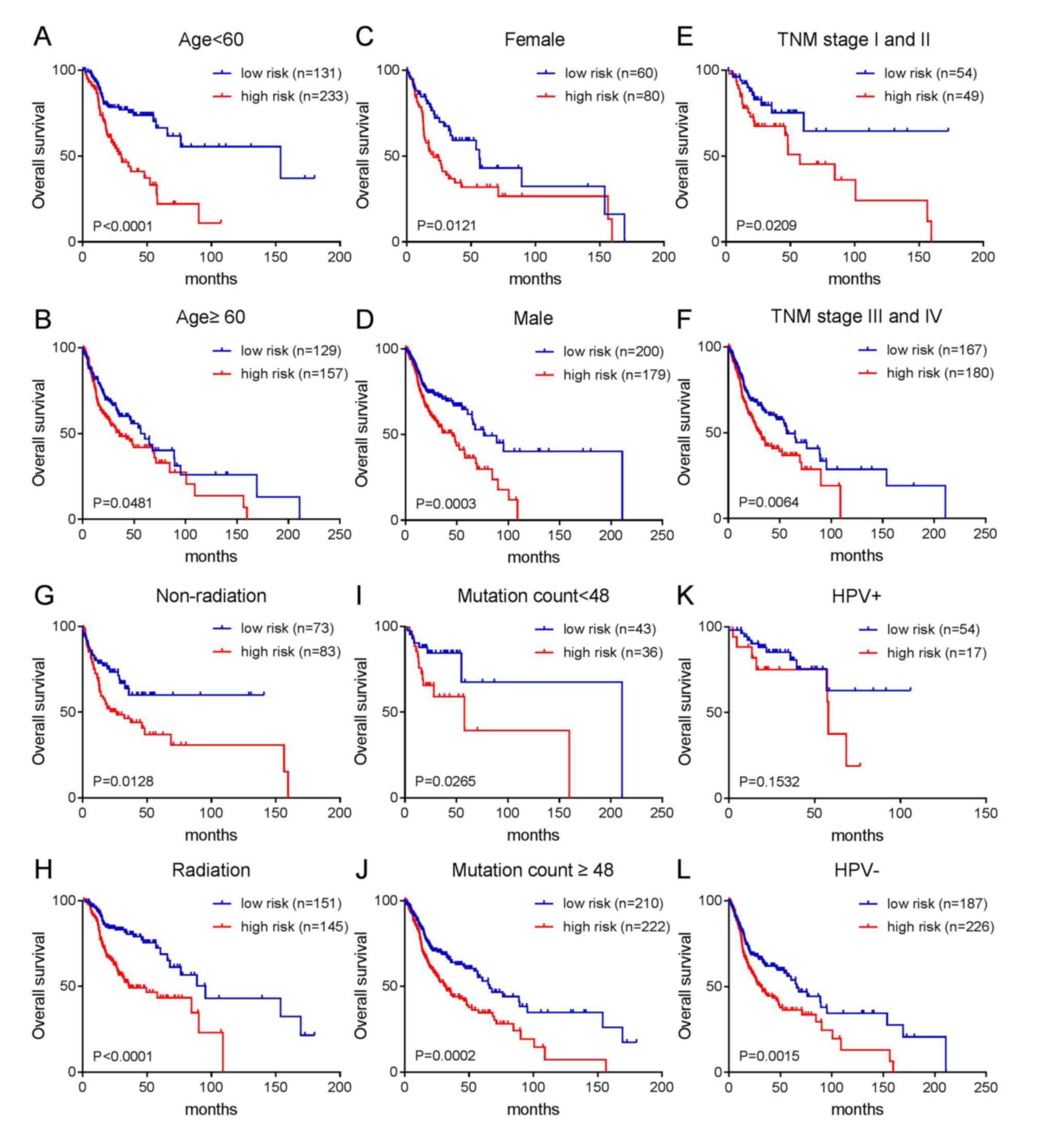
According to the aforementioned results, age,
gender, TNM stage, radiotherapy, HPV status and mutation number can
affect the prognosis of patients with HNC. Therefore, a further
stratified analysis was conducted to analyze whether these
clinicopathological characteristics can interfere with the
prediction effect of the risk score. The results are shown in
Fig. 4. With the exception of the
HPV-positive infection group, patients with a high-risk score in
each group had a poor prognosis, while patients with a low-risk
score had an improved prognosis. As shown in Fig. 4A-D, patients with high-risk scores
had a poor prognosis regardless of age (≥60 or <60) or gender
(female or male). As shown in Fig.
4E-H, patients with high-risk scores had short overall survival
regardless of whether the patients were in the early or late TNM
stage or whether they had received radiotherapy. As shown in
Fig. 4I and J, the two groups with
more or less mutations (divided by median mutation number) were
divided into two groups according to prognosis (good prognosis and
poor prognosis) by risk score. Similarly, patients with a high-risk
score in the HPV-negative group had a short survival time (Fig. 4L). Although the P-value in the
HPV-positive group was not significant, it can be seen from
Fig. 4K that patients with a
high-risk score had a shorter survival period, while those with a
low-risk score had a longer survival time.
Signaling pathways associated with HNC
prognosis
To explore the signaling pathways related to the
aforementioned microRNAs identified, the HNC cases in the TCGA
database were divided into high-risk and low-risk groups according
the risk score, calculated by the risk equation. A total of 556
differentially expressed genes, including 356 upregulated genes and
200 downregulated genes, were found between the high-risk and
low-risk groups through differential gene expression analysis
(logFC >1; P<0.05). The differentially expressed genes
between HNC tissues and adjacent cancer tissues in the TCGA
database (logFC >1; P<0.05) were also analyzed. The
upregulated genes in HNC tissues were intersected with the
upregulated genes of the high-risk group, and 106 upregulated genes
were obtained. Similarly, the downregulated genes in HNC tissues
were intersected with the downregulated genes of the high-risk
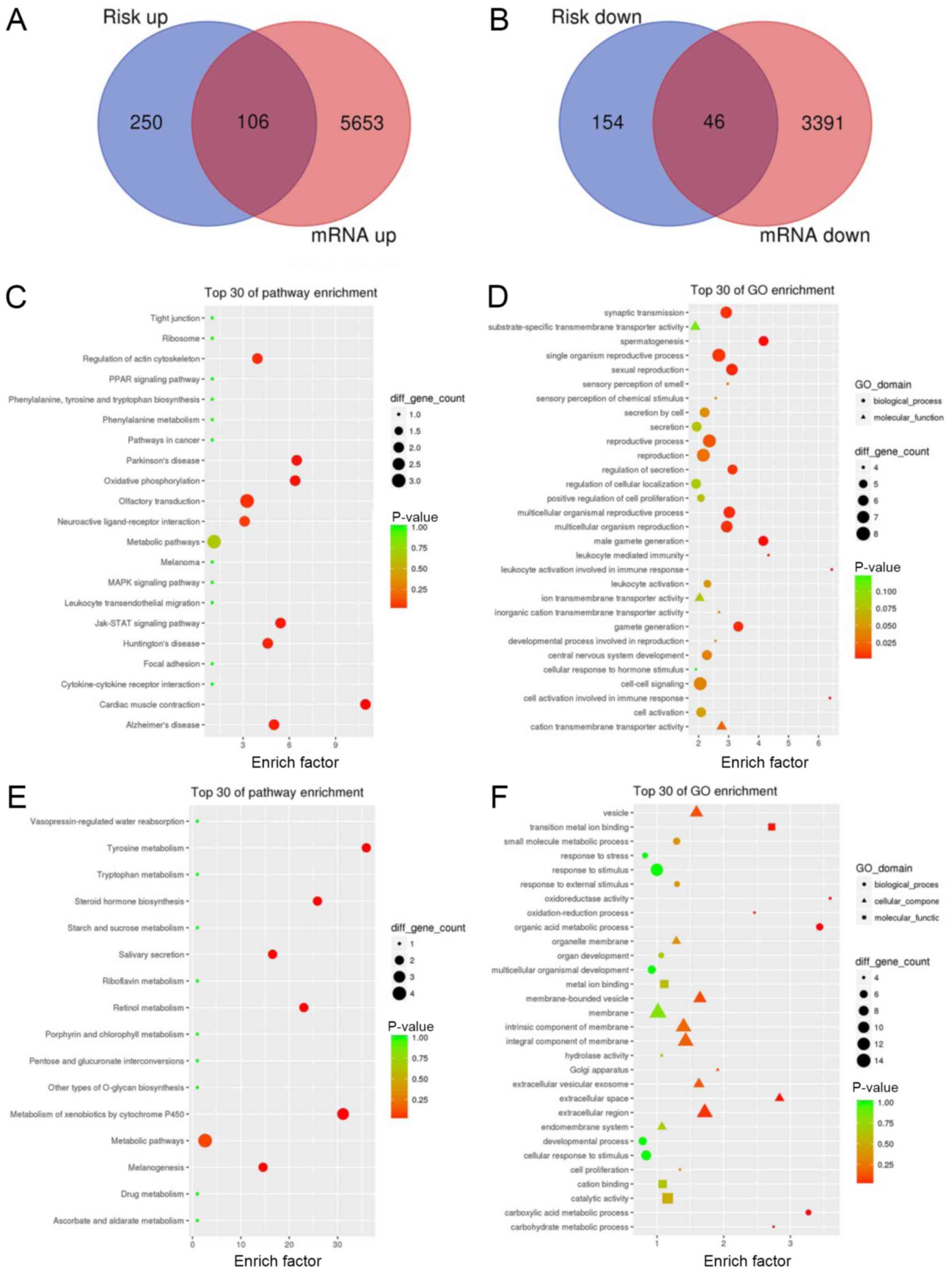
group, and 46 downregulated genes were obtained, as shown in
Fig. 5A and B. Then, KEGG and GO
analyses of the intersecting upregulated and downregulated genes
were performed, as shown in Fig.
5C-F. The results showed that genes in the high-risk group were
mainly enriched in ‘Regulation of action cytoskeleton’,
‘Parkinson's disease’, ‘JAK-STAT signaling pathway’ and others. The
genes in the low-risk group were mainly enriched in ‘Tyrosine
metabolism’, ‘hormone biosynthesis’ and others, which suggested
that The JAK-STAT signaling pathway and some metabolic pathways,
such as tryptophan metabolism may associated with HNC
prognosis.
Discussion
MicroRNAs are non-coding RNA 18–25 nucleotides in
length that can pair with a complementary base of a specific mRNA,
causing degradation or dysfunction of the mRNA (17). MicroRNAs play an important role in
the regulation of gene expression and can affect the
differentiation, proliferation, apoptosis, migration and invasion
of tumor cells (18–21). Numerous studies have shown that the
initiation, progression and metastasis of HNC are often accompanied
by the abnormal expression of specific microRNAs. Tran et al
found that 33 microRNAs were upregulated and 22 microRNAs were
downregulated in HNC cell lines compared with normal cell lines
(22). The present study further
explored microRNA expression profiles in HNC tissue specimens based
on the TCGA database, providing more sensitive, specific and
independent prognostic biomarkers for HNC. Therefore, the present
study provided new clues for individual treatment, which may
improve the prognosis of patients with HNC.
The TCGA project began in 2006 and aimed to create a
comprehensive ‘atlas’ of the cancer genome through large-scale
genome sequencing. To date, the TCGA has included multidimensional
genomics, epigenomics and proteomics data of ≥20 types of cancer
(15). The TCGA database is freely
accessible to the public, providing big data support for
researchers to explore new microRNAs and to study their functions.
The present study identified three microRNAs, namely, hsa-miR-99a,
hsa-miR-499a and hsa-miR-1911, which are significantly related to
the prognosis of patients with HNSCC. These findings were based on
microRNA expression profiles in the TCGA database, Cox regression
analyses and other bioinformatics analyses, and have also been
reported in other tumors (23).
hsa-miR-99a is a member of the miR-99 family, and
studies have shown that it can significantly inhibit the invasion
and migration of lung cancer and oral cancer cells (24,25).
However, the role of miR-99a in the invasion and migration of HNC
cells is rarely reported in the literature (26) In addition, it has been reported
that miR-99b in the miR-99 family may play an important role in the
initiation and progression of cervical cancer. The upregulated
expression of miR-99b can inhibit proliferation and induce
apoptosis of cervical cancer cells, and may serve as a therapeutic
target in cervical cancer. Studies have also reported that
hsa-miR-499a and hsa-miR-1911 are associated with cancer
susceptibility and prognosis, as in glioma (27,28).
Yang and Wang (29) found that
hsa-miR-1911 plays a key role in the occurrence and development of
glioma. Therefore, hsa-miR-99a, hsa-miR-499a and hsa-miR-1911 may
all play important roles in the genesis and development of HNSCC,
and an accurate clarification of their functions will be beneficial
for HNC treatment.
In conclusion, the present study screened three
microRNAs related to HNSCC prognosis by integrating genomic and
transcriptomic information in TCGA, and provided insight for
subsequent analysis and research. Moreover, it was found that age,
gender, TNM stage, radiotherapy, HPV status and mutation number can
affect the prognosis of patients with HNC by effectively analyzing
and summarizing clinicopathological data through stratified and
survival analyses. This study also had some limitations. In the
screening process of the data model, there was a certain extent of
false positive and false negative results, which need to be further
verified by similar experimental data to provide a reliable basis
for tumor research.
Overall, the present study revealed that
hsa-miR-99a, hsa-miR-499a and hsa-miR-1911 were closely associated
with HNC prognosis, and their combined risk equation may be more
effective in predicting HNC prognosis. The specific roles and
biological functions of the aforementioned miRNAs in the initiation
and progression of HNC require further investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81072197, 81470758,
81272368 and 81471755).
Availability of data and materials
The datasets generated and analyzed during the
current study are available in TCGA database (http://cancergenome.nih.gov).
Authors' contributions
MZ and CD designed the research. XW, ZY and YZ
carried out the research and drafted the manuscript. MH analyzed
and interpreted the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HNC
|
head and neck cancer
|
|
TCGA
|
The Cancer Genome Atlas
|
|
ROC
|
receiver operating characteristic
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
GO
|
Gene Ontology
|
|
HNSCC
|
head and neck squamous cell
carcinoma
|
|
HPV
|
human papillomavirus
|
References
|
1
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar
|
|
2
|
Siemianowicz K, Likus W, Dorecka M, Wilk
R, Dziubdziela W and Markowski J: Chemoprevention of head and neck
cancers: Does it have only one face? Biomed Res Int.
2018:90518542018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Porceddu SV and Haddad RI: Management of
elderly patients with locoregionally confined head and neck cancer.
Lancet Oncol. 18:e274–e83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lubek JE: Head and neck cancer research
and support foundations. Oral Maxillofac Surg Clin North Am.
30:459–469. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Budach V and Tinhofer I: Novel prognostic
clinical factors and biomarkers for outcome prediction in head and
neck cancer: A systematic review. Lancet Oncol. 20:e313–e26. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fan YH, Ye MH, Wu L, Lv SG, Wu MJ, Xiao B,
Liao CC, Ji QK, Chai Y and Zhu XG: Overexpression of miR-98
inhibits cell invasion in glioma cell lines via downregulation of
IKKepsilon. Eur Rev Med Pharmacol Sci. 19:3593–3604.
2015.PubMed/NCBI
|
|
7
|
Medina PP, Nolde M and Slack FJ: OncomiR
addiction in an in vivo model of microRNA-21-induced pre-B-cell
lymphoma. Nature. 467:86–90. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu Z, Yang Q, Zhang B, Wu W, Yuan F and
Zhu Z: miR-106b promotes metastasis of early gastric cancer by
Targeting ALEX1 in vitro and in vivo. Cell Physiol Biochem.
52:606–616. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen L, Hu W, Li G, Guo Y, Wan Z and Yu J:
Inhibition of miR-9-5p suppresses prostate cancer progress by
targeting StarD13. Cell Mol Biol Lett. 24:20–32. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Z, Zhang W, Mao J, Xu Z and Fan M:
miR-186-5p functions as a tumor suppressor in human osteosarcoma by
targeting FOXK1. Cell Physiol Biochem. 52:553–564. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen X, Cai S, Li B, Zhang X, Li W, Liang
H, Cao X, Wang L and Wu Z: MicroRNA21 regulates the biological
behavior of esophageal squamous cell carcinoma by targeting RASA1.
Oncol Rep. 41:1627–1637. 2019.PubMed/NCBI
|
|
12
|
Yu HX, Wang XL, Zhang LN, Zhang J and Zhao
W: MicroRNA-384 inhibits the progression of esophageal squamous
cell carcinoma through blockade of the LIMK1/cofilin signaling
pathway by binding to LIMK1. Biomed Pharmacother. 109:751–761.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res. 44:e712016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hoadley KA, Yau C, Hinoue T, Wolf DM,
Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et
al: Cell-of-origin patterns dominate the molecular classification
of 10,000 tumors from 33 types of cancer. Cell. 73:291–304.e6.
2018. View Article : Google Scholar
|
|
15
|
Deng M, Brägelmann J, Schultze JL and
Perner S: Web-TCGA: An online platform for integrated analysis of
molecular cancer data sets. BMC Bioinformatics. 17:722016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pouyanrad S, Rahgozar S and Ghodousi ES:
Dysregulation of miR-335-3p, targeted by NEAT1 and MALAT1 long
non-coding RNAs, is associated with poor prognosis in childhood
acute lymphoblastic leukemia. Gene. 692:35–43. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang R, Sun Y, Yu W, Yan Y, Qiao M, Jiang
R, Guan W and Wang L: Downregulation of miRNA-214 in
cancer-associated fibroblasts contributes to migration and invasion
of gastric cancer cells through targeting FGF9 and inducing EMT. J
Exp Clin Cancer Res. 38:20–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie X, Pan J, Han X and Chen W:
Downregulation of microRNA-532-5p promotes the proliferation and
invasion of bladder cancer cells through promotion of
HMGB3/Wnt/β-catenin signaling. Chem Biol Interact. 300:73–81. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu Y, Yin W, Yu ZH, Zhou YJ, Chi JR, Ge J
and Cao XC: miR-190 enhances endocrine therapy sensitivity by
regulating SOX9 expression in breast cancer. J Exp Clin Cancer Res.
38:222019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tran N, McLean T, Zhang X, Zhao CJ,
Thomson JM, O'Brien C and Rose B: MicroRNA expression profiles in
head and neck cancer cell lines. Biochem Biophys Res Commun.
358:12–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pérez Sayáns M, Chamorro Petronacci CM,
Lorenzo Pouso AI, Padín Iruegas E, Blanco Carrión A, Suárez
Peñaranda JM and García García A: Comprehensive genomic review of
TCGA head and neck squamous cell carcinomas (HNSCC). J Clin Med.
8:E18962019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun D, Lee YS, Malhotra A, Kim HK, Matecic
M, Evans C, Jensen RV, Moskaluk CA and Dutta A: miR-99 family of
MicroRNAs suppresses the expression of prostate-specific antigen
and prostate cancer cell proliferation. Cancer Res. 71:1313–1324.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi Y, Bo Z, Pang G, Qu X, Bao W, Yang L
and Ma Y: MiR-99a-5p regulates proliferation, migration and
invasion abilities of human oral carcinoma cells by targeting NOX4.
Neoplasma. 64:666–673. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mei LL, Qiu YT, Huang MB, Wang WJ, Bai J
and Shi ZZ: MiR-99a suppresses proliferation, migration and
invasion of esophageal squamous cell carcinoma cells through
inhibiting the IGF1R signaling pathway. Cancer Biomark. 20:527–537.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Margolis LM and Rivas DA: Potential role
of MicroRNA in the anabolic capacity of skeletal muscle with aging.
Exerc Sport Sci Rev. 46:86–91. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yagi Y, Ohkubo T, Kawaji H, Machida A,
Miyata H, Goda S, Roy S, Hayashizaki Y, Suzuki H and Yokota T:
Next-generation sequencing-based small RNA profiling of
cerebrospinal fluid exosomes. Neurosci Lett. 636:48–57. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang H and Wang Y: Five miRNAs considered
as molecular targets for predicting neuroglioma. Tumour Biol.
37:1051–1059. 2016. View Article : Google Scholar : PubMed/NCBI
|