Introduction
Schizophrenia (SCZ) has a strong genetic component
with a heritability of up to 80% (1,2).
Tardive dyskinesia (TD) is presented by repetitive and jerking
movements in the face, neck and tongue and is considered to be a
serious side effect associated with the use of specific
antipsychotic medications that are used to treat SCZ and other
mental illnesses. TD may demonstrate a comorbidity of 20–30% with
SCZ (3,4). Recent studies have reported that
complex interactions of genetic, environmental and epigenetic
factors play important roles in SCZ and TD (4–7). DNA
methylation is an epigenetic mechanism that influences CG
dinucleotides by adding a methyl group (CH3) to them.
Previously, several studies have reported the association between
DNA methylation and SCZ (8–12)
and TD (13,14).
Insulin resistance has reportedly been associated
with atherosclerosis and type 2 diabetes (15). The insulin receptor substrate 1
(IRS1) gene is located at 2q36.3 (16–18)
and it may be involved in the regulation of critical metabolic and
signaling and secretion pathways (19–22).
A genome wide association study in a French sample showed that one
single nucleotide polymorphism (SNP) rs2943641, within 500 kb
upstream of the IRS1 gene, was associated with type 2
diabetes, insulin resistance and hyperinsulinemia (23). Furthermore, the IRS1 gene
DNA methylation was associated with high body mass index (BMI)
scores and obesity (24,25). However, there was no significant
association between the methylation of the IRS1 gene and
type 2 diabetes (26). Gunnell
et al (27) attempted to
evaluate the SNP rs1801278 in the IRS1 gene with SCZ but
could not identify any significant association. Recently,
IRS1 gene methylation has been found to be associated with
SCZ (9,28). Pyrosequencing provides a
quantitative analysis of DNA methylation levels (29–31)
and has been employed to detect DNA methylation levels of the
IRS1 gene in type 2 diabetes (26) and human islets of type 2 diabetes
(32).
A pilot study using methylated DNA
immunoprecipitation coupled with next-generation sequencing (our
unpublished data) revealed that the IRS1 gene was
hypomethylated in TD compared with SCZ groups. The present study
aimed to quantify DNA methylation levels of the IRS1 gene
via pyrosequencing to examine the association of the IRS1
gene DNA methylation levels with SCZ or TD. The general linear
model (GLM) was used to examine the differences in the DNA
methylation levels among diagnostic groups.
Materials and methods
Participants
The present study recruited 10 SCZ patients with TD
and 10 without TD (NTD) from the Beijing Huilongguan Hospital
(China) between January 2016 and June 2017. SCZ was diagnosed using
the Diagnostic and Statistical Manual of Mental Disorders version
IV (DSM-IV) (33) and TD was
confirmed by two well-trained psychiatrists with extensive clinical
experiences, based on the criteria of Schooler and Kane (34). The 20 SCZ patients were
administered with one of the following: Risperidone, paliperidone
or olanzapine. The TD patients were typically between 18 and 40
years of age and demonstrated an abnormal involuntary movement
scale (AIMS) score that was >3 in at least one part or >2 in
two or more parts (35). The same
criteria were used for NTD patients with AIMS=0. The exclusion
criteria included: Patients with severe physical or organic
encephalopathy, drug or alcohol abuse history (except tobacco),
pregnant or lactating women, patients administered with
neurotrophic agents or free radical metabolism drugs within 12
weeks prior to participation, and other mental illnesses
demonstrating a diagnosis of DSM-IV Axis I (14). The severity of TD symptoms was
assessed using AIMS, with an inter-rater correlation coefficient
(ICC)>0.80. The patients' psychotic symptoms were evaluated
using the positive and negative syndrome scale (PANSS) (36), with an ICC>0.85, which was
maintained for the PANSS total score after the scale training. A
total of 10 healthy controls (HCs) matched for age, sex and
education were subsequently recruited from the local community.
Ethical approval was received from the Ethics Review Board of
Beijing Huilongguan Hospital, China, and written informed consent
was obtained from all participants or their guardians.
DNA extraction, bisulfite treatment
and pyrosequencing
Fasting venous blood (5 ml) from a forearm vein was
obtained from each participant at 7:00 a.m. the next morning after
the day of clinical assessment. DNA was extracted from the above
blood samples using a standard genomic DNA sample kit (Illumina,
Inc.), according to the manufacturer's protocol. DNA concentration
and purity were detected by NanoDrop spectrophotometer (NanoDrop
Technologies, Thermo Fisher Scientific, Inc.) and the integrity was
tested using 1% agarose gel electrophoresis (14). Subsequently, 500 ng of each sample
was treated with bisulfite, by employing the Epitect Bisulfite Kit
(Qiagen GmbH), according to the manufacturer's protocol. Parts of
the CpG islands in the promoter region within the IRS1 gene
were amplified with the help of PCR assays. DNA fragments were
typically amplified using the PyroMark PCR kit (Qiagen GmbH),
according to the manufacturer's protocol, from 2 µl
bisulfite-treated genomic DNA. Sample preparation and
pyrosequencing reactions were subsequently carried out using the
PyroMark Q96 ID (Qiagen GmbH). The pyrosequencing assays were
performed for all the study samples on both Pyro Mark Q24 MDx and
PyroMarkQ96 ID, while using Pyro Mark Gold reagents (Qiagen GmbH).
The Pyro Mark Assay Design software version 2.0 (Qiagen GmbH) was
used to generate the primers for the IRS1, targeting four
CpGs in the gene promoter (Table
I). Percentage of each CpG site and the mean methylation
percentage of the 4 CpGs quantitatively revealed the methylation
levels.
 | Table I.Primer sequences used in the
pyrosequencing analysis. |
Table I.
Primer sequences used in the
pyrosequencing analysis.
| Primer type | Primer
sequence | CpG Sites | Position 5′-3′ |
|---|
| Forward |
5′-AGTGGTTATAGAGTTTGATGTTTATTAGT-3′ | 4 | 146–174 |
| Reverse |
5′-CCTAAAACCCAAAAACCTAAATCA-3′ |
| 294–271 |
| Sequencing |
5′-GTTTGATGTTTATTAGTTGTAGTA-3′ |
| 158–181 |
Statistical analysis
The χ2 test was used to detect gender
differences among TD, NTD and HC groups. Pearson's correlation
analysis was concurrently used to examine the correlations among
methylation levels among the 4 CpG sites, average level of the 4
CpG sites and severity of SCZ. Differences among the continuous
variables including age and education were evaluated by Fisher's F
test in the GLM. Normality of DNA methylation levels was tested by
the Shapiro-Wilk test. Differences of DNA methylation levels among
the 3 groups, TD, NTD and HC, for individual CpG sites and the mean
of 4 CpGs were detected using GLM. All the analyses were performed
using SAS version 9.4 software (SAS Institute, Inc.). The PROC
POWER statement in SAS was used to compute power in the present
study. P<0.05 was considered to indicate a statistically
significant difference.
Results
Demographics and clinical
characteristics
The demographic factors in the TD, NTD and HC groups
are presented in Table II. No
statistical significances were observed in age (P=0.910), sex
(P=1.00) and educational levels (P=0.832) among the 3 groups. There
were no significant differences between TD and NTD groups in the
disease duration and treatment, drug dose quantized using CPZ
equivalents or drug types. Additionally, no significant difference
was observed between TD and NTD groups in the PANSS total,
positive, negative or general scores (Table III).
 | Table II.Descriptive characteristics of
patients and controls. |
Table II.
Descriptive characteristics of
patients and controls.
| Variable | TH group
(n=10) | NTH group
(n=10) | HC group
(n=10) | P-value |
|---|
| Male/female | 5/5 | 5/5 | 5/5 | 1.000 |
| Age (years) |
31.6±11.8 |
33.8±11.0 |
33.2±11.9 | 0.910 |
| Education
(years) | 10.6±3.2 | 10.9±2.6 | 11.4±3.3 | 0.832 |
| Duration of disease
(years) |
11.1±11.1 | 10.2±9.2 | n/a | 1.000 |
| Duration of
treatment (months) |
25.6±22.1 |
24.5±26.1 | n/a | 0.570 |
| CPZ equivalents
(mg) |
662.1±431.4 |
498.4±275.4 | n/a | 0.474 |
| Drug type |
|
|
| 0.584 |
| 1
typical antipsychotic | 1 | 0 | n/a |
|
| 1
atypical antipsychotic | 6 | 7 | n/a |
|
| 2
atypical antipsychotics | 3 | 3 | n/a |
|
 | Table III.Clinical parameters of TD and NTD
groups. |
Table III.
Clinical parameters of TD and NTD
groups.
| Variable | TH group
(n=10) | NTH group
(n=10) | P-value |
|---|
| PANSS total
score |
74.0±21.6 | 73.4±9.7 | 0.633 |
| PANSS |
|
|
|
|
Positive | 17.0±7.2 | 18.7±4.6 | 0.489 |
|
Negative | 23.6±8.5 | 18.4±6.2 | 0.184 |
|
General | 33.6±8.6 | 35.2±7.0 | 0.458 |
| Abnormal
involuntary movement scale | 13.4±5.3 | 0 | <0.0001 |
Normality test
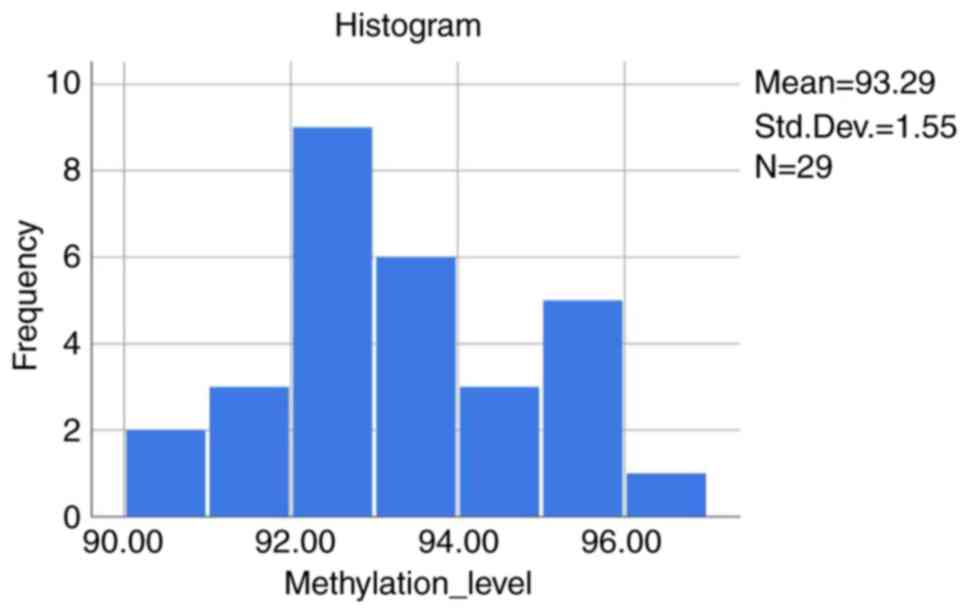
The average DNA methylation levels of the 4 CpG
sites followed normal distribution (Fig. 1) based on the Shapiro-Wilk test
(P>0.150).
Correlation analyses
Table IV
demonstrated that no significant correlations were found among the
individual methylation levels of the 4 CpG sites (all P-values
>0.05); however, CpG sites 1 and 4 demonstrated a strong
correlation to the mean value of the 4 CpG sites (P<0.0001).
Furthermore, there were no obvious correlations among CpG sites and
the severity of SCZ measured by PANSS scores.
 | Table IV.Pearson correlation coefficients
among methylation levels and clinical characters. |
Table IV.
Pearson correlation coefficients
among methylation levels and clinical characters.
| Variable | IRS1 Site 1 | IRS1 Site 2 | IRS1 Site 3 | IRS1 Site 4 | IRS1 Average | PANSST | PANSSP | PANSSN | PANSSG |
|---|
| IRS1 Site 1 | 1.000 | −0.345 | 0.299 | 0.285 | 0.821a | −0.323 | −0.499 | −0.219 | −0.121 |
| IRS1 Site 2 |
| 1.000 | −0.055 | −0.222 | −0.276 | −0.092 | 0.013 | 0.122 | 0.037 |
| IRS1 Site 3 |
|
| 1.000 | 0.076 | 0.310 | 0.198 | 0.013 | 0.266 | 0.111 |
| IRS1 Site 4 |
|
|
| 1.000 | 0.770a | 0.059 | −0.052 | −0.051 | 0.153 |
| IRS1 Average |
|
|
|
| 1.000 | −0.160 | −0.366 | −0.128 | 0.008 |
| PANSST |
|
|
|
|
| 1.000 | 0.521b | 0.832a | 0.836a |
| PANSSP |
|
|
|
|
|
| 1.000 | 0.184 | 0.177 |
| PANSSN |
|
|
|
|
|
|
| 1.000 | 0.623c |
| PANSSG |
|
|
|
|
|
|
|
| 1.000 |
GLM analyses
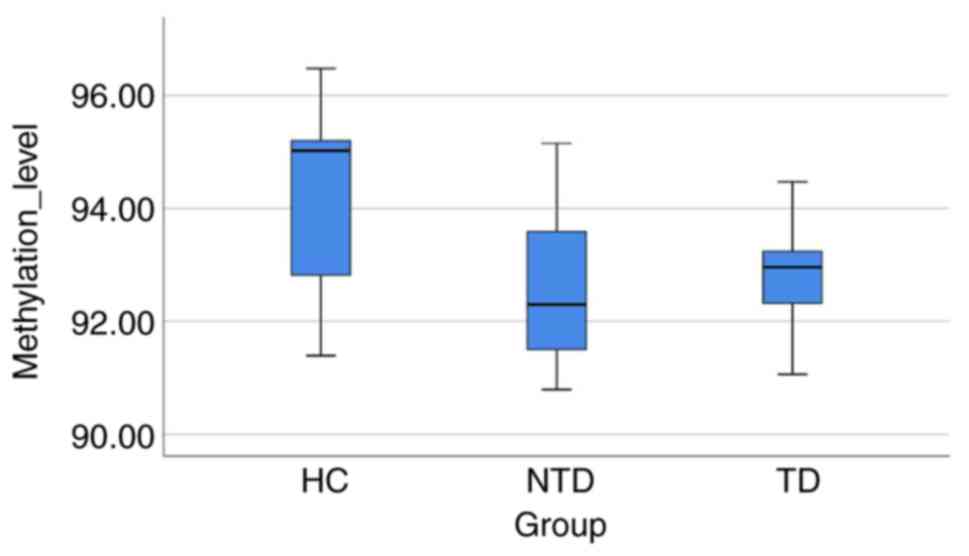
The linear GLM revealed significant differences with
regard to the CpG site 1 and the mean value of the 4 CpG sites
(P=0.0001 and P=0.0126, respectively; Table V and Fig. 2), among the 3 groups. Furthermore,
the HC group demonstrated higher methylation levels in the CpG site
1 compared with those in TD, NTD and TD + NTD (P=0.0003,
P<0.0001 and P<0.0001, respectively) and the average of 4 CpG
sites (P=0.0176, P=0.0063 and P=0.003, respectively).
 | Table V.General linear model analysis of DNA
methylation levels. |
Table V.
General linear model analysis of DNA
methylation levels.
| Group |
Mean/F/t/P-value | Site 1 | Site 2 | Site 3 | Site 4 | Average |
|---|
| TD | Mean ± standard
deviation | 84.3±3.8 | 100.0±0.0 | 100.0±0.0 | 86.9±1.3 | 92.8±1.0 |
| NTD |
| 84.0±3.4 |
99.7±0.7 |
99.8±0.7 | 86.8±2.9 | 92.6±1.4 |
| HC |
| 90.2±1.4 |
99.8±0.7 | 100.0±0.0 | 87.8±5.6 | 94.4±1.6 |
| TD vs. NTD vs
HC | F value | 13.14 | 0.51 | 0.95 | 0.14 | 5.20 |
|
| P-value | 0.0001 | 0.6052 | 0.4011 | 0.8659 | 0.0126 |
| TD vs. HC | t value | −4.22 | 0.92 | 0.0 | −0.42 | −2.53 |
|
| P-value | 0.0003 | 0.3677 | 1.0000 | 0.6758 | 0.0176 |
| NTD vs. HC | t value | −4.59 | 0.07 | −1.20 | 0.06 | −2.97 |
|
| P-value | <0.0001 | 0.9433 | 0.2403 | 0.9524 | 0.0063 |
| TD + NTD vs.
HC | t value | −5.21 | 0.56 | −0.72 | −0.54 | −3.26 |
|
| P-value | <0.0001 | 0.5822 | 0.4762 | 0.5903 | 0.0030 |
| TD vs. NTD | t value | 0.25 | 0.85 | 1.17 | −0.50 | 0.36 |
|
| P-value | 0.8072 | 0.4048 | 0.2527 | 0.6237 | 0.7220 |
Discussion
To the best of our knowledge, this is the first
study to quantitatively analyze DNA methylation using
pyrosequencing and determine the IRS1 gene promoter
methylation levels of the 4 CpG sites among the TD, NTD and HC
groups. GLM analyses revealed lower methylation levels in CpG site
1 and the mean value of the 4 CpG sites of the TD, NTD and TD + NTD
groups compared with the control group.
The methylation status of the IRS1 gene has
been found to be associated with SCZ through post-mortem analysis
of human brain tissue from 24 patients with SCZ and 24 unaffected
controls using the Illumina Infinium HumanMethylation450 Bead Chip
(9). Another study using blood
samples and Illumina HumanMethylation450 BeadChip reported that
IRS1 was associated with SCZ (28). The present study used
pyrosequencing to reveal that the DNA methylation level in SCZ
patients (NTD group) was significantly lower compared with healthy
controls with regard to the CpG site 1 and average values of the 4
CpG sites in a Chinese sample (Table
V). Furthermore, it was observed that the TD group demonstrated
significantly lower DNA methylation levels compared with healthy
controls (Table V).
Studies have previously indicated that insulin may
act as a metabolic signal and the IRS1 gene may influence
body weight control and glucose homeostasis (37–39).
Furthermore, several studies have also reported associations of
IRS1 polymorphisms with cancer, diabetes, glucose levels and
obesity (40–44). Furthermore, insulin/insulin
receptor signaling and the insulin-like growth factor (IGF) pathway
may have different functions in the central nervous system in brain
development, blood glucose regulation, dendritic growth and
neuronal apoptosis (37,45–48).
However, Che et al (47)
was unable to identify any association between the IRS1 gene
and epilepsy in a Chinese sample but suggested that the genes
associated with the insulin signaling pathway may affect the
therapeutic response of temporal lobe epilepsy. Previous
epidemiologic studies have demonstrated that SCZ and other mental
illnesses may increase the risk of developing type 2 diabetes and
other metabolic disorders (49,50).
The co-morbidity of type 2 diabetes and SCZ may partially be due to
concurrent biological susceptibility of these two conditions
(51,52). As an example, the TCF7L2
gene is associated with both type 2 diabetes and type 2 diabetes
with SCZ or the schizoaffective disorder observed in African
American patients (50), while the
rs7903146 in the TCF7L2 gene is associated with SCZ
(53). Furthermore, insulin-like
growth factor II mRNA-binding protein 2 gene (IGF2BP2) may
be associated with SCZ (54).
Additionally, insulin signaling, among other type 2
diabetes-related pathways, could act as a bridge between SCZ and
type 2 diabetes (55). However,
one previous study could not associate rs1801278 in the IRS1
gene with SCZ (27). Therefore,
the present study provided further evidence of the involvement of
IRS1 gene with regard to the development of SCZ or TD
(Table IV).
A previous study examined the DNA methylation levels
of three CpG sites in the IRS1 gene with type 2 diabetes
using pyrosequencing (26);
however, there was no significant difference in the methylation
levels of the 3 CpG sites in the IRS1 gene between the
controls and type 2 diabetes patients, which suggested that the DNA
methylation levels of the IRS1 gene did not play a major
role in the occurrence of type 2 diabetes (26). The present study revealed that both
TD and NTD groups had significantly lower methylation levels in the
CpG site 1 and mean values of the 4 CpG sites in the IRS1
gene compared with healthy controls (Table V). One primary difference may be
that there were 3 CpG sites in the above type 2 diabetes study
(26), while there were 4 in the
present study; however, it is possible that these CpG sites may be
different. Notably, the present study found numerous similarities,
which included the fact that DNA methylation levels in the diabetes
groups were slightly lower compared with those in the non-diabetes
groups, specifically in the CpG sites 1 and 2 in the entire sample.
Furthermore, in the above type 2 diabetes study, a similar trend
was noted among the groups of men and women, despite the fact that
the differences did not reach a 5% significant level (26).
Previous studies have suggested a role of
IRS1 in cognitive impairment and Alzheimer's disease
(56–59). The present study and the previous
reports related to the role of IRS1 gene methylation in SCZ
(9,28) may provide adequate evidence to
demonstrate that SCZ and Alzheimer's disease may share a common
network of dysregulation (60).
Furthermore, epidemiologic studies have reported a possible
association between the insulin resistance of type 2 diabetes
mellitus and increased incidence of Alzheimer's disease (61,62).
As a matter of fact, insulin resistance results in a diminished
glucose uptake in similar regions of the brain in Alzheimer's
disease and type 2 diabetes mellitus (58,59).
Therefore, the IRS1 gene may have a pleiotropic effect on
Alzheimer's disease, type 2 diabetes and SCZ.
There are a number of strengths in the current
study. First, this is the first study to conduct a quantitative
analysis of DNA methylation levels of the IRS1 gene in SCZ
and TD, while previous studies focused on the methylation status in
SCZ (9,28). Second, the present study applied a
GLM to examine the methylation levels of the 4 CpG sites within the
IRS1 gene among TD, NTD and HC groups. Third, the present
study tested the normality of the average methylation levels of the
4 CpG sites and observed that the average DNA methylation levels of
these sites followed normal distribution. Although correlation
analysis was performed, significant correlations among the
methylation percentages of the four CpG sites with the severity of
SCZ was not observed. However, the present study has certain
limitations: First, a peripheral blood sample was used in view of
the difficulty in obtaining brain tissues to study the disorders of
the central nervous system. Second, the sample size of the three
groups in the methylation study was relatively small because the
prevalence of TD in general population is typically low. Based on
the PROC POWER determined in SAS 9.4, the power level was 63.3%
with 30 individuals on comparing overall means in DNA methylation
levels for the four CpG sites; however, it was possible to increase
the power up to 99.1% while testing the CpG site 1. Third, the
present study examined and included a limited number of sites
associated with IRS1 gene methylation (only 4 sites), which
highlighted the importance of increasing the number of sites in
future investigations. Additionally, there may be variations
between medications, which could not have been detected by the
present study.
In conclusion, pyrosequencing demonstrated that the
DNA methylation levels of the IRS1 gene in TD and NTD groups
were significantly lower compared with the healthy control group.
However, the DNA methylation levels in TD did not demonstrate any
significant differences compared with those in the NTD group. This
is the first study to compare CpG methylation levels of TD and NTD
with healthy controls and the findings demonstrated adequate
evidence of the possible roles of IRS1-associated DNA methylation
in SCZ and TD. In the future, it will be worthy to detect age and
gender effects using a large sample and perform the functional
study of these 4 CpG sites of the IRS1 gene to evaluate the
role of this gene in the pathogenesis of SCZ and TD.
Acknowledgements
The authors would like to thank KangChen Bio-Tech
Co., Ltd. for experimental assistance in MeDIP sequencing and
Beijing Liuhe Huada Gene Technology Co., Ltd. for experiment
assistance in pyrosequencing.
Funding
Dr Yunlong Tan received support from the Beijing
Natural Science Foundation (grant no. 7151005) and the National
Science Foundation of China (grant no. 81771452) for the present
study.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding authors on reasonable
request.
Authors' contributions
YLL, FY and YT planned and managed the project. YLL,
JH, ZW, FY and ST were involved in designing the study and
collecting the data. YLL, PZ, ZW and ST performed recruitment and
clinical assessment. KW, PZ, YLL, YL, YKL and YT conducted
statistical analyses, interpreted the results, searched the
literatures and wrote parts of the manuscript. All authors
performed a final review of the manuscript and all authors approved
the submission of this manuscript.
Ethics approval and consent to
participate
The current study was approved by the Ethics Review
Board of Beijing Huilongguan Hospital (China). Written informed
consent was obtained from all participants enrolled in our study
and/or from their guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sullivan PF, Kendler KS and Neale MC:
Schizophrenia as a complex trait: Evidence from a meta-analysis of
twin studies. Arch General Psychia. 60:1187–1192. 2003. View Article : Google Scholar
|
|
2
|
Gejman PV, Sanders AR and Duan J: The role
of genetics in the etiology of schizophrenia. Psychiatr. Clin North
Am. 33:35–66. 2010. View Article : Google Scholar
|
|
3
|
Tarsy D, Lungu C and Baldessarini RJ:
Epidemiology of tardive dyskinesia before and during the era of
modern antipsychotic drugs. J Handb Clin Neurol. 100:601–616. 2011.
View Article : Google Scholar
|
|
4
|
Correll CU, Kane JM and Citrome LL:
Epidemiology, prevention, and assessment of tardive dyskinesia and
advances in treatment. J Clin Psychiatry. 78:1136–1147. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Csoka AB and Szyf M: Epigenetic
side-effects of common pharmaceuticals: A potential new field in
medicine and pharmacology. Med Hypotheses. 73:770–780. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee HJ and Kang SG: Genetics of tardive
dyskinesia. Int Rev Neurobiol. 98:231–264. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lanning RK, Zai CC and Müller DJ:
Pharmacogenetics of tardive dyskinesia: An updated review of the
literature. Pharmacogenomics. 17:1339–1351. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishioka M, Bundo M, Kasai K and Iwamoto
K: DNA methylation in schizophrenia: Progress and challenges of
epigenetic studies. Genome Med. 4:962012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wockner LF, Noble EP, Lawford BR, Young
RM, Morris CP, Whitehall VL and Voisey J: Genome-wide DNA
methylation analysis of human brain tissue from schizophrenia
patients. Transl Psychiatry. 4:e3392014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hannon E, Dempster E, Viana J, Burrage J,
Smith AR, Macdonald R, St Clair D, Mustard C, Breen G, Therman S,
et al: An integrated genetic-epigenetic analysis of schizophrenia:
Evidence for co-localization of genetic associations and
differential DNA methylation. Genome Biol. 17:1762016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee SA and Huang KC: Epigenetic profiling
of human brain differential DNA methylation networks in
schizophrenia. BMC Med Genomics. 9 (Suppl 3):S682016. View Article : Google Scholar
|
|
12
|
Pries LK, Gülöksüz S and Kenis G: DNA
Methylation in Schizophrenia. Adv Exp Med Biol. 978:211–236. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang P, Li YL, An HM and Tan YL:
Preliminary construction of DNA methylation profiles of
schizophrenia patients with tardive dyskinesia. Chin J Psychiatry.
51:13–19. 2018.
|
|
14
|
Li Y, Wang KS, Zhang P, Huang J, An H,
Wang N, Yang F, Wang Z, Tan S, Chen S and Tan YL: Quantitative DNA
methylation analysis of DLGAP2 gene using pyrosequencing in
schizophrenia with tardive dyskinesia: A linear mixed model
approach. Sci Rep. 8:174662018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Abe H, Yamada N, Kamata K, Kuwaki T,
Shimada M, Osuga J, Shionoiri F, Yahagi N, Kadowaki T, Tamemoto H,
et al: Hypertension, hypertriglyceridemia, and impaired
endothelium-dependent vascular relaxation in mice lacking insulin
receptor substrate-1. J Clin Invest. 101:1784–1788. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun XJ, Rothenberg P, Kahn CR, Backer JM,
Araki E, Wilden PA, Cahill DA, Goldstein BJ and White MF: Structure
of the insulin receptor substrate IRS-1 defines a unique signal
transduction protein. Nature. 352:73–77. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stoffel M, Espinosa R III, Keller SR,
Lienhard GE, Le Beau MM and Bell GI: Human insulin receptor
substrate-1 gene (IRS1): Chromosomal localization to 2q35-q36.1 and
identification of a simple tandem repeat DNA polymorphism.
Diabetologia. 36:335–337. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nishiyama M, Inazawa J, Ariyama T,
Nakamura Y, Matsufuji S, Furusaka A, Tanaka T, Hayashi S and Wands
JR: The human insulin receptor substrate-1 gene (IRS1) is localized
on 2q36. Genomics. 20:139–141. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Myers MG Jr, Sun XJ and White MF: The
IRS-1 signaling system. Trends Biochem Sci. 19:289–293. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kulkarni RN, Winnay JN, Daniels M, Brüning
JC, Flier SN, Hanahan D and Kahn CR: Altered function of insulin
receptor substrate-1-deficient mouse islets and cultured beta-cell
lines. J Clin Invest. 104:R69–R75. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kido Y, Burks DJ, Withers D, Bruning JC,
Kahn CR, White MF and Accili D: Tissue-specific insulin resistance
in mice with mutations in the insulin receptor, IRS-1, and IRS-2. J
Clin Invest. 105:199–205. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim JK, Fillmore JJ, Sunshine MJ, Albrecht
B, Higashimori T, Kim DW, Liu ZX, Soos TJ, Cline GW, O'Brien WR, et
al: PKC-theta knockout mice are protected from fat-induced insulin
resistance. J Clin Invest. 114:823–827. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rung J, Cauchi S, Albrechtsen A, Shen L,
Rocheleau G, Cavalcanti-Proença C, Bacot F, Balkau B, Belisle A,
Borch-Johnsen K, et al: Genetic variant near IRS1 is associated
with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat
Genet. 41:1110–1115. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rönn T, Volkov P, Gillberg L, Kokosar M,
Perfilyev A, Jacobsen AL, Jørgensen SW, Brøns C, Jansson PA,
Eriksson KF, et al: Impact of age, BMI and HbA1c levels on the
genome-wide DNA methylation and mRNA expression patterns in human
adipose tissue and identification of epigenetic biomarkers in
blood. Hum Mol Genet. 24:3792–3813. 2015.PubMed/NCBI
|
|
25
|
Fradin D, Boëlle PY, Belot MP, Lachaux F,
Tost J, Besse C, Deleuze JF, De Filippo G and Bougnères P:
Genome-wide methylation analysis identifies specific epigenetic
marks in severely obese children. Sci Rep. 7:463112017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma J, Cheng J, Wang L, Wang H, Xu L, Liu
P, Bu S, Zhang L, Le Y, Ye M, et al: No association between IRS-1
promoter methylation and type 2 diabetes. Mol Med Rep. 8:949–953.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gunnell D, Lewis S, Wilkinson J, Georgieva
L, Davey GS, Day IN, Holly JM, O'Donovan MC, Owen MJ, Kirov G and
Zammit S: IGF1, growth pathway polymorphisms and schizophrenia: A
pooling study. Am J Med Genet B Neuropsychiatr Genet. 144B:117–120.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Montano C, Taub MA, Jaffe A, Briem E,
Feinberg JI, Trygvadottir R, Idrizi A, Runarsson A, Berndsen B, Gur
RC, et al: Association of DNA methylation differences with
schizophrenia in an epigenome-wide association study. JAMA
Psychiatry. 73:506–514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tost J and Gut IG: DNA methylation
analysis by pyrosequencing. Nat Protoc. 2:2265–2275. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mikeska T, Felsberg J, Hewitt CA and
Dobrovic A: Analysing DNA methylation using bisulphite
pyrosequencing. Methods Mol Biol. 791:33–35. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fakruddin M and Chowdhury A:
Pyrosequencing-An alternative to traditional Sanger sequencing. Am
J Biochem Biotech. 8:14–20. 2012. View Article : Google Scholar
|
|
32
|
Dayeh TA, Olsson AH, Volkov P, Almgren P,
Rönn T and Ling C: Identification of CpG-SNPs associated with type
2 diabetes and differential DNA methylation in human pancreatic
islets. Diabetologia. 56:1036–1046. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
American Psychiatric Association.
Diagnostic and Statistical Manual of Mental Disorders (DSM-IV)
Washington, DC: American Psychiatric Association; 1994
|
|
34
|
Schooler NR and Kane JM: Research
diagnoses for tardive dyskinesia. Arch Gen Psychiatry. 39:486–487.
1982. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fan B: Abnormal involuntary movement
rating scale (AIMS). Shanghai Arch Psychiat. 80–81. 1984.
|
|
36
|
Kay SR, Fiszbein A and Opler LA: The
positive and negative syndrome scale (PANSS) for schizophrenia.
Schizophr Bull. 13:261–276. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
White MF: Insulin signaling in health and
disease. Science. 302:1710–1711. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Burks DJ and White MF: IRS proteins and
β-cell function. Diabetes. 50:140–145. 2001. View Article : Google Scholar
|
|
39
|
Withers DJ, Burks DJ, Towery HH, Altamuro
SL, Flint CL and White MF: IRS-2 coordinates IGF-1
receptor-mediated beta-cell development and peripheral insulin
signalling. Nat Genet. 23:32–40. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lautier C, El Mkadem SA, Renard E, Brun
JF, Gris JC, Bringer J and Grigorescu F: Complex haplotypes of
IRS2 gene are associated with severe obesity and reveal
heterogeneity in the effect of Gly1057Asp mutation. Hum Genet.
113:34–43. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Slattery ML, Samowitz W, Curtin K, Ma KN,
Hoffman M, Caan B and Neuhausen S: Associations among IRS1, IRS2,
IGF1, and IGFBP3 genetic polymorphisms and colorectal cancer.
Cancer Epidemiol Biomark Prev. 13:1206–1214. 2004.
|
|
42
|
Neuhausen SL, Brummel S, Ding YC, Singer
CF, Pfeiler G, Lynch HT, Nathanson KL, Rebbeck TR, Garber JE, Couch
F, et al: Genetic variation in insulin-like growth factor signaling
genes and breast cancer risk among BRCA1 and BRCA2 carriers. Breast
Cancer Res. 11:R762009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng X, Tucker KL, Parnell LD, Shen J, Lee
YC, Ordovas JM, Ling WH and Lai CQ: Insulin receptor substrate 1
(IRS1) variants confer risk of diabetes in the Boston Puerto Rican
Health Study. Asia Pac J Clin Nutr. 22:150–159. 2013.PubMed/NCBI
|
|
44
|
Winder T, Giamas G, Wilson PM, Zhang W,
Yang D, Bohanes P, Ning Y, Gerger A, Stebbing J and Lenz HJ:
Insulin-like growth factor receptor polymorphism defines clinical
outcome in estrogen receptor-positive breast cancer patients
treated with tamoxifen. Pharmacogen J. 14:28–34. 2014. View Article : Google Scholar
|
|
45
|
Jones JI and Clemmons DR: Insulin-like
growth factors and their binding proteins: Biological actions.
Endocr Rev. 16:3–34. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Laban C, Bustin SA and Jenkins PJ: The
GH-IGF-I axis and breast cancer. Trends Endocrinol Metab. 14:28–34.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Che F, Fu Q, Li X, Gao N, Qi F, Sun Z, Du
Y and Li M: Association of insulin receptor H1085H C>T, insulin
receptor substrate 1 G972R and insulin receptor substrate 2 1057G/A
polymorphisms with refractory temporal lobe epilepsy in Han
Chinese. Seizure. 25:178–180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Park HJ, Kim SK, Kang WS, Park JK, Kim YJ,
Nam M, Kim JW and Chung JH: Association between IRS1 gene
polymorphism and autism spectrum disorder: A pilot case-control
study in Korean males. Int J Mol Sci. 17:E12272016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Suvisaari J, Perälä J, Saarni SI, Härkänen
T, Pirkola S, Joukamaa M, Koskinen S, Lönnqvist J and Reunanen A:
Type 2 diabetes among persons with schizophrenia and other
psychotic disorders in a general population survey. Eur Arch
Psychiatry Clin Neurosci. 258:129–136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Irvin MR, Wiener HW, Perry RP, Savage RM
and Go RC: Genetic risk factors for type 2 diabetes with
pharmacologic intervention in African-American patients with
schizophrenia or schizoaffective disorder. Schizophr Res.
114:50–56. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bellivier F: Schizophrenia, antipsychotics
and diabetes: Genetic aspects. Eur Psychiatry. 20 (Suppl
4):S335–S339. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lin PI and Shuldiner AR: Rethinking the
genetic basis for comorbidity of schizophreniaand type 2 diabetes.
Schizophr Res. 123:234–243. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hansen T, Ingason A, Djurovic S, Melle I,
Fenger M, Gustafsson O, Jakobsen KD, Rasmussen HB, Tosato S,
Rietschel M, et al: At-risk variant in TCF7L2 for type II diabetes
increases risk of schizophrenia. Biol Psychiatry. 70:59–63. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang X, Hui L, Liu Y, Wang ZQ, You Y,
Miao LN, Sun SL, Guan SL, Xiang Y, Kosten TR and Zhang XY: The type
2 diabetes mellitus susceptibility gene IGF2BP2 is associated with
schizophrenia in a Han Chinese population. J Clin Psychiatry.
74:e287–e292. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liu Y, Li Z, Zhang M, Deng Y, Yi Z and Shi
T: Exploring the pathogenetic association between schizophreniaand
type 2 diabetes mellitus diseases based on pathway analysis. BMC
Med Genomics. 6 (Suppl 1):S172013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Talbot K, Wang HY, Kazi H, Han LY, Bakshi
KP, Stucky A, Fuino RL, Kawaguchi KR, Samoyedny AJ, Wilson RS, et
al: Demonstrated brain insulin resistance in Alzheimer's disease
patients is associated with IGF-1 resistance, IRS-1 dysregulation,
and cognitive decline. J Clin Invest. 122:1316–1338. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yarchoan M, Toledo JB, Lee EB, Arvanitakis
Z, Kazi H, Han LY, Louneva N, Lee VM, Kim SF, Trojanowski JQ and
Arnold SE: Abnormal serine phosphorylation of insulin receptor
substrate 1 is associated with tau pathology in Alzheimer's disease
and tauopathies. Acta Neuropathol. 128:679–689. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kapogiannis D, Boxer A, Schwartz JB, Abner
EL, Biragyn A, Masharani U, Frassetto L, Petersen RC, Miller BL and
Goetzl EJ: Dysfunctionally phosphorylated type 1 insulin receptor
substrate in neural-derived blood exosomes of preclinical
Alzheimer's disease. FASEB J. 29:589–596. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tanokashira D, Fukuokaya W and Taguchi A:
Involvement of insulin receptor substrates in cognitive impairment
and Alzheimer's disease. Neural Regen Res. 14:1330–1334. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Douaud G, Groves AR, Tamnes CK, Westlye
LT, Duff EP, Engvig A, Walhovd KB, James A, Gass A, Monsch AU, et
al: A common brain network links development, aging, and
vulnerability to disease. Proc Natl Acad Sci USA. 111:17648–17653.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Schrijvers EM, Witteman JC, Sijbrands EJ,
Hofman A, Koudstaal PJ and Breteler MM: Insulin metabolism and the
risk of Alzheimer disease: The Rotterdam Study. Neurology.
75:1982–1987. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Qiu C, Sigurdsson S, Zhang Q, Jonsdottir
MK, Kjartansson O, Eiriksdottir G, Garcia ME, Harris TB, van Buchem
MA, et al: Diabetes, markers of brain pathology and cognitive
function: The Age, Gene/Environment Susceptibility-Reykjavik Study.
Ann Neurol. 75:138–146. 2014. View Article : Google Scholar : PubMed/NCBI
|