Introduction
Cardiovascular disease is one of the leading causes
of death worldwide (1). During
cardiac surgery, myocardial ischemia/reperfusion (I/R) injury often
leads to adverse cardiovascular outcomes, including acute heart
failure due to severe hypoxia (2).
Multiple signaling pathways are involved in protective mechanisms
against myocardial I/R injury, including the PI3K/AKT and the
mitogen-activated protein kinase (MAPK) signaling pathways. The
PI3K/AKT pathway is an important intracellular signaling pathway
for the regulation of the cell cycle. Previous studies have
suggested that the PI3K/AKT signaling pathway mediates a protection
mechanism against myocardial I/R injury in rats (3,4).
Inhibition of the cardiac p38-MAPK pathway has been reported to
delay ischemic cell death and protect cardiac mitochondria from I/R
injury (5,6).
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNA molecule with a length of >200 nucleotides.
lncRNAs represent the most prevalent class of non-coding RNA, with
the majority of the human genome transcribed into lncRNAs (7). lncRNAs can act as important
regulators in a number of biological processes, including stem cell
lineage differentiation (8),
cancer development and metastasis (9,10)
and angiogenesis (11,12). The lncRNA H19 (H19) is located on
human chromosome 11 and expresses a 2.3 kb lncRNA transcribed from
the maternal inherited allele (13). Increased expression of H19 was
detected in rats following surgically induced myocardial I/R,
suggesting that H19 may have a role in the protective mechanisms
against I/R injury (14). Previous
studies have also indicated that H19 exerts protective effects
against hypoxia-induced injury in cardiomyocytes (15,16).
Nevertheless, to date, the functional role of H19 in myocardial
hypoxic injury has not been fully elucidated. The present study
aimed to investigate the effects and the molecular mechanism of H19
in response to hypoxia-induced I/R injury using a myocardial cell
model.
Materials and methods
Cell lines and culture
The rat cardiomyoblast cell line, H9c2, and 293T
cells were purchased from BeNa Culture Collection. Cells were
maintained in DMEM (Hyclone; GE Healthcare Life Sciences)
containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100
U/ml penicillin and 100 µg/ml streptomycin at 37°C and 5%
CO2 in an incubator.
Design and construction of H19
expressing/silencing lentivirus
To construct an H19 expressing lentiviral vector,
the H19 cDNA sequence (NR_002196.2) was synthesized by Sangon
Biotech Co., Ltd. and cloned into the pLVX-Puro vector following
the Lentivector User Manual (Takara Bio, Inc.). The following PCR
reaction mixture was used: 25 µl 2× Super Pfx MasterMix (Beijing
CoWin Biotech Co., Ltd.), 2.5 µl 10 µM forward primer, 2.5 µl 10 µM
reverse primer, 2.0 µl DNA template, 18 µl ddH2O. The
PCR was performed as follows: 98°C for 1 min, 30 cycles of 98°C for
5 sec, 58°C for 30 sec, and 72°C for 30 sec, followed by 72°C for 5
min. The pLVX-Puro vector was used as a negative control. To
construct an H19 silencing lentiviral vector, the following
oligonucleotides encoding a short hairpin RNA (shRNA) against H19
were designed and synthesized by Genscript and cloned into the
pGreenPuro™ vector (Addgene, Inc.): Sense,
5′-GATCCTGAATATGCTGCACTTTACAACTCGAGTTGTAAAGTGCAGCATATTCATTTTTG-3′
and antisense,
5′-AATTCAAAAAATGAATATGCTGCACTTTACAACTCGAGTTGTAAAGTGCAGCATATTCAG-3′.
The positive-sense and antisense strands harboring BamH and
EcoR digestion sites annealed to form a double-stranded
structure, which was then cloned into the pGreenPuro plasmid. The
lentiviral vector containing a non-silencing sequence was used as a
negative control. The sequence of constructs was confirmed by PCR
and Sanger sequencing.
To package lentiviruses, 293T packaging cells were
plated in a 10 cm plate. At 70% confluency, the cells were
co-transfected with 2.5 µg of the appropriate lentiviral vector
(pLVX-Puro-H19 vector, pLVX-Puro vector, pGreenPuro™ shH19 vector
or pGreenPuro™ non-silencing vector), 5 µg of psPAX and 5 µg of
pMD2G (Addgene, Inc.) using a Lipofectamine® 3000
transfection kit (Invitrogen; Thermo Fisher Scientific, Inc.). The
viral supernatants were harvested after 48 h and filtered with a
0.45 µm filter. The titer of the lentivirus was determined using a
lentivirus titration kit (Applied Biological Materials), following
the manufacturer's instructions.
Cell culture and infection
H9c2 cells were cultured in 6-well plates and
divided into six groups: Normal control, model control, H19
expression, blank expression, H19 interference and blank
interference groups. Cells in the H19 expression, blank expression,
H19 interference and blank interference groups were infected with
H19 overexpression, blank expression, H19-targeting shRNA and blank
interference lentiviruses, respectively, at a multiplicity of
infection of 1:100. Cells in the normal control group were then
cultured under normoxia (21% O2, 5% CO2 and
74% N2). Cells in all other groups were incubated in an incubator
containing 94% N2, 5% CO2 and 1%
O2 to stimulate hypoxia injury. After 48 h, cells in
each group were collected and subjected to subsequent analyses.
Apoptosis assay
At 48 h after transfection, apoptosis was determined
using a FITC Annexin V Apoptosis Detection kit I (BD Biosciences).
Briefly, cells were resuspended in 1X binding buffer, stained with
5 µl FITC Annexin V (BD Biosciences) and 10 µl of propidium iodide
(PI; BD Biosciences) in the dark for 10 min. The fluorescence of
cells was detected using a NovoCyte 2060R flow cytometer (ACEA
Biosciences, Inc.) at 488 nm within 1 h and analyzed using
NovoExpress software version 1.3.0 (ACEA Biosciences, Inc.).
Measurement of mitochondrial membrane
potential
At 48 h after transfection, the mitochondrial
membrane potential of cells was determined using a JC-1
mitochondrial membrane potential assay kit (Abcam). Briefly, cells
were resuspended in 500 µl of 1X incubation buffer and stained with
1 µl JC-1 at 37°C, 5% CO2 for 10 min. Cells were
collected by centrifugation at 420 × g at room temperature for 5
min and washed twice with 1X incubation buffer. The fluorescence of
cells was analyzed using a NovoCyte 2060R flow cytometer (ACEA
Biosciences, Inc.) at 488 nm within 1 h and analyzed using
NovoExpress software version 1.3.0 (ACEA Biosciences, Inc.).
Cell cycle analysis
At 48 h after transfection, the cell cycle was
analyzed using flow cytometry. Briefly, cells were fixed with 70%
ethanol for 24 h at 4°C and stained with 100 µl of PI (30 ng/ml)
for 30 min at 37°C in the dark. The percentage of the population in
G1, S or G2/M phase was analyzed using a NovoCyte 2060R flow
cytometer (ACEA Biosciences, Inc.) at 488 nm within 1 h and
analyzed using NovoExpress software version 1.3.0 (ACEA
Biosciences, Inc.).
Western blot analysis
Cells were collected at 48 h after transfection.
Total protein was extracted using RIPA lysis buffer (Beijing CoWin
Biotech Co., Ltd.) and quantified using bicinchoninic acid protein
assay kit (Beijing CoWin Biotech Co., Ltd.). Protein extracts (60
µg) were separated by SDS-PAGE using 10% gels and transferred onto
PVDF membranes. The membranes were blocked with 5% skim milk for 30
min at room temperature and incubated overnight at 4°C with the
following antibodies: β-actin (1:2,000; cat. no. TA-09; OriGene
Technologies, Inc.), AKT (1:2,000; cat. no. bs-0115R; BIOSS),
ERK1/2 (1:1,000; cat. no. bs-0022R; BIOSS), p38 (1:2,000; cat. no.
bs-0637R; BIOSS), PI3K (1:2,000; cat. no. ab191606; Abcam),
phosphorylated (p-)AKT (1:1,000; cat. no. ab38449; Abcam), p-p38
(1:1,000; cat. no. ab47363; Abcam), p-ERK1/2 (1:1,000; cat. no.
ab214362; Abcam) and p-PI3K (1:1,000; cat. no. ab127617; Abcam).
The membranes were washed with PBS and incubated with horseradish
peroxidase-conjugated goat anti-mouse (1:2,000; cat. no. ZB-2305;
OriGene Technologies, Inc.) or anti-rabbit (1:2,000; cat. no.
ZB-2305; OriGene Technologies, Inc.) secondary antibodies for 2 h
at room temperature. Protein bands were visualized using an ECL
Western blotting detection system (GE Healthcare). The relative
expression of proteins was quantified using ImageJ software version
1.52e (National Institutes of Health) with β-actin used as the
internal control.
Reverse transcription-quantitative PCR
(RT-qPCR)
Cells were collected 48 h after transfection. Total
RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and reverse transcribed into cDNA using
SuperScript II (Invitrogen; Thermo Fisher Scientific, Inc.) at
42°C. qPCR was performed using UltraSYBR mixture (Takara
Biotechnology Co., Ltd.) in an ABI 7500 System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The reactions conditions were as
follows: 95°C for 10 min followed by 40 cycles of 95°C for 10 sec,
58°C for 30 sec and 72°C for 30 sec. The following primers were
synthesized by Invitrogen (Thermo Fisher Scientific, Inc.) and used
in the PCR: H19 forward, 5′-GTGGGACACTGCCGTAGAA-3′ and reverse,
5′-CAGGAAAGGAGGAAGAAGAAAA-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Data was analyzed using the 2−ΔΔCq method (17). The relative expression of lncRNA
H19 was calculated using U6 as the internal control.
Statistical analysis
All experiments were performed in triplicate. Data
are presented as the mean ± SD and were analyzed using SPSS 19.0
software (IBM Corp.). Differences among groups were compared using
one-way ANOVA followed by Tukey's post-hoc test. Ratios were
compared using the χ2 tests. P<0.05 was considered to
indicate a statistically significant difference.
Results
Hypoxia-induced apoptosis is
alleviated by H19 overexpression and is aggravated by H19
interference
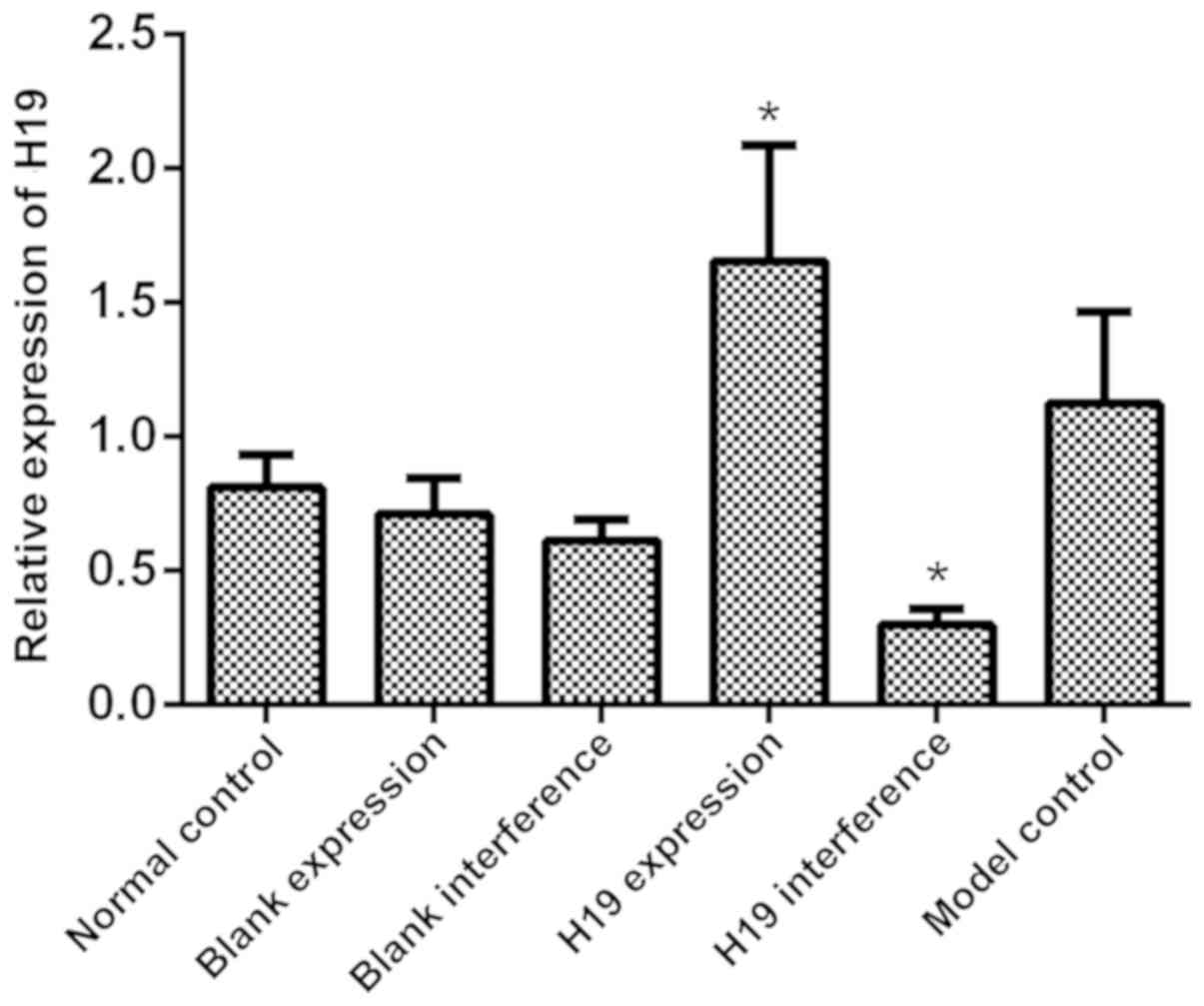
The expression of H19 in all groups was determined
using RT-qPCR. As shown in Fig. 1,
the model control, blank expression, blank interference and normal
control groups had a similar level of H19 expression (all
P>0.05). H19 expression was significantly increased in the H19
expression group (P=0.015) and significantly reduced in the H19
interference group (P=0.023) compared with the model control group.
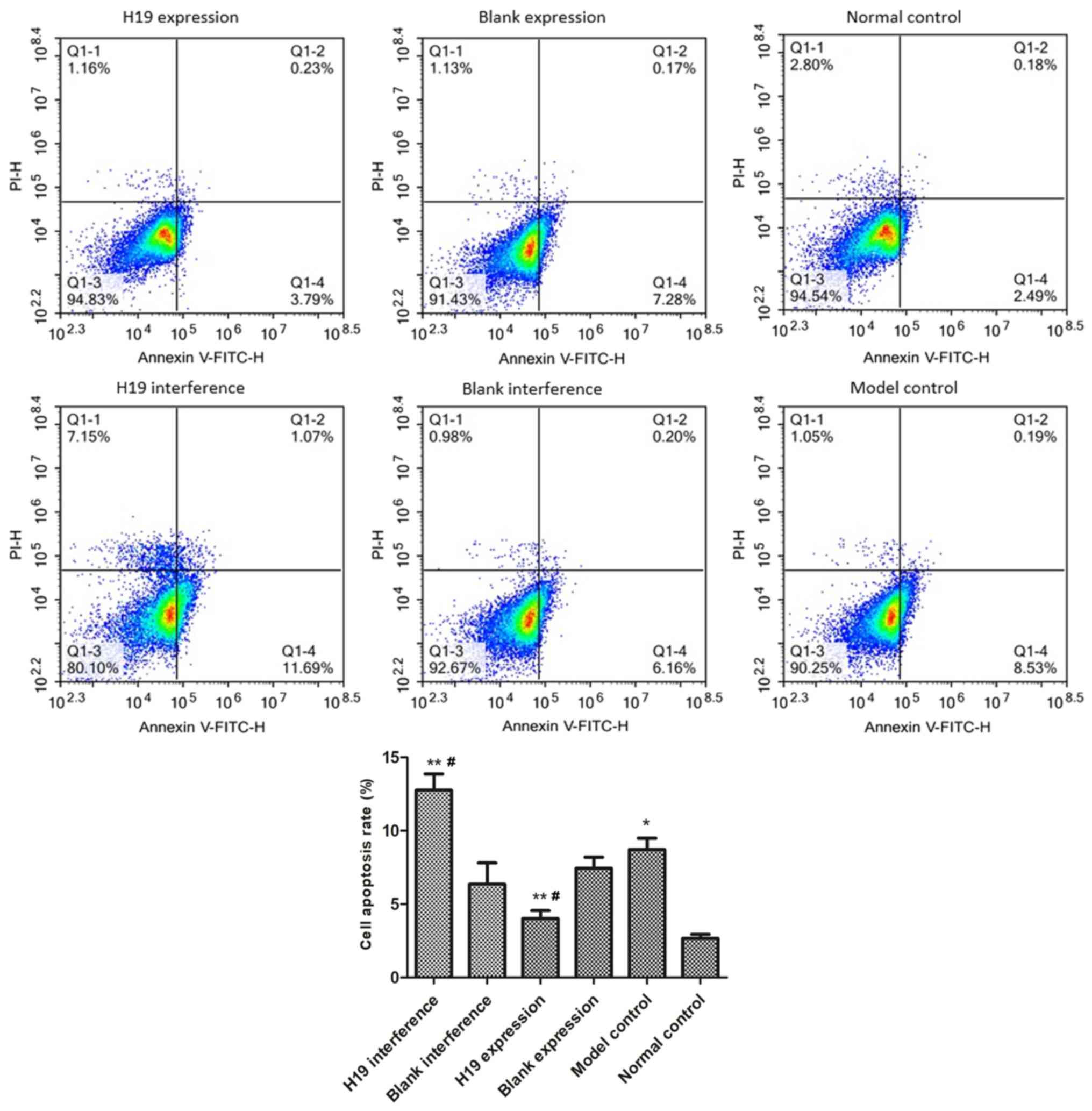
As shown in Fig. 2, the rate of
apoptosis in the model group was significantly higher than in
normal control group (P=0.021), indicative of hypoxia-induced
myocardial cell injury. The rate of apoptosis was significantly
reduced in the H19 expression group (P=0.016) and was significantly
increased in the H19 interference group compared with the model
group (P=0.022). The blank expression and blank interference groups
had similar rates of apoptosis as the model group (both
P>0.05).
Hypoxia-induced G1 phase cell cycle
arrest is attenuated by H19 overexpression and is aggravated by H19
interference
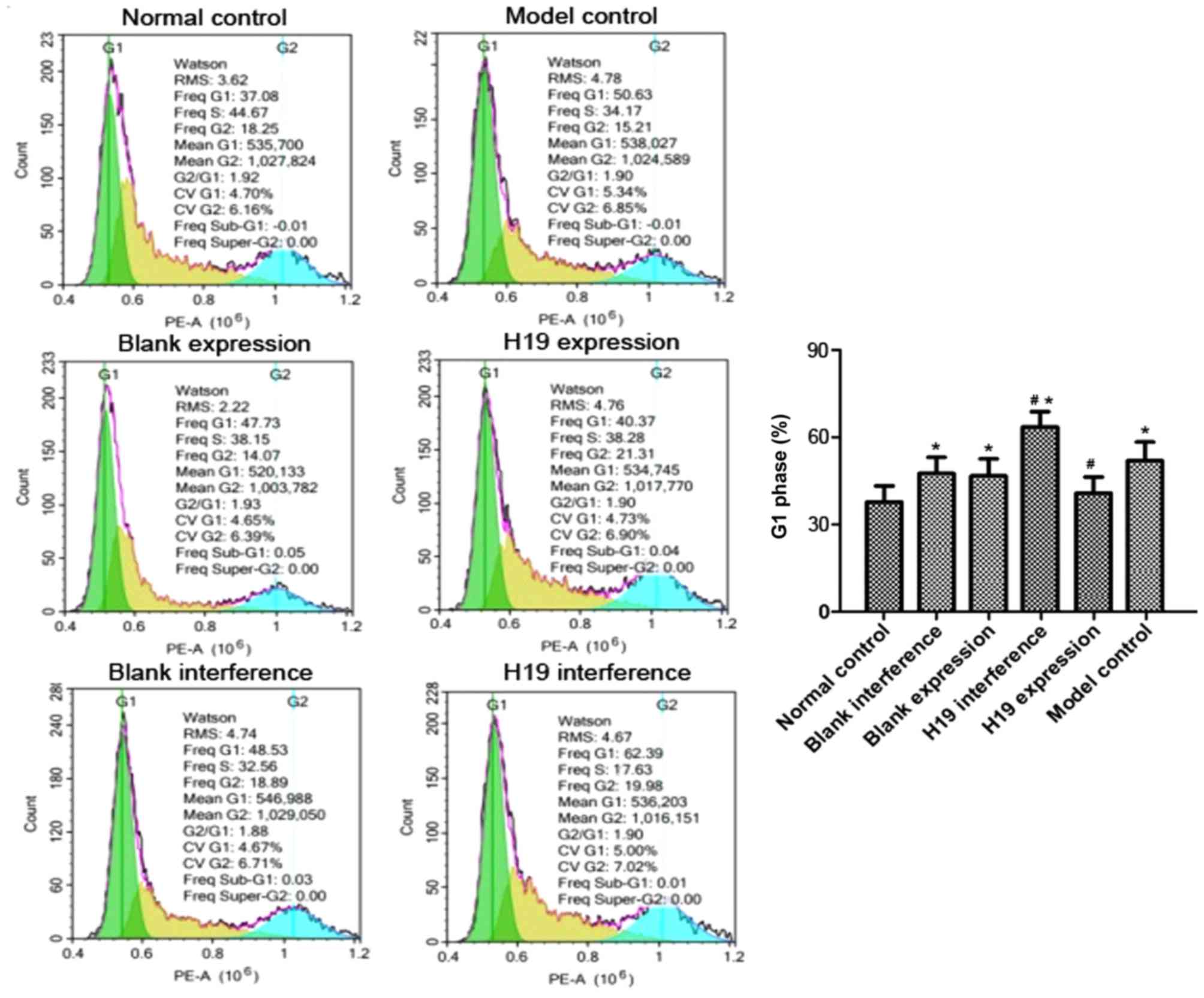
The cell cycle distribution was analyzed using flow
cytometry. As shown in Fig. 3, the
proportion of H9c2 cells in the G1 phase in the model group was
significantly higher compared with the normal control group
(P=0.020) and the percentage of S phase cells was significantly
lower (P=0.043), suggesting that hypoxia induced a G1 phase cell
cycle arrest. The H19 expression and normal control groups had a
similar proportion of H9c2 cells in G1 phase (P>0.05). The H19
interference group had a significantly higher G1 population
compared with the normal control group (P=0.012). When compared
with the model group, the H19 expression group had a significantly
lower G1 and a higher S phase population (P=0.018 and 0.031,
respectively), whereas the H19 interference group had a
significantly higher G1 population and a smaller S phase population
(P=0.029 and 0.045, respectively). The G1 and S phase populations
in the blank expression and blank interference groups were similar
to the model group (all P>0.05).
Hypoxia-induced mitochondrial membrane
depolarization is reduced by H19 overexpression, but is enhanced by
H19 interference
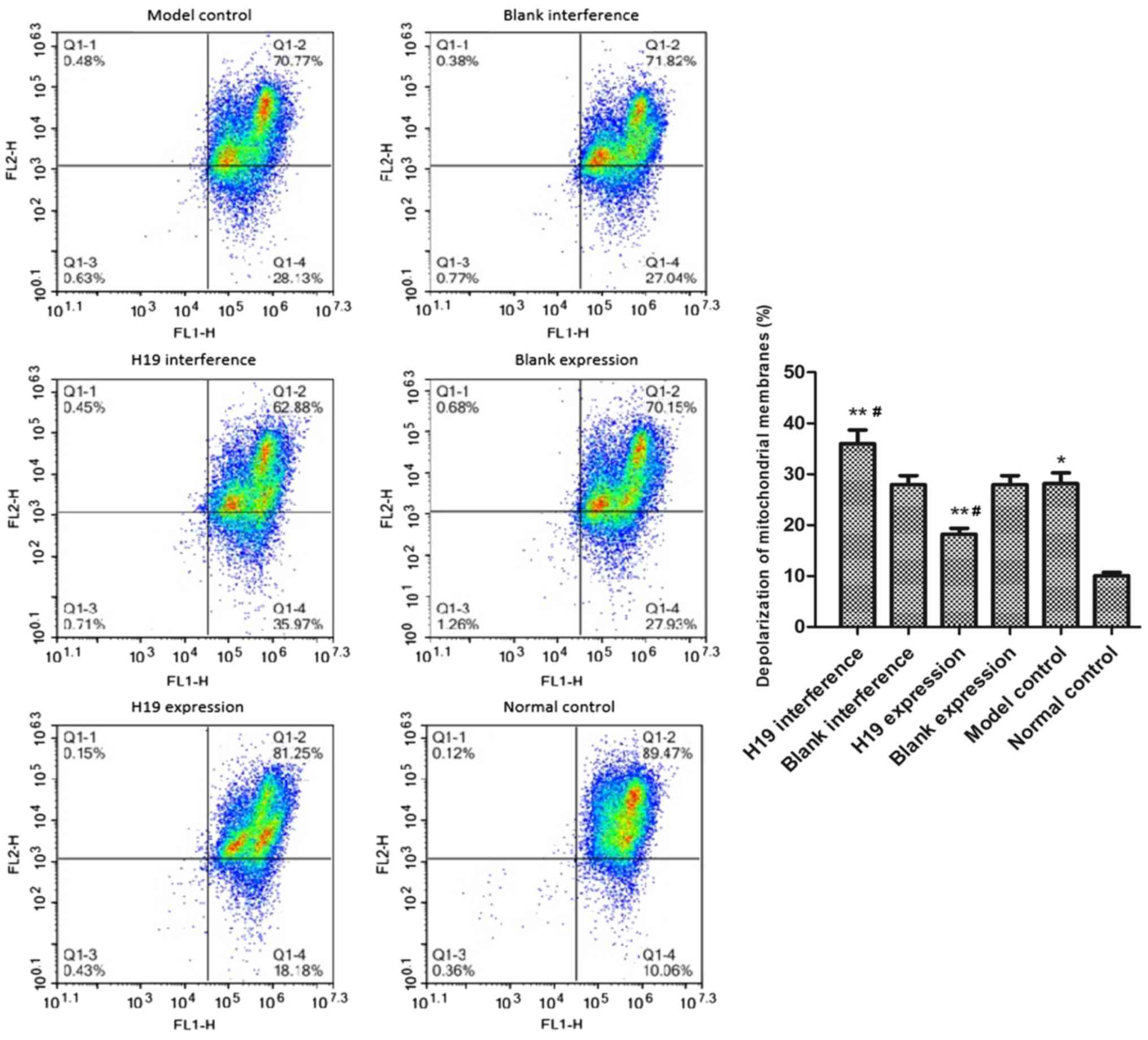
Under hypoxic treatment, the depolarization rate of
the mitochondrial membrane potential in the model group was
increased compared with the normal control group (P=0.033; Fig. 4). The mitochondrial depolarization
rate was significantly reduced in the H19 expression group
(P=0.036) and was increased in the H19 interference group (P=0.012)
compared with the model group. By contrast, the blank expression
and blank interference groups had similar depolarization rates as
the model group (both P>0.05).
PI3K/AKT and ERK/p38 pathways are
activated by H19 overexpression, but are inhibited by H19
interference
The underlying mechanism for the H19-asssociated
regulation of apoptosis, the cell cycle and mitochondrial membrane
potential was analyzed by western blotting. The results showed that
the levels of p-PI3K, p-AKT and p-p38 in the model group were
higher compared with the normal control group (all P>0.05).
p-ERK1/2 expression was significantly lower in the model group
compared with the normal control (P<0.05). The overexpression of
H19 increased the levels of p-PI3K, p-ERK1/2 and p-p38 compared
with the model group (all P<0.05). p-AKT level in the H19
overexpression group was similar as compared to the model group
(P>0.05). The effects of H19 interference were the opposite to
H19 overexpression in regulating the expression of p-p38 and p-PIK3
(all P<0.05, Fig. 5). H19
interference also significantly reduced the p-AKT expression as
compared to model group (P<0.05). H19 interference and model
groups had similar p-ERK1/2 expression (P>0.05). These findings
indicated that the overexpression of H19 may attenuate
hypoxia-induced cell injury by regulating the PI3K/AKT and ERK/p38
pathways. Nevertheless, it is worth noting that there are some
variation among the model and blank expression/interference groups,
indicating that some other pathways may also be involved in the H19
regulatory process.
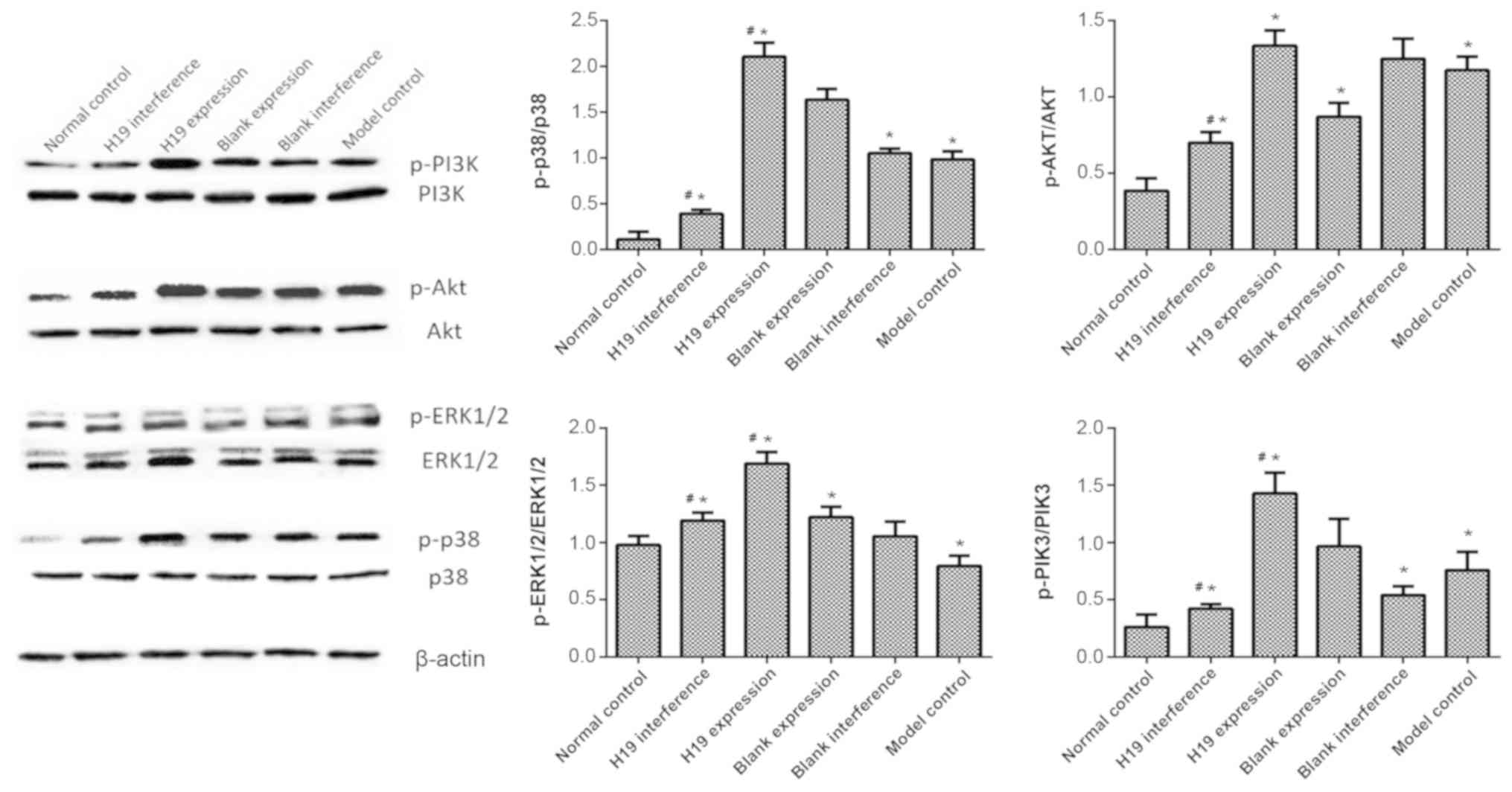 | Figure 5.PI3K/AKT and ERK/p38 pathways were
regulated by H19. Cells in the normal control group were cultured
under normoxia. Cells in the model control and different infection
groups (H19 expression, blank expression, H19 interference and
blank interference) were collected after 48 h of culture in hypoxic
conditions. The expression levels of p-PI3K, p-AKT, p-ERK1/2,
p-p38, PI3K, AKT, ERK1/2 and p38 were determined using western
blotting with β-actin as the internal control. All samples were
measured in triplicate. *P<0.05 vs. normal group;
#P<0.05 vs. model group. H19, long non-coding RNA
H19; p-, phosphorylated. |
Discussion
Hypoxia is commonly used to induce a myocardial cell
injury model (18). In the present
study, H9c2 cells were exposed to hypoxic conditions to induce cell
injury. As a result, these cells exhibited an increased level of
apoptosis, G1 cell cycle arrest and depolarization of the
mitochondrial membrane potential. The role of H19 has been
previously studied in several cancers; however, its role remains
controversial. For instance, H19 overexpression has been reported
to enhance carcinogenesis and metastasis in gastric cancer
(19). Contrary to this, it was
found that H19 does not affect the proliferation or cell cycle
distribution of breast cancer cells (20). There are few studies reporting the
effects of H19 in hypoxia-injured myocardial cells (16). In the present study, H19 expression
was found to be increased in hypoxia-treated cells, suggesting the
possible involvement of H19 in protection against hypoxia-induced
cell injury. H9c2 cells were transfected with H19 expressing or
silencing lentiviruses to investigate the role of H19 in
hypoxia-injured cells. It was found that H19 overexpression
alleviated hypoxia-induced injury mediated apoptosis, cell cycle
arrest and mitochondrial membrane potential depolarization;
however, these processes were aggravated by H19 knockdown. These
results suggested that H19 attenuated hypoxia-induced myocardial
cell injury, which is consistent with previous studies (15,16).
The findings of this previous report (16) and the present study are consistent
with each other in regard to the molecular mechanism of H19 in
protecting against myocardial I/R injury. Nevertheless, it is worth
noting that the two studies have different experimental designs.
Specifically, the present study examined the role of H19 regulation
on the cell cycle and mitochondrial changes in cells in addition to
cell apoptosis, which more directly reveals the
cellular/intracellular responses during the process of myocardial
I/R injury, whereas the reference only examined the cell behavior,
including cell viability, migration and apoptosis.
The PI3K/AKT signal pathway is an important pathway
for protecting myocardial cells against hypoxia-induced injury
(21,22). Xiao et al (23) reported that hydrogen sulfide
protects myocardial cells against hypoxia-induced injury via mTOR
activation. Chen et al (24) reported that lipoxin A4-induced heme
oxygenase-1 protects cardiomyocytes against hypoxia/reoxygenation
injury via p38 MAPK activation and the nuclear factor erythroid
2-related/antioxidant responsive element complex. IT has been
previously reported that AKT activation reduces myocardial cell
apoptosis by upregulating the expression of Bcl-2 (25). The relationship between the Bcl-2
family proteins and mitochondria-mediated apoptosis pathway is
well-established (26). Thus, it
is speculated that H19 may regulate mitochondrial apoptosis by
activating the PI3K/AKT/mTOR pathway. In the present study the
effects of H19 on mitochondrial membrane potential were assessed,
as was the activation of the PI3K/AKT/mTOR signaling pathway and
MAPK activation, in order to elucidate the mechanisms underlying
the possible protective effects of H19 against hypoxia-induced cell
injury. It was demonstrated that the overexpression of H19
stabilized the mitochondrial membrane potential and upregulated the
PI3K/AKT/mTOR pathway in hypoxia-treated H9c2 cells, while H19
interference had the opposite effect, indicating that H19
alleviated hypoxia-induced cell injury by reducing the
mitochondrial apoptosis pathway and activating the PI3K/AKT and
MAPK pathways.
In conclusion, the present study demonstrated that
the overexpression of H19 decreased hypoxia-induced cell injury by
increasing cell viability and decreasing apoptosis, whereas
knockdown of H19 had the opposite effects. Furthermore, it was
found that the overexpression of H19 may protect H9c2 cells from
hypoxia-induced injury by activating the PI3K/AKT/mTOR and MAPK
pathways. The present study may provide new insights into the
prevention and treatment of acute myocardial infarction.
Nevertheless, the present study was limited by the use of only one
cell line. Future experiments should be conducted with other types
of cardiomyocytes (for example, AC16 human cardiomyocyte and HCFB
human cardiac fibroblasts) to verify the findings of the present
study. Additionally, in-depth mechanistic studies should be
performed to further elucidate the protective effects of H19.
Findings from future studies should also be verified in animal
models.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LiY performed the majority of experiments and
drafted the paper. LeY, JZ, ZZ and CL helped with experiments. BZ
and XH analyzed the data and drafted part of the paper. GX and YT
conceived the study, supervised the experiments and edited the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mc Namara K, Alzubaidi H and Jackson JK:
Cardiovascular disease as a leading cause of death: How are
pharmacists getting involved? Integr Pharm Res Pract. 8:1–11. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hearse DJ and Bolli R: Reperfusion induced
injury: Manifestations, mechanisms, and clinical relevance.
Cardiovasc Res. 26:101–108. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu LN, Yu J, Zhang FJ, Yang MJ, Ding TT,
Wang JK, He W, Fang T, Chen G and Yan M: Sevoflurane
postconditioning reduces myocardial reperfusion injury in rat
isolated hearts via activation of PI3K/Akt signaling and modulation
of Bcl-2 family proteins. J Zhejiang Univ Sci B. 11:661–672. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Q, Xu T, Li D, Pan D, Wu P, Luo Y, Ma
Y and Liu Y: JNK/PI3K/Akt signaling pathway is involved in
myocardial ischemia/reperfusion injury in diabetic rats: Effects of
salvianolic acid A intervention. Am J Transl Res. 8:2534–2548.
2016.PubMed/NCBI
|
|
5
|
Zhao MM, Yang JY, Wang XB, Tang CS, Du JB
and Jin HF: The PI3K/Akt pathway mediates the protection of SO(2)
preconditioning against myocardial ischemia/reperfusion injury in
rats. Acta Pharmacol Sin. 34:501–506. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vassalli G, Milano G and Moccetti T: Role
of mitogen-activated protein kinases in myocardial
ischemia-reperfusion injury during heart transplantation. J
Transplant. 2012:9289542012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Furuno M, Pang KC, Ninomiya N, Fukuda S,
Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, et al:
Clusters of internally primed transcripts reveal novel long
noncoding RNAs. PLoS Genet. 2:e372006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hou J, Zhou C, Long H, Zheng S, Guo T, Wu
Q, Wu H, Zhong T and Wang T: Long noncoding RNAs: Novel molecules
in cardiovascular biology, disease and regeneration. Exp Mol
Pathol. 100:493–501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li L, Feng T, Lian Y, Zhang G, Garen A and
Song X: Role of human noncoding RNAs in the control of
tumorigenesis. Proc Natl Acad Sci USA. 106:12956–12961. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Whitehead J, Pandey GK and Kanduri C:
Regulation of the mammalian epigenome by long noncoding RNAs.
Biochim Biophys Acta. 1790:936–947. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kumar MM and Goyal R: LncRNA as a
therapeutic target for angiogenesis. Curr Top Med Chem.
17:1750–1757. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J,
Zhou Z, Zhang W, Qin L and Huang G: LncRNA PVT1 promotes
angiogenesis via activating the STAT3/VEGFA axis in gastric cancer.
Oncogene. 37:4094–4109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gabory A, Jammes H and Dandolo L: The H19
locus: Role of an imprinted non-coding RNA in growth and
development. Bioessays. 32:473–480. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rajagopalan V, Zhang Y, Pol C, Costello C,
Seitter S, Lehto A, Savinova OV, Chen YF and Gerdes AM: Modified
low-dose triiodo-L-thyronine therapy safely improves function
following myocardial ischemia-reperfusion injury. Front Physiol.
8:2252017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Cheng L, Xu L, Zhang Y, Yang Y,
Fu Q, Mi W and Li H: The Lncrna, H19 mediates the protective effect
of hypoxia postconditioning against hypoxia-reoxygenation injury to
senescent cardiomyocytes by targeting microRNA-29b-3p. Shock.
52:249–256. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu HM and Deng L: Evaluation of
cardiomyocyte hypoxia injury models for the pharmacological study
in vitro. Pharm Biol. 50:167–174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gong LC, Xu HM, Guo GL, Zhang T, Shi JW
and Chang C: Long non-coding RNA H19 protects H9c2 cells against
hypoxia-induced injury by targeting microRNA-139. Cell Physiol
Biochem. 44:857–869. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lottin S, Adriaenssens E, Dupressoir T,
Berteaux N, Montpellier C, Coll J, Dugimont T and Curgy JJ:
Overexpression of an ectopic H19 gene enhances the tumorigenic
properties of breast cancer cells. Carcinogenesis. 23:1885–1895.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li C, Tian J, Li G, Jiang W, Xing Y, Hou
J, Zhu H, Xu H, Zhang G, Liu Z and Ye Z: Asperosaponin VI protects
cardiac myocytes from hypoxia-induced apoptosis via activation of
the PI3K/Akt and CREB pathways. Eur J Pharmacol. 649:100–107. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin KH, Kuo WW, Jiang AZ, Pai P, Lin JY,
Chen WK, Day CH, Shen CY, Padma VV and Huang CY:
Tetramethylpyrazine ameliorated hypoxia-induced myocardial cell
apoptosis via HIF-1α/JNK/p38 and IGFBP3/BNIP3 inhibition to
upregulate PI3K/Akt survival signaling. Cell Physiol Biochem.
36:334–344. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiao J, Zhu X, Kang B, Xu J, Wu L, Hong J,
Zhang Y, Ni X and Wang Z: Hydrogen sulfide attenuates myocardial
hypoxia-reoxygenation injury by inhibiting autophagy via mTOR
activation. Cell Physiol Biochem. 37:2444–2453. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen XQ, Wu SH, Zhou Y and Tang YR:
Lipoxin A4-induced heme oxygenase-1 protects cardiomyocytes against
hypoxia/reoxygenation injury via p38 MAPK activation and Nrf2/ARE
complex. PLoS One. 8:e671202013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang B, Shravah J, Luo H, Raedschelders K,
Chen DD and Ansley DM: Propofol protects against hydrogen
peroxide-induced injury in cardiac H9c2 cells via Akt activation
and Bcl-2 up-regulation. Biochem Biophys Res Commun. 389:105–111.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Edlich F: BCL-2 proteins and apoptosis:
Recent insights and unknowns. Biochem Biophys Res Commun.
500:26–34. 2018. View Article : Google Scholar : PubMed/NCBI
|