Introduction
Tongue squamous cell carcinoma (TSCC) is the most
common type of oral squamous cell carcinoma comprising ~41% of all
cases (1), and its frequency is
increasing (2). The 5 year
survival rate of TSCC is 30–50%, and it is influenced by the high
rate of proliferation and early lymph node metastasis (2). Current standard treatments for TSCC
include surgical resection combined with postoperative radiotherapy
and chemotherapy (2). Although
advances have been made in recent years, the identification of a
treatment remains a challenge for TSCC. A large proportion of
patients with TSCC suffer from postoperative oral defects and lymph
node metastasis in multiple regions (3). The lack of effective targets to
control and monitor the disease is one of the principal problems in
TSCC treatment (3). Therefore, it
would be beneficial to identify novel biological targets to control
tumors effectively and reduce the impairment of oral functions.
Circular RNAs (circRNAs) are a type of non-coding
RNAs, which form covalently closed continuous loops with the 3′-
and 5′- ends joined (4). Due to
the distinctive molecular structure, circRNAs are resistant to the
degradation from RNAses, resulting in a stable and conserved
expression in the cytoplasm (5).
circRNAs were thought to be the results of defective RNA splicing
events and have been only recently identified (6). A number of previous studies have
reported that circRNAs play important roles in carcinogenesis and
cancer development by acting as microRNA (miRNA) sponges,
regulating gene expression at the transcriptional and translational
levels (6,7). The circular transcript ciRS-7 was
found to bind miR-7, resulting in the inhibition of miR-7 (8). In addition, miR-7 served as an
epidermal growth factor receptor (EGFR) suppressor by binding to
the EGFR mRNA 3′-untranslated region (8). The overexpression of ciRS-7 was
observed to increase the expression levels of EGFR and activate its
downstream signaling pathway through inhibiting miR-7 expression
(7). circRNAs also regulate cancer
cell biological features by altering the genomic DNA (9). Therefore, identifying the functions
of circRNAs may improve the understanding of the molecular
mechanisms underlying cancer development and may provide potential
targets for cancer treatment. Notably, the expression patterns and
potential roles of circRNAs in TSCC remain poorly understood.
In the present study, the expression profile of
various circRNAs was investigated in TSCC and was compared with the
expression profile of circRNAs in paired adjacent normal tissues
using microarray analysis. The identification of the differentially
expressed circRNAs in TSCC may provide novel insights into the
diagnosis and treatment of TSCC.
Materials and methods
Patients
The study was approved by The Ethics Committee of
Peking University School and Hospital of Stomatology (approval no.
PKUSSIRB-2013009). The patient consents were acquired before tissue
collection.
The present study included 14 patients (males, 8;
females, 6; mean age, 50.7 years; age range, 39–73 years) who
underwent surgery at Peking University School and Hospital of
Stomatology to pathologically confirm primary TSCC, between January
2017 and December 2017. None of the patients had received any
anti-cancer therapy before sample collection.
Part of the resected tumor tissues and paired
adjacent normal tissues (≥2 cm from the tumor margins) were
collected. Of the tissues collected, four pairs of tumor and
adjacent normal tissues were used for microarray analysis and ten
pairs of tumor and adjacent normal tissues were used for
validation. The tissue pairs selected for each analysis occurred
randomly based on the patient inclusion criteria. Tumor clinical
stage and histological grading were classified based on the 8th
edition of the TNM classification of the Union for International
Cancer Control (10).
Histological evaluation
Paraffin embedded consecutive tissue sections (5 µm)
were fixed in 4% paraformaldehyde at room temperature for 24 h and
subsequently stained with 0.5% hematoxylin solution for 30 min at
room temperature and 0.5% eosin solution for 10 min at room
temperature. The sections were observed and photographed using an
Olympus CKX41 light microscope (magnification ×10; Olympus
Corporation).
RNA extraction
Total RNA from the ten pairs of tissue samples was
isolated using TRIzol® reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instruction. RNA
quantity and quality were assessed using the NanoDrop ND-100
spectrometer (Thermo Fisher Scientific, Inc.), and RNA integrity
was tested by agarose gel electrophoresis.
Labeling and hybridization and
microarray analysis
Sample labeling and array hybridization were
performed based on the manufacturer's instruction (Arraystar,
Inc.). The acquired hybridized array data were analyzed by Agilent
Feature Extraction software (version 11.0.1.1; http://www.agilent.com/en/product/mirna-microarray-platform/mirna-microarray--software/feature-extraction-software-228496/download-software--feature-extraction-software)
(11). Quantile normalization of
raw data and subsequent data processing were conducted using R
software limma package (version 3.1.2; http://www.bioconductor.org/packages/release/bioc/html/limma.html).
When comparing two groups (TSCC vs. paracancer), the fold change
between the groups for each circRNA was calculated. The statistical
significance of the difference was estimated by paired Student's
t-test. circRNAs with fold changes ≥2 or ≤0.5 and P<0.05 were
selected as differentially expressed circRNAs. CircRNA/miRNA
interactions were predicted using the TargetScan (release 7.2;
http://www.targetscan.org) and miRanda (release
2010; http://www.microrna.org) databases.
Quantitative (q)PCR
cDNA samples were prepared from total RNA of TSCC
samples using the PrimeScript™ RT reagent kit (Takara Bio, Inc.),
according to the manufacturer's protocol. The following conditions
were used for the reverse transcription: 37°C for 15 min and 85°C
for 10 sec. In total, two upregulated circRNAs and five
downregulated circRNAs were detected on a PikoReal Real-Time PCR
System (Thermo Fisher Scientific, Inc.) using the SYBR®
Green qPCR kit (Thermo Fisher Scientific, Inc.). The primer
sequences were listed in Table I.
The following thermocycling conditions were used for the qPCR:
Initial denaturation at 95°C for 10 min; 35 cycles of 95°C for 10
sec and 60°C for 40 sec; and a final extension at 72°C for 10 min.
The relative expression of circRNAs was calculated using the
2−ΔΔCq method (12),
β-actin was used as the internal reference gene to normalize the
expression data.
 | Table I.circRNA primers for quantitative PCR
analysis. |
Table I.
circRNA primers for quantitative PCR
analysis.
| Gene | Primer sequence
(5′→3′) | Product size (base
pairs) |
|---|
| β-actin (Human) | F:
GTGGCCGAGGACTTTGATTG | 73 |
|
| R:
CCTGTAACAACGCATCTCATATT |
|
|
hsa_circRNA_000780 | F:
TAGGAAACCTGCTGTGGAGTG-3 | 108 |
|
| R:
AAGGGAACTATACAAGGAAATGC |
|
|
hsa_circRNA_102039 | F:
CTATCATTCACAAAGGGAAAACTAC | 164 |
|
| R:
CCATAACTGGAGTAACCGCTG |
|
|
hsa_circRNA_003251 | F:
GGAGAAGACGACGACCCACTA | 113 |
|
| R:
TAGGACAGGGCCTTCTTTGAC |
|
|
hsa_circRNA_045179 | F:
GCTGCTGTGCAAGAAACGG | 139 |
|
| R:
CACCTGGCTGAACTTCTGTGACT |
|
|
hsa_circRNA_081069 | F:
CTGGACTTCCTGGCTTCAAA | 164 |
|
| R:
TCCTCTATCTCCGGCTGGG |
|
|
hsa_circRNA_087212 | F:
TTCAATACCATCCTTACCACC | 113 |
|
| R:
CTCTGATTTCTTTGTTAGTTCTTG |
|
|
hsa_circRNA_000317 | F:
AAAGGGCCAGAGGTAGACAT | 79 |
|
| R:
GCAAATCAAAGTCAGGCATAG |
|
Cell culture and transfection
Human TSCC cell line, SCC15 and SCC25, were
purchased from the American Type Culture Collection. Cells were
cultured in DMEM/F12 medium (Gibco; Thermo Fisher Scientific, Inc.)
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with
5% CO2 for 3 days before use.
Silencing RNA (siRNA) against circ_081069
(sicirc_081069; cat. no. siBDM2500) and siRNA control (cat. no.
siBDM2500) were purchased from Guangzhou RiboBio Co., Ltd. miR-665
mimic (ACCAGGAGGCUGAGGCCCCU; cat. no. 4464066) and negative control
(cat. no. 4464066) were purchased from Thermo Fisher Scientific,
Inc. and 100 nM of each was transfected into TSCC cells using
Lipofectamine® 3000 reagent (Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Cells were
collected for future experiments following 24 h of
transfection.
Migration assay
Cell migration was measured by Transwell assay. A
total of 1×104 SCC15 and SCC25 cells were seeded in a
24-well cell culture chamber (pore size, 8 µm; Corning, Inc.) in
200 µl serum-free DMEM/F12 medium. The lower chamber was plated in
DMEM/F12 medium supplemented with 20% FBS. After 24 h incubation at
37°C and 5% CO2, cells on the lower side of the membrane
were fixed in 95% ethanol at room temperature for 10 min and
stained with 0.5% hematoxylin at room temperature for 30 min
followed by 0.5% eosin staining at room temperature for 30 min. The
whole membrane was photographed using an Olympus CKX41 light
microscope (magnification ×20; Olympus Corporation) and the cells
on the membrane were quantified using ImageJ version 1.44 software
(National Institutes of Health), as previously described (13). All experiments were performed in
triplicate and cells transfected with control siRNA acted as the
control group.
Proliferation assay
Proliferation of SCC15 and SCC25 cells were
evaluated by a Cell Counting Kit-8 (CCK8) cell viability assay
(Dojindo Molecular Technologies, Inc.), according to the
manufacturer's protocol. For the CCK-8 assay, 5×103
cells were seeded into 96-well plates. Following incubation at 37°C
for 48 and 72 h, cell viability was measured at a wavelength of 450
nm using a spectrophotometric plate reader.
Flow cytometry
For cell apoptosis analysis, 1×104 cells
were seeded into 6-well plates. After transfection for 48 h, cells
were resuspended with 100 µl binding buffer after centrifugation at
200 × g at room temperature for 5 min. Annexin V-FITC (20 µg/ml;
Dojindo Molecular Technologies, Inc.) and propidium (50 µg/ml;
Dojindo Molecular Technologies, Inc.) were added to cells and
incubated for 15 min at room temperature. The cell apoptotic rate
was measured using a BD FACSCalibur™ flow cytometer (BD
Biosciences) and analyzed with CellQuest™ version 7.5.3 software
(BD Biosciences).
Dual-luciferase reporter assay
The sequences containing the binding site of miR-665
in circRNA_081069 (416–435 bp) and the mutant sequences
(TCCTGG>CGGCCA) were synthesized by Shanghai Generay Biotech
Co., Ltd. and inserted into the luciferase reporter vector
psiCHECK-2 (Promega Corporation). Both plasmids were transfected
into cells using Lipofectamine® 3000 reagent (Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
Luciferase activity was measured using the Dual-Luciferase Reporter
Assay kit (Promega Corporation), according to the manufacturer's
instructions following 24 h of transfection. Relative luciferase
activity was normalized to Renilla luciferase activity.
Statistical analysis
Data are presented as the mean ± SEM. The
statistical significance of microarray data was analyzed by fold
change using a paired Student's t-test and the false discovery rate
was calculated to correct the P-value using SPSS Statistics version
20.0 software (IBM Corp.). Fold change ≥2.0 or ≤0.5 and P<0.05
were applied to find the differentially expressed circRNAs. The
remaining statistical analysis was performed using SPSS Statistics
version 20.0 software (IBM Corp.). Differences were determined by a
paired Student's t-test for paracancer and TSCC groups. P<0.05
was considered to indicate a statistically significant
difference.
Results
Patient characteristics
The clinical characteristics of the 14 patients with
pathologically confirmed primary TSCC are presented in Table II. Tissue samples of four patients
were used for microarray analysis and the remaining ten pairs of
tissue specimens were used for further validation. In total, eight
patients with TSCC were male and six patients were female.
Moreover, eight patients were classified as T1 and T2 stage, and
six patients were classified as T3 and T4 stage. Lymph node
metastasis was present in ten patients. Histopathologic
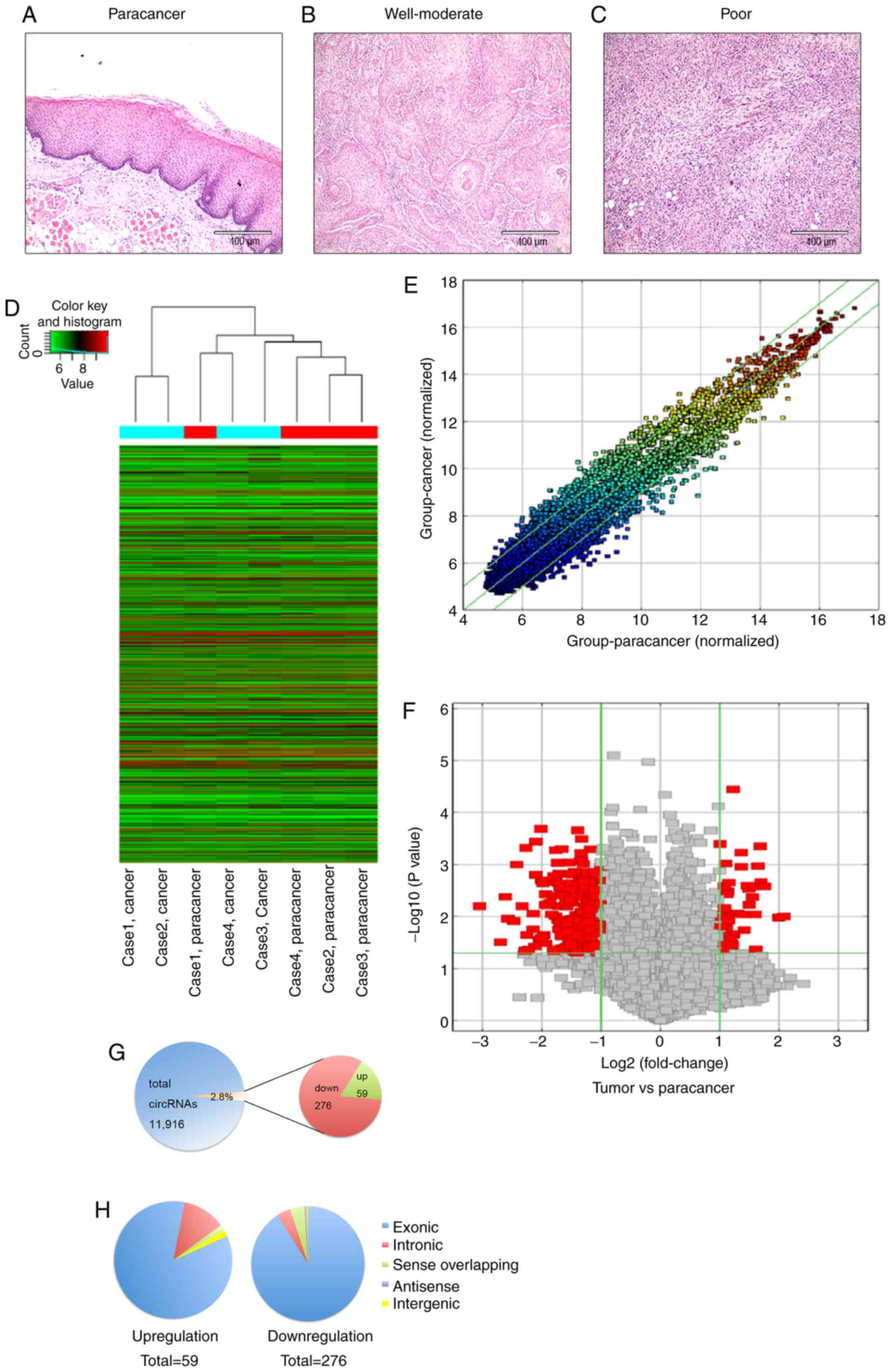
characteristics were assessed by hematoxylin & eosin staining
(Fig. 1A-C).
 | Table II.Clinical characteristics of the 14
patients with tongue squamous cell carcinoma. |
Table II.
Clinical characteristics of the 14
patients with tongue squamous cell carcinoma.
| Case no. | Age (years) | Sex | Histologic
differentiation | TNM stage | Admission date |
|---|
| 1 | 40 |
Male |
Moderately | T4aN1M0 | 26/04/2017 |
| 2 | 44 |
Male |
Well-moderately | T2N2M0 | 30/05/2017 |
| 3 | 53 |
Male |
Poorly | T4aN2M0 | 17/06/2017 |
| 4 | 49 |
Male |
Moderately | T1N0M0 | 17/06/2017 |
| 5 | 64 |
Male |
Well-moderately | T4aN1M0 | 30/06/2017 |
| 6 | 39 |
Male |
Moderately | T4aN1M0 | 30/06/2017 |
| 7 | 40 |
Female |
Well-moderately | T2N1M0 | 07/07/2017 |
| 8 | 55 |
Female |
Well-moderately | T2N0M0 | 13/07/2017 |
| 9 | 47 |
Male |
Well-moderately | T1N0M0 | 13/07/2017 |
| 10 | 62 |
Female |
Well-moderately | T1N1M0 | 13/07/2017 |
| 11 | 73 |
Female |
Poorly | T4aN2M0 | 31/08/2017 |
| 12 | 49 |
Female |
Moderately | T1N2M0 | 11/12/2017 |
| 13 | 55 |
Male |
Moderately | T2N0M0 | 15/02/2017 |
| 14 | 40 |
Female |
Well-moderately | T4aN1M0 | 20/01/2017 |
Analysis of differentially expressed
circRNAs
Arraystar human circRNA microarray was used to
screen 11,916 circRNAs, and differentially expressed circRNAs were
identified between TSCC and paracancer tissues. Hierarchical
clustering was performed to group circRNAs according to the
expression levels among samples (Fig.
1D). The threshold was set as fold change ≥2.0 or ≤0.5 and
P<0.05 (Fig. 1E and F). The
results showed that 335 circRNAs were differentially expressed,
including 59 upregulated circRNAs and 276 downregulated circRNAs
(Fig. 1G). The top 15 upregulated
and top 15 downregulated circRNAs, combined with the detailed
molecular information, are presented in Tables III and IV. Among the upregulated circRNAs, there
were 50 exonic, seven intronic, one intergenic and one sense
overlapping. Among the downregulated circRNAs, there were 251
exonic, 11 intronic, 11 sense overlapping, two antisense and one
intergenic (Fig. 1H).
 | Table III.Top 15 upregulated circRNAs between
tongue squamous cell carcinoma tissues and paired paracancer
tissues. |
Table III.
Top 15 upregulated circRNAs between
tongue squamous cell carcinoma tissues and paired paracancer
tissues.
| circRNA | P-value | Fold change | Regulation | circRNA type | Chromosome
number | Strand | Best
transcript | Gene symbol |
|---|
|
hsa_circRNA_045179 | 0.009971374 | 4.2212226 | Up | Exonic | Chr17 | + | NM_025185 | TANC2 |
|
hsa_circRNA_014280 | 0.010356383 | 3.9734682 | Up | Exonic | Chr1 | − | NM_004515 | ILF2 |
|
hsa_circRNA_081069 | 0.002628269 | 3.379894 | Up | Exonic | Chr7 | + | NM_000089 | COL1A2 |
|
hsa_circRNA_402089 | 0.002161838 | 3.2279163 | Up | Exonic | Chr19 | + | NM_018443 | ZNF302 |
|
hsa_circRNA_067209 | 0.000443419 | 3.1954049 | Up | Exonic | Chr3 | + | NM_021937 | EEFSEC |
|
hsa_circRNA_404474 | 0.001060291 | 3.1198996 | Up | Exonic | Chr1 | − | NM_018207 | TRIM62 |
|
hsa_circRNA_101323 | 0.042507978 | 3.0498428 | Up | Exonic | Chr14 | − | NM_002797 | PSMB5 |
|
hsa_circRNA_404905 | 0.002648443 | 3.0176954 | Up | Intronic | Chr11 | + |
ENST00000298229 | INPPL1 |
|
hsa_circRNA_103121 | 0.009017454 | 3.0172602 | Up | Exonic | Chr21 | − | NM_000819 | GART |
|
hsa_circRNA_067130 | 0.004192175 | 2.9217705 | Up | Exonic | Chr3 | − | NM_025112 | ZXDC |
|
hsa_circRNA_006226 | 0.005920498 | 2.8194496 | Up | Intronic | Chr3 | + |
ENST00000295874 | PTPRG |
|
hsa_circRNA_407284 | 0.002908595 | 2.8054966 | Up | Intronic | Chrx | + |
ENST00000329236 | RBM10 |
|
hsa_circRNA_100053 | 0.00255781 | 2.7576059 | Up | Exonic | Chr1 | + | NM_014874 | MFN2 |
|
hsa_circRNA_062557 | 0.018213635 | 2.6570433 | Up | Exonic | Chr22 | − | NM_213720 | CHCHD10 |
|
hsa_circRNA_036186 | 0.009051291 | 2.6020041 | Up | Exonic | Chr15 | − | NM_002654 | PKM |
 | Table IV.Top 15 downregulated circRNAs between
tongue squamous cell carcinoma tissues and paired paracancer
tissues. |
Table IV.
Top 15 downregulated circRNAs between
tongue squamous cell carcinoma tissues and paired paracancer
tissues.
| circRNA | P-value | Fold change | Regulation | Chromosome
number | Strand | circRNA type | Best
transcript | Gene symbol |
|---|
|
hsa_circRNA_000317 | 0.006309622 | 8.2823629 | Down | Chr11 | − | Intronic |
ENST00000257247 | AHNAK |
|
hsa_circRNA_087212 | 0.030852306 | 6.4938933 | Down | Chr9 | + | Exonic | NM_000700 | ANXA1 |
|
hsa_circRNA_000780 | 0.004147446 | 6.1215191 | Down | Chr10 | − | Intronic |
ENST00000181796 | FAM107B |
|
hsa_circRNA_000320 | 0.010885129 | 5.9447551 | Down | Chr11 | − | Intronic |
ENST00000257247 | AHNAK |
|
hsa_circRNA_000319 | 0.010916423 | 5.9139973 | Down | Chr11 | − | Intronic |
ENST00000257247 | AHNAK |
|
hsa_circRNA_404013 | 0.012111731 | 5.5509098 | Down | Chr8 | − | Exonic | NM_018310 | BRF2 |
|
hsa_circRNA_102039 | 0.000991458 | 5.3535917 | Down | Chr17 | + | Exonic | NM_003487 | TAF15 |
|
hsa_circRNA_082680 | 0.006278996 | 5.03148 | Down | Chr7 | − | Exonic | NM_022740 | HIPK2 |
|
hsa_circRNA_101001 | 0.045188085 | 4.8819295 | Down | Chr12 | − | Exonic | NM_001038 | SCNN1A |
|
hsa_circRNA_401782 | 0.000479405 | 4.8091228 | Down | Chr17 | + | Exonic | NM_003487 | TAF15 |
|
hsa_circRNA_103809 | 0.044401398 | 4.8006013 | Down | Chr5 | − | Exonic | NM_016107 | ZFR |
|
hsa_circRNA_075650 | 0.022393199 | 4.7640369 | Down | Chr6 | − | Exonic | NM_005493 | RANBP9 |
|
hsa_circRNA_026358 | 0.042942656 | 4.6710087 | Down | Chr12 | + | Exonic | NM_005556 | KRT7 |
|
hsa_circRNA_100191 | 0.017573345 | 4.4330842 | Down | Chr1 | + | Exonic | NM_002840 | PTPRF |
|
hsa_circRNA_003251 | 0.008740651 | 4.4168694 | Down | Chr12 | + | Exonic | NM_014823 | WNK1 |
qPCR validation
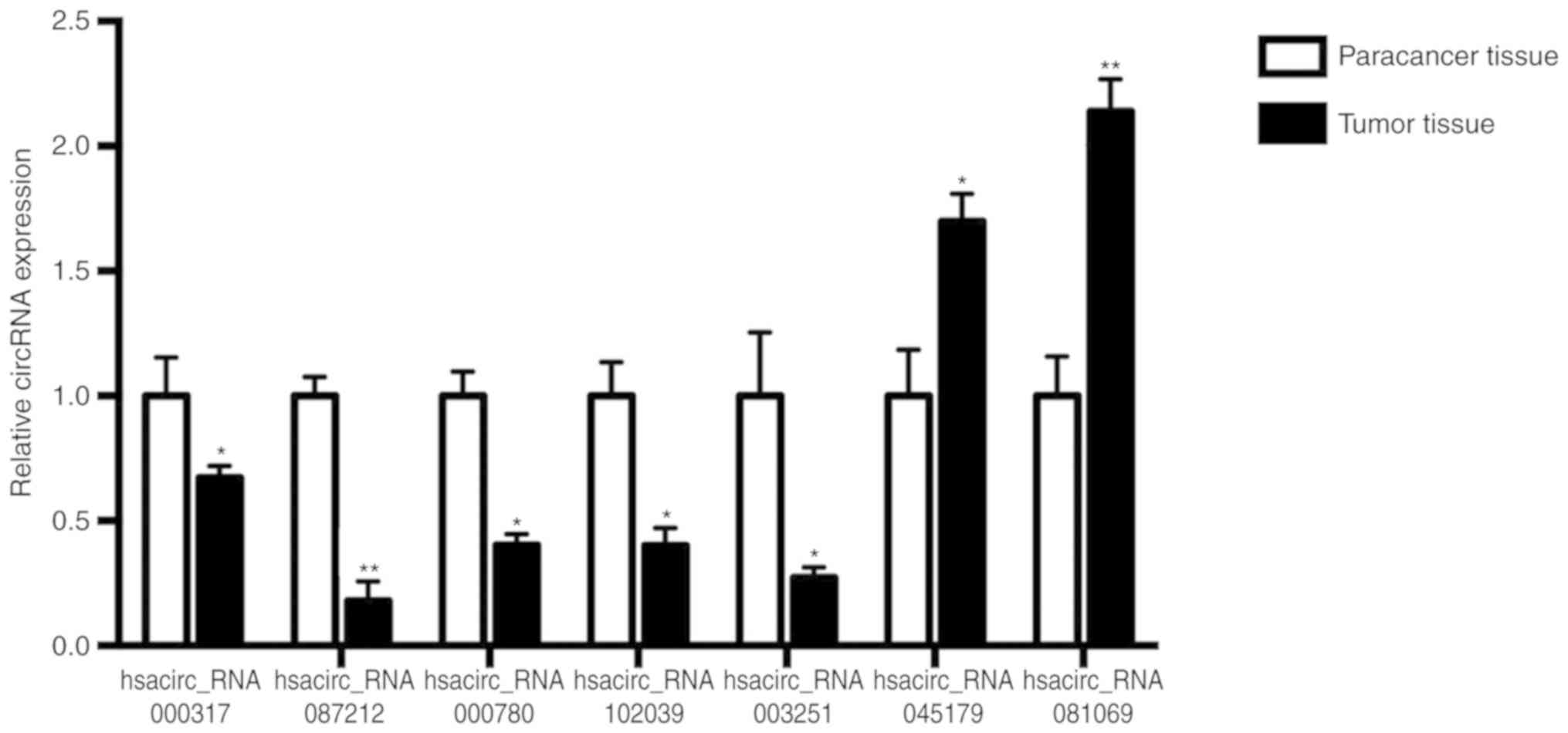
To validate the microarray results, two upregulated
circRNAs (circ_045179 and circ_081069) and five downregulated
circRNAs (circ_000317, circ_087212, circ_000780, circ_102039 and
circ_003251) were selected from the differentially expressed
circRNAs in the microarray. In total, ten pairs of TSCC tissues and
paracancer tissues were examined by qPCR to confirm the expression
levels of the selected circRNAs. The expression patterns of
selected circRNAs showed the same consistency with the microarray
results (Fig. 2). Of them,
circ_081069 and circ_087212 were the most upregulated and
downregulated circRNA, respectively (Fig. 2).
circRNA and miRNA interaction
analysis
Previous studies have reported that circRNAs mainly
function as miRNA sponges and serve important roles in gene
regulation (6,7). The circRNA/miRNA interaction has been
shown to be involved in cancer progression, and could be exploited
as a potential therapeutic target. To analyze the potential
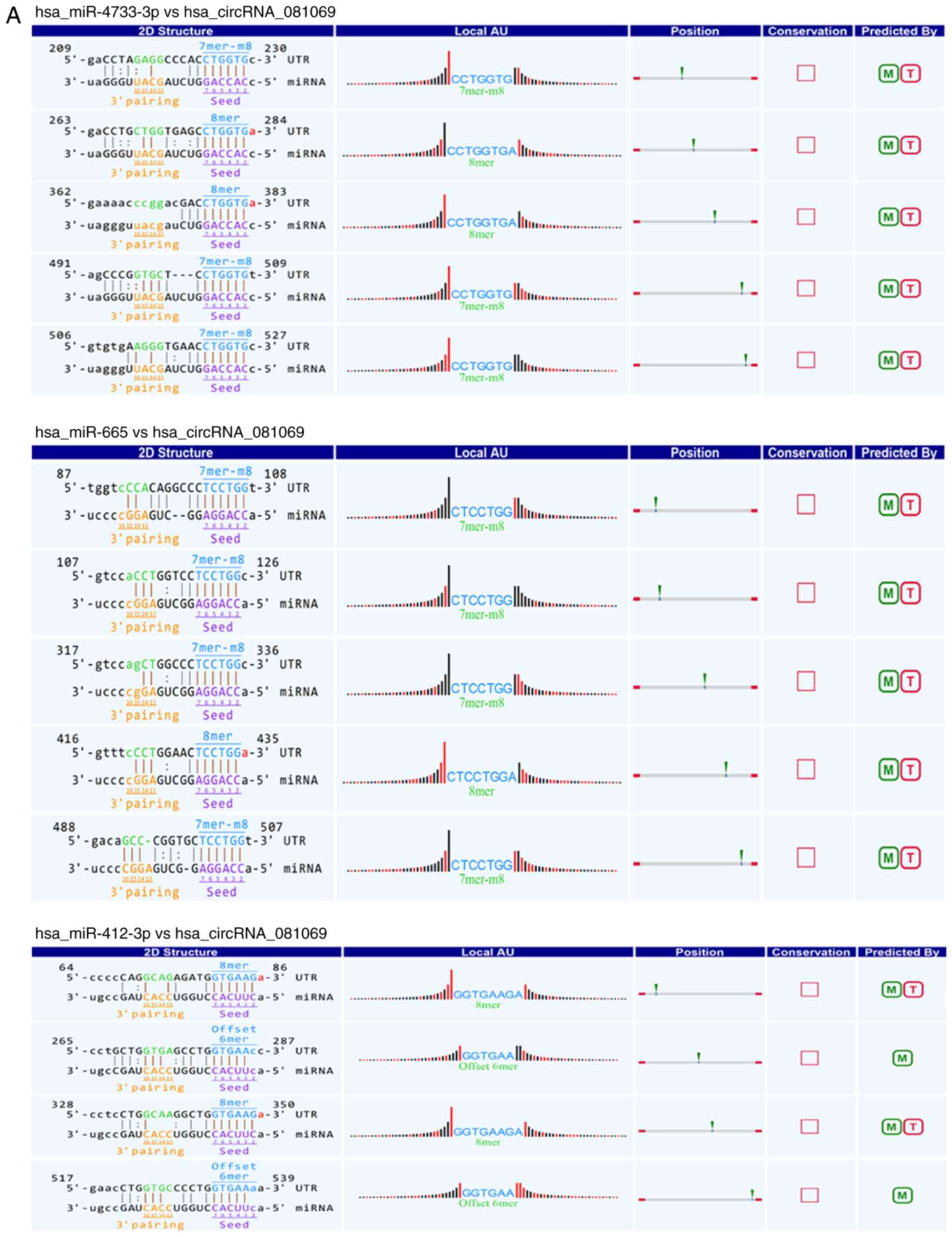
functions of the selected circRNAs, circRNA/miRNA interactions were
predicted using TargetScan (release 7.2; http://www.targetscan.org/) and miRanda (release 2010;
http://www.microrna.org). A total of two circRNAs
(circ_081069 and circ_087212) were examined in detail using the
circRNA/miRNA interaction analysis (Fig. 3). The potential miRNA targets of
circ_081069 included miR-4733-3p, miR-665 and miR-412-3p. The
potential miRNAs interacting with circ_087212 included miR-29a-5p,
miR-6873-3p, miR-6809-3p, miR-6515-3p and miR-5584-3p. A previous
study showed that miR-665 suppresses osteosarcoma cell
proliferation and invasion by inhibiting the small GTPase protein
Rab23 (14). In addition, miR-29a
regulates the expression levels of p21, p53 and survivin in cancer
cells, resulting in enhanced cell proliferation (15). However, the function of the
remaining miRNAs on cancer development remains unclear.
miR-665 is a target of
circ_081069
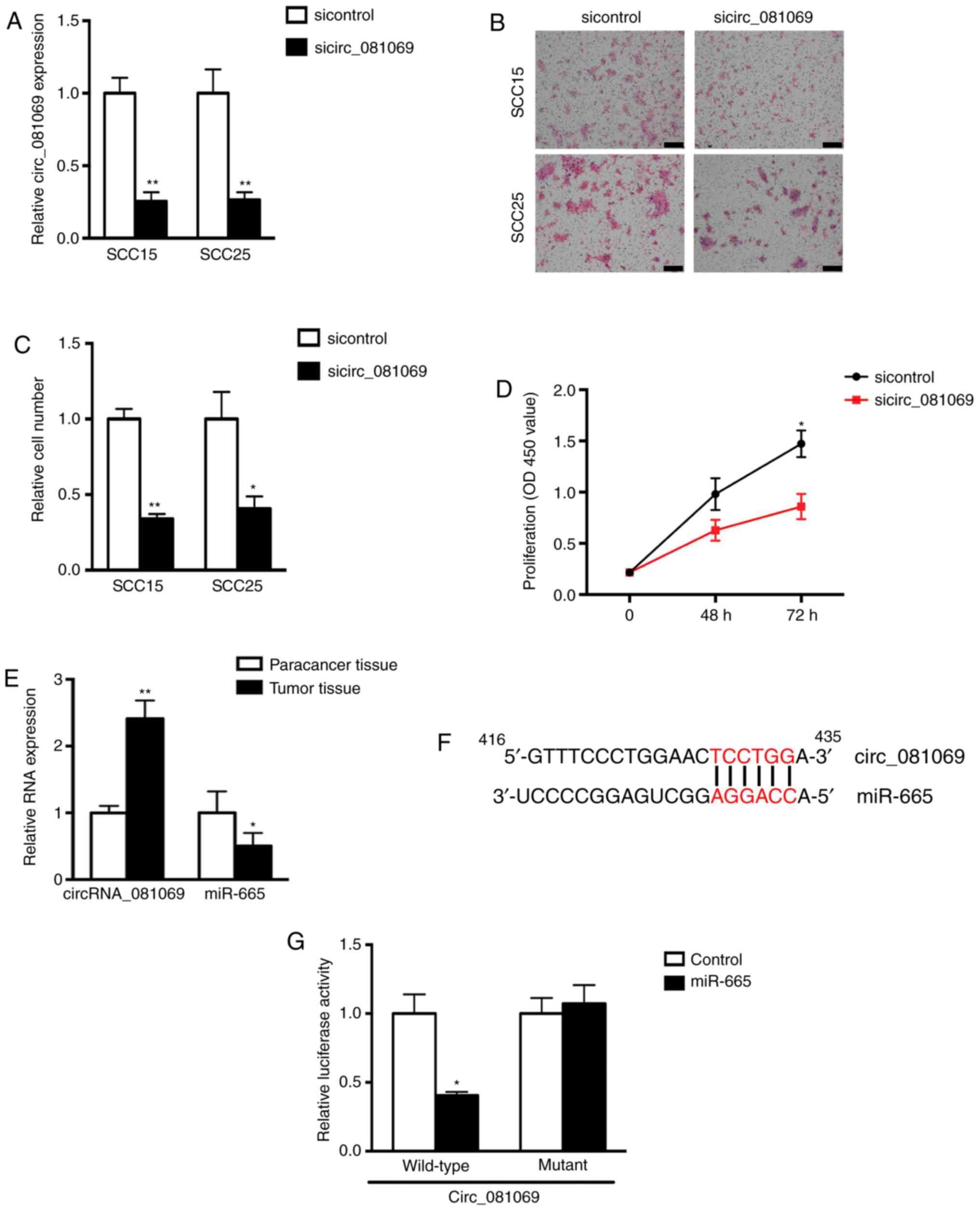
In the present study, the most upregulated circRNA
in the microarray analysis, circ_081069, was selected to
investigate its potential biological role in TSCC and identify its
potential miRNA targets. After knockdown of circ_081069 in TSCC
cells by siRNA (Fig. 4A), the
migratory and proliferative ability were suppressed significantly
(Fig. 4B-D); however, apoptosis
was not affected (Fig. S1). The
present results suggested that circ_081069 may exert oncogenic
effects by regulating cell migration and proliferation. Using
bioinformatic prediction methods circ_081069 was predicted to
interact with miR-665, possibly acting as a competing endogenous
RNA. Using qPCR, the present study found that miR-665 levels were
decreased in four TSCC tissues (Fig.
4E). In order to further investigate the direct interaction
between circ_081069 and miR-665, a luciferase reporter assay was
performed. The present results showed that overexpression of
miR-665 inhibited the wild-type circ_081069 reporter, whereas no
significant difference was found between the cells overexpressing
miR-665 and the negative control in the mutant circ_081069 reporter
(Fig. 4F and G). These results
suggested that circ_081069 promoted TSCC cell migration and
proliferation. Furthermore, miR-665 was able to interact with
circ_081069, in line with the bioinformatic prediction results.
Discussion
In the present study, microarray analysis of
differentially expressed circRNAs was conducted by comparing the
transcriptome profiles of TSCC and adjacent normal tissues. A total
of 11,196 circRNAs were examined. After screening, 59 upregulated
and 276 downregulated circRNAs were identified and their
characteristics were analyzed. The results of the present study
analyzed the circRNAs expression in TSCC, which may facilitate the
complicated molecular mechanisms in TSCC development.
circRNAs represent a distinctive type of endogenous
non-coding RNA. Due to their distinctive molecular structure,
circRNAs have higher stability in different body fluids and
exosomes compared with other types of RNAs, which makes them useful
as novel diagnostic and prognostic biomarkers (16–18).
circRNAs are categorized into three types: i) Exonic circRNAs; ii)
intronic circRNAs; and iii) exon-intro circRNAs (EIciRNAs)
(19). Exonic circRNAs consist of
two or more exons, that function as microRNA sponges, and are
mainly localized in cytoplasm (19). Intronic circRNAs and EIciRNAs are
localized in the nucleus and function as transcriptional regulators
of the parent gene (20,21). CircRNAs can regulate gene
expression at different stages, including transcription and
translation (21). Previous
studies have reported the important roles of circRNAs in cancer
(22,23). The altered expression of circRNAs
has been confirmed to be associated with the malignant features of
cancer cells, including their dysregulated proliferation, migration
and invasion. circ_0001649 was found to be downregulated in several
cancer types, including gastric cancer, hepatocellular cancer, and
cholangiocarcinoma (22–24), and was found to act as a tumor
suppressor and a diagnostic marker. circ_0008039 is upregulated in
breast cancer, and can promote cell proliferation and migration by
serving as a competing endogenous RNA of miR-432-5p (25). circHIPK3 was found to be
significantly upregulated in hepatocellular cancer and bladder
cancer, and its dysregulation affects the proliferation, migration
and invasion of cancer cells by serving as a sponge for multiple
miRNAs, including miR-124, miR-558 and miR-379 (26). circRNAs are also involved in the
progression of oral cancer. Specifically, circ_100290 is
upregulated in oral cancer tissues and functions as a competing
endogenous RNA by sponging miR-29 family members, upregulating CDK6
expression, which accelerates cancer progression (27). There are still multiple circRNAs
that remain to be investigated. Furthermore, the biogenesis,
cellular locations, functions and molecular mechanisms of circRNAs
require further investigation.
To the best of our knowledge, the expression profile
of circRNAs in TSCC has not been previously investigated. In the
present study, a number of dysregulated circRNAs were identified.
Among these circRNAs, circRNA_081069 and circRNA_087212 were the
most significantly upregulated and downregulated, respectively. To
the best of our knowledge, there is no detailed research of the
function and underlying mechanisms of these circRNAs. In the
present study, it was found that knockdown of circ_081069
suppressed the migratory and proliferative ability of TSCC cells,
suggesting that this circRNA may have an oncogenic effect. Since
circRNAs can act as miRNA sponges, bioinformatic analysis was
conducted to predict the possible interacting miRNAs, and it was
predicted that circ_081069 was potentially able to interact with
miR-665. miR-665 was found to suppress osteosarcoma cell
proliferation and invasion by inhibiting the small G protein Rab23
(14). In the present study, the
potential interaction between circ_081069 and miR-665 was
investigated, and luciferase activity assay showed that miR-665
directly targeted circ_081069. In addition, miR-29a was predicted
to interact with circ_087212. miR-29a was identified to regulate
malignant cell features and drug resistance in several cancer
types, including hepatocellular cancer and lung cancer by targeting
Sirtuin 1 and NRAS (15,28). Based on the present results,
identifying novel approaches that may target and affect the
stability of the interaction between circRNA and miRNAs may be
important for inhibiting cancer metastasis, invasiveness and
recurrence. However, additional studies are required to further
clarify the circRNA-miRNA network in TSCC.
The present study identified circRNAs profiles in
TSCC and the possible circRNA-miRNA interactions. The present
findings may increase the understanding of TSCC carcinogenesis and
development, facilitating the development of novel approaches for
the diagnosis and treatment of TSCC.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Xin Cong
(Department of Physiology and Pathophysiology, Peking University
Health Science Center and Key Laboratory of Molecular
Cardiovascular Sciences) for her kind suggestions.
Funding
The present work was supported by The Peking
University School of Stomatology Postdoctoral Fund (grant no.
YS0203), The Research Foundation of Peking University School and
Hospital of Stomatology (grant no. PKUSS20190102) and The Doctoral
Start Up Fund of Beijing Hospital (grant no. bj-2018-022).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TW performed the cell experiment and wrote the
manuscript. PY collected the tissue samples and analyzed the data.
GYY designed the study. ZYZ designed the experimental study and
revised the manuscript. All the authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by The Ethics Committee of
Peking University School and Hospital of Stomatology (approval no.
PKUSSIRB-2013009). The patient consents were acquired before tissue
collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Patel RS, Clark JR, Dirven R, Wyten R, Gao
K and O'Brien CJ: Prognostic factors in the surgical treatment of
patients with oral carcinoma. ANZ J Surg. 79:19–22. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yuen PW, Lam KY, Chan AC, Wei WI and Lam
LK: Clinicopathological analysis of local spread of carcinoma of
the tongue. Am J Surg. 175:242–244. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rodriguez T, Altieri A, Chatenoud L,
Gallus S, Bosetti C, Negri E, Franceschi S, Levi F, Talamini R and
Lavecchia C: Risk factors for oral and pharyngeal cancer in young
adults. Oral Oncol. 40:207–213. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Mo Y, Gong Z, Yang X, Yang M,
Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al: Circular RNAs in
human cancer. Mol Cancer. 16:252017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL
and Yang L: CircRNA-derived pseudogenes. Cell Res. 26:747–750.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brierley JD, Gospodarowicz MK and
Wittekind C: TNM Classification of Malignant Tumours. 8th. Wiley;
Chichester: 2017
|
|
11
|
Wu HJ, Zhang CY, Zhang S, Chang M and Wang
HY: Microarray expression profile of circular RNAs in heart tissue
of mice with myocardial infarction-induced heart failure. Cell
Physiol Biochem. 39:205–216. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo XH, Wang JY, Gao Y, Gao M, Yu GY,
Xiang RL, Li L, Yang NY, Cong X, Xu XY, et al: Decreased
adiponectin level is associated with aggressive phenotype of tongue
squamous cell carcinoma. Cancer Sci. 104:206–213. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong C, Du Q, Wang Z, Wang Y, Wu S and
Wang A: MicroRNA-665 suppressed the invasion and metastasis of
osteosarcoma by directly inhibiting RAB23. Am J Transl Res.
8:4975–4981. 2016.PubMed/NCBI
|
|
15
|
Zhang Y, Yang L, Wang S, Liu Z and Xiu M:
MiR-29a suppresses cell proliferation by targeting SIRT1 in
hepatocellular carcinoma. Cancer Biomark. 22:151–159. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dou Y, Cha DJ, Franklin JL, Higginbotham
JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG
and Zhang B: Circular RNAs are down-regulated in KRAS mutant colon
cancer cells and can be transferred to exosomes. Sci Rep.
6:379822016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lasda E and Parker R: Circular RNAs
co-precipitate with extracellular vesicles: A possible mechanism
for circRNA clearance. PLoS One. 11:e01484072016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Has circ 0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L,
Cui Y and Jiang X: Downregulated circular RNA has circ 0001649
regulates proliferation, migration and invasion in
cholangiocarcinoma cells. Biochem Biophys Res Commun. 496:455–461.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li WH, Song YC, Zhang H, Zhou ZJ, Xie X,
Zeng QN, Guo K, Wang T, Xia P and Chang DM: Decreased expression of
has circ 00001649 in gastric cancer and its clinical significance.
Dis Markers. 2017:45876982017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Lu C, Zhou Y, Zhang Z and Sun L:
Circular RNA has circ 0008039 promotes breast cancer cell
proliferation and migration by regulating miR-432-5p/E2F3 axis.
Biochem Biophys Res Commun. 3:358–363. 2018. View Article : Google Scholar
|
|
26
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: CircRNA 100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu X, Lv X, Yang Q, Jin H, Zhou W and Fan
Q: MicroRNA-29a functions as a tumor suppressor and increases
cisplatin sensitivity by targeting NRAS in lung cancer. Technol
Cancer Res Treat. 17:15330338187589052018. View Article : Google Scholar : PubMed/NCBI
|