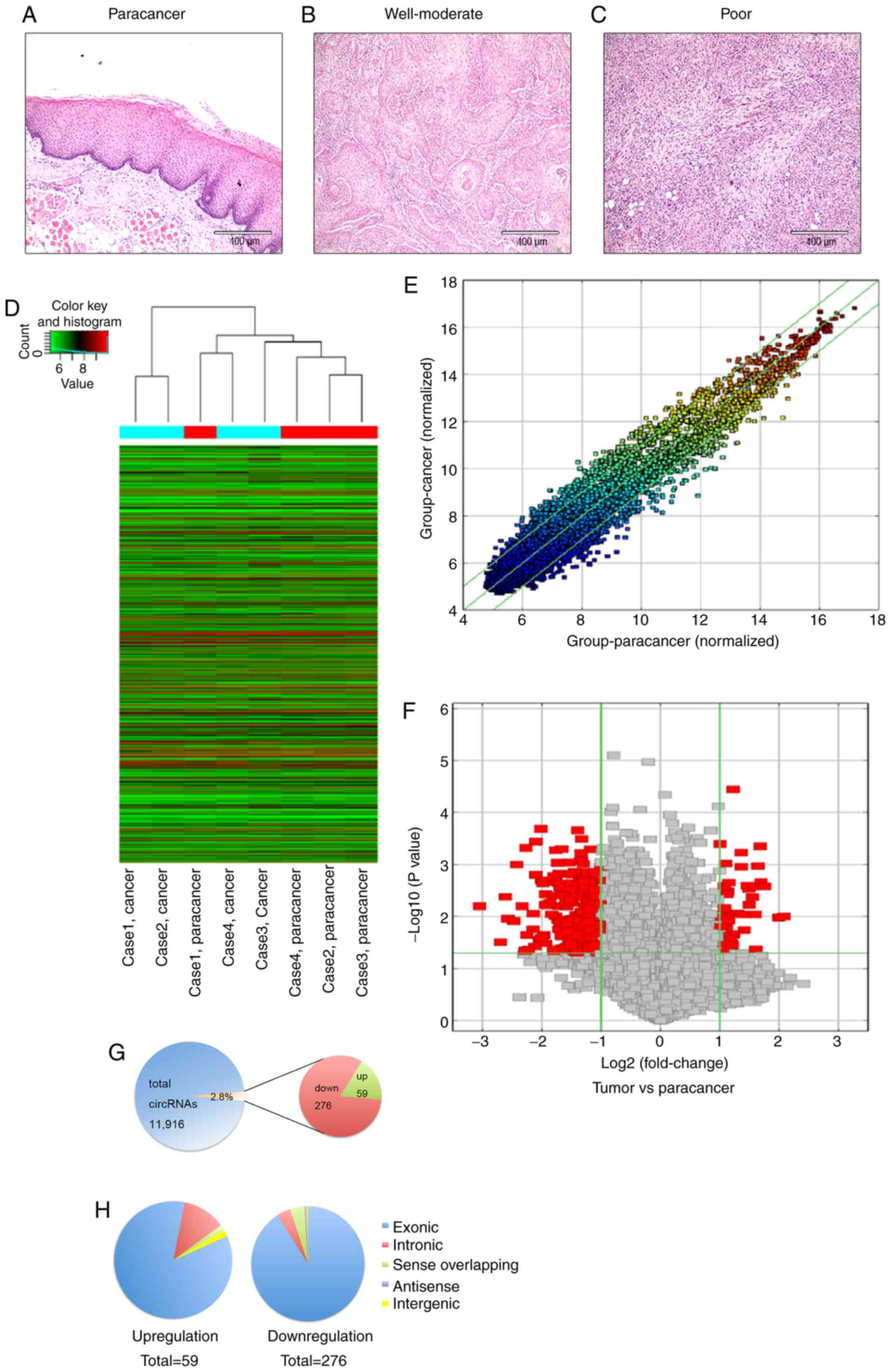
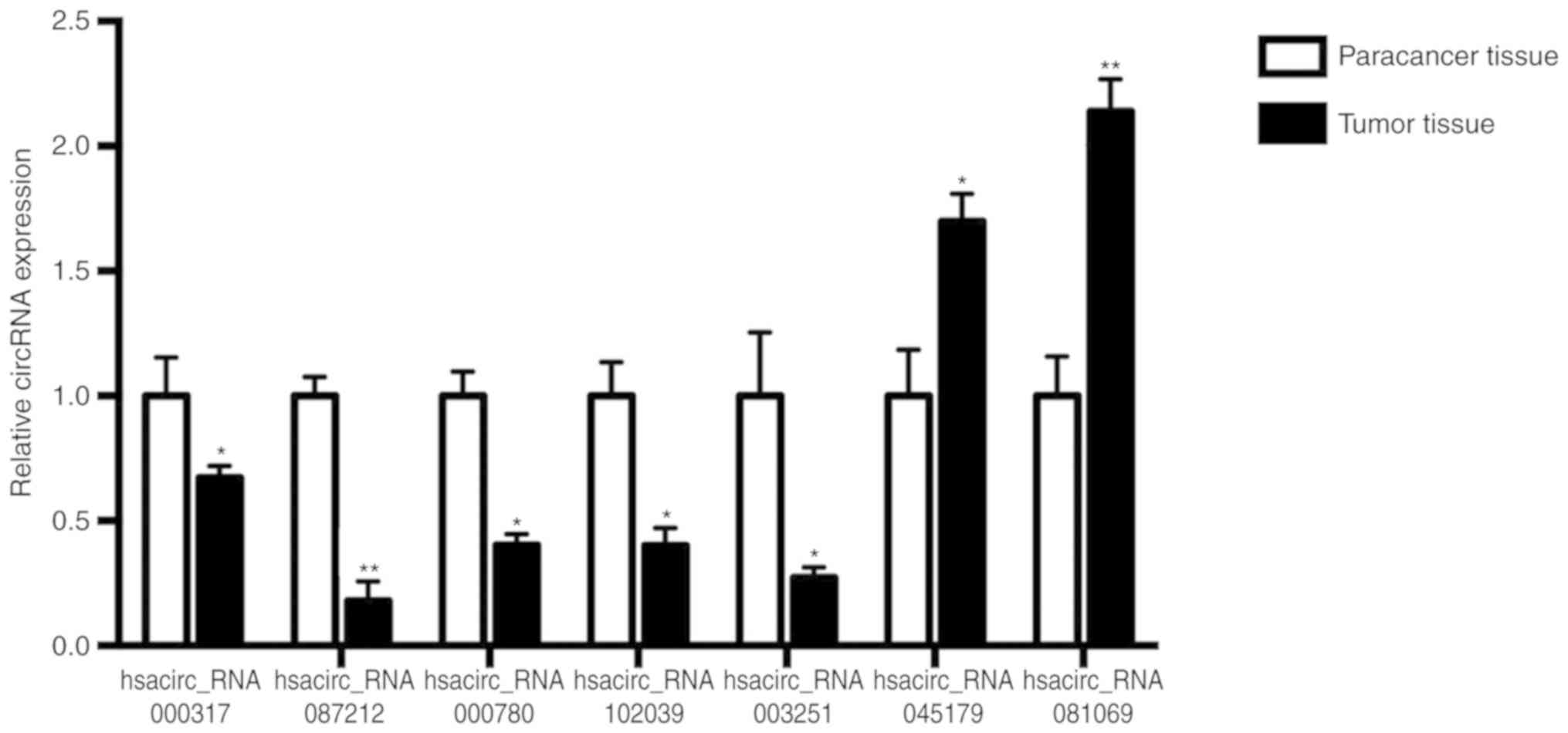
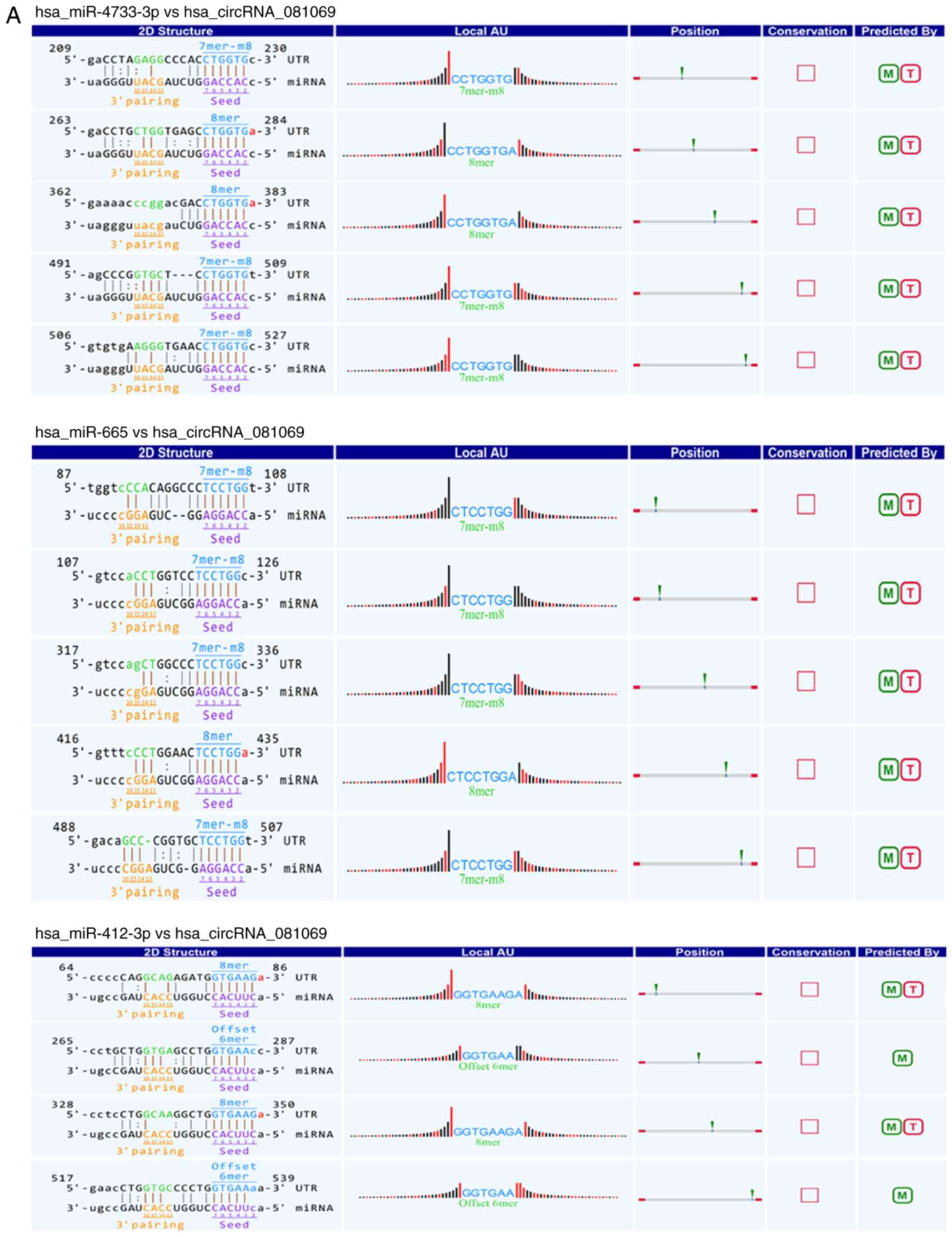
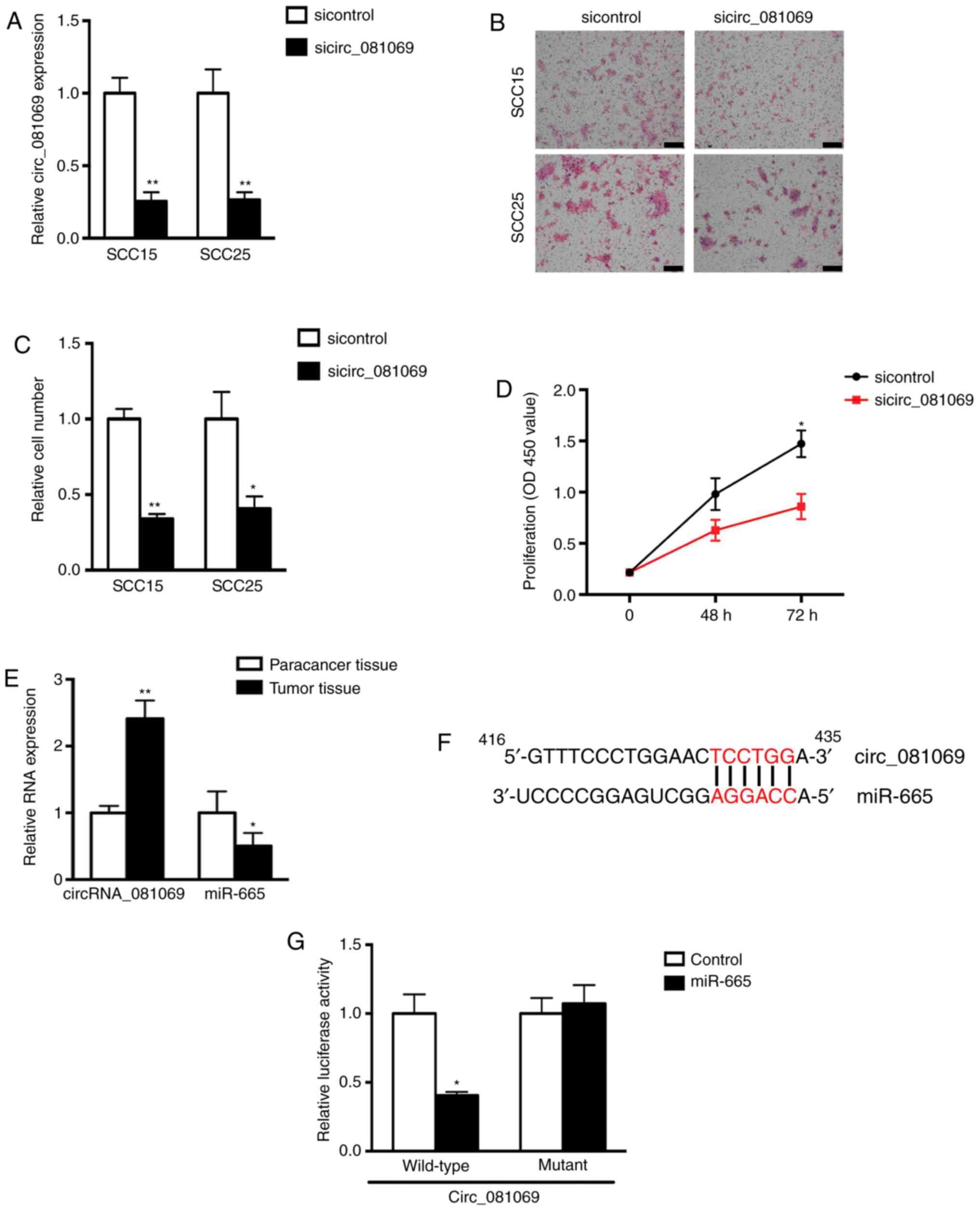
|
1
|
Patel RS, Clark JR, Dirven R, Wyten R, Gao
K and O'Brien CJ: Prognostic factors in the surgical treatment of
patients with oral carcinoma. ANZ J Surg. 79:19–22. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yuen PW, Lam KY, Chan AC, Wei WI and Lam
LK: Clinicopathological analysis of local spread of carcinoma of
the tongue. Am J Surg. 175:242–244. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rodriguez T, Altieri A, Chatenoud L,
Gallus S, Bosetti C, Negri E, Franceschi S, Levi F, Talamini R and
Lavecchia C: Risk factors for oral and pharyngeal cancer in young
adults. Oral Oncol. 40:207–213. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Mo Y, Gong Z, Yang X, Yang M,
Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al: Circular RNAs in
human cancer. Mol Cancer. 16:252017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL
and Yang L: CircRNA-derived pseudogenes. Cell Res. 26:747–750.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brierley JD, Gospodarowicz MK and
Wittekind C: TNM Classification of Malignant Tumours. 8th. Wiley;
Chichester: 2017
|
|
11
|
Wu HJ, Zhang CY, Zhang S, Chang M and Wang
HY: Microarray expression profile of circular RNAs in heart tissue
of mice with myocardial infarction-induced heart failure. Cell
Physiol Biochem. 39:205–216. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo XH, Wang JY, Gao Y, Gao M, Yu GY,
Xiang RL, Li L, Yang NY, Cong X, Xu XY, et al: Decreased
adiponectin level is associated with aggressive phenotype of tongue
squamous cell carcinoma. Cancer Sci. 104:206–213. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong C, Du Q, Wang Z, Wang Y, Wu S and
Wang A: MicroRNA-665 suppressed the invasion and metastasis of
osteosarcoma by directly inhibiting RAB23. Am J Transl Res.
8:4975–4981. 2016.PubMed/NCBI
|
|
15
|
Zhang Y, Yang L, Wang S, Liu Z and Xiu M:
MiR-29a suppresses cell proliferation by targeting SIRT1 in
hepatocellular carcinoma. Cancer Biomark. 22:151–159. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dou Y, Cha DJ, Franklin JL, Higginbotham
JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG
and Zhang B: Circular RNAs are down-regulated in KRAS mutant colon
cancer cells and can be transferred to exosomes. Sci Rep.
6:379822016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lasda E and Parker R: Circular RNAs
co-precipitate with extracellular vesicles: A possible mechanism
for circRNA clearance. PLoS One. 11:e01484072016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Has circ 0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L,
Cui Y and Jiang X: Downregulated circular RNA has circ 0001649
regulates proliferation, migration and invasion in
cholangiocarcinoma cells. Biochem Biophys Res Commun. 496:455–461.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li WH, Song YC, Zhang H, Zhou ZJ, Xie X,
Zeng QN, Guo K, Wang T, Xia P and Chang DM: Decreased expression of
has circ 00001649 in gastric cancer and its clinical significance.
Dis Markers. 2017:45876982017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Lu C, Zhou Y, Zhang Z and Sun L:
Circular RNA has circ 0008039 promotes breast cancer cell
proliferation and migration by regulating miR-432-5p/E2F3 axis.
Biochem Biophys Res Commun. 3:358–363. 2018. View Article : Google Scholar
|
|
26
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: CircRNA 100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu X, Lv X, Yang Q, Jin H, Zhou W and Fan
Q: MicroRNA-29a functions as a tumor suppressor and increases
cisplatin sensitivity by targeting NRAS in lung cancer. Technol
Cancer Res Treat. 17:15330338187589052018. View Article : Google Scholar : PubMed/NCBI
|