Introduction
Epigenetic processes, including DNA methylation,
histone modification and non-coding RNA regulation, are inheritable
changes that can affect gene expression without altering the DNA
sequence (1). In mammals,
methylation at the carbon 5 position of cytosine [5-methyl-cytosine
(5mC)] is the most common DNA modification, and it is involved in
various biological processes, including cell differentiation,
development and proliferation (2).
5mC is associated with the regulation of a number of
pathophysiological processes and states, including gene expression
regulation, genomic imprinting, X-chromosome inactivation,
repression of transposable elements, embryonic development and cell
differentiation (2–5).
The ten-eleven translocation (TET) family, which
consists of Fe(II)- and 2-oxoglutarate-dependent dioxygenases, has
attracted increasing attention due to its key role in the reversal
of DNA methylation (3). It has
been reported that TET proteins (TET1, TET2 and TET3) sequentially
oxidize 5mC to 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine
(5fC) and 5-carboxylcytosine (5caC) in DNA (2,6,7).
Previous studies have reported that 5fC and 5caC may be converted
into cytosine by a thymine-DNA glycosylase/base excision
repair-dependent pathway (8–10).
In addition to mediating the formation of 5-hmC, TET proteins are
involved in gene transcription repression via histone
de-acetylation, which represents a DNA methylation-independent
epigenetic mechanism (11).
Increasing evidence suggested that 5-hmC, which is
regulated by TET2, plays an important role in various diseases,
such as immune system disorders, hematopoietic diseases and cancers
(5,6,8). Li
et al (12), reported a
predominant role for TET2 in the differentiation of adult neural
stem cells by increasing the levels of 5-hmC. Sun et al
(13), indicated that inhibiting
the TET2 protein decreases the expression of genes associated with
cell death by mediating the formation of 5-hmC, which aggravated
ischemic damage in a rat model of spinal cord injury. In addition,
decreased 5-hmC levels are associated with the progression and poor
survival of numerous malignancies, including melanoma, renal cell
carcinoma, and gastric and ovarian cancer (14).
The skin provides the first line of defense against
skin injury and invading pathogens, and keratinocytes are the main
active cell type in the epidermis (15,16).
Recent studies indicated that keratinocytes actively regulate
cutaneous immune reactions, which may play a role in inflammatory
diseases affecting the skin, including psoriasis and atopic
dermatitis (15,17). The present study suggested that
TET2 knockdown in HaCaT keratinocytes promoted inflammation.
Therefore, the effects of TET2 on epigenetic modifications and
cellular functions were investigated in the present study. The
results of the present study indicated that the TET2
protein-mediated formation of 5-hmC increased the proportion of
apoptotic cells and the expression of inflammatory factors in HaCaT
cells.
Materials and methods
Cell culture
The human keratinocyte HaCaT cell line (cat. no.
GDC106; China Center for Type Culture Collection) was identified by
STR profiling performed by Cobioer Biosciences Co., Ltd. HaCaT
cells were maintained in minimal essential medium (HyClone; GE
Healthcare Life Sciences) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) and 1% penicillin-streptomycin (HyClone;
GE Healthcare Life Sciences) at 37°C with 5% CO2 in a
humidified incubator. The medium was replaced every 3 days.
Keratinocytes from generations 3 to 4 were used for subsequent
experiments. At 60–80% confluency, cells were transfected. For
small interfering (si)RNA transfection, HaCaT cells were randomly
divided into four groups: i) Negative control (NC) group
(transfected with control siRNA); ii) si-TET2-01 group (transfected
with a siRNA targeting TET2); iii) si-TET2-02 group (transfected
with a second siRNA targeting TET2), and iv) si-TET2-03 group
(transfected with a third siRNA targeting TET2). For plasmid DNA
transfection, HaCaT cells were randomly divided into two groups: i)
NC group (transfected with the empty GV230 vector), and ii) TET2
group (transfected with GV230 containing the coding sequence of
TET2).
siRNA transfection
siRNAs were designed and obtained from Guangzhou
RiboBio Co., Ltd. The sequences of the siRNAs were as follows:
si-TET2-01, 5′-CCAGAATAGTCGTGTGAGT-3′; si-TET2-02,
5′-GCTCTGAACGGTATTTAAA-3′; si-TET2-03, 5′-CGAGACTCATAATGTCCAA-3′;
and NC siRNA, 5′-UUCUCCGAACGUGUCACGU-3′. TET2 and control siRNAs
(40 pmol/ml) were transfected into HaCaT cells
(5×105/ml) using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. After 48 h, at 37°C in a humidified
atmosphere with 5% CO2, cells were collected and used
for subsequent experiments.
Plasmid DNA transfection
The TET2 coding sequence was synthesized according
to the NM_017628 transcript and inserted into the GV230 plasmid
(GeneChem, Inc.). At 70% confluency, HaCaT cells
(5×105/ml) were transfected with the GV230 plasmids (1
µg/ml) using Lipofectamine 3000. The empty GV230 vector was used as
the negative control. After 48 h, at 37°C in a humidified
atmosphere with 5% CO2, cells were collected and used
for subsequent experiments.
Dot blot analysis
Genomic DNA from transfected HaCaT cells was
extracted using a DNeasy Blood and Tissue kit (cat. no. 69504;
Qiagen, Inc.), according to the manufacturer's protocol. DNA
concentration (50 ng/µl) was quantified using a micro
ultraviolet-visible spectrophotometer. DNA denaturation was
performed at 95°C for 10 min. Denatured DNA samples were
immediately placed in an ice water bath for 5 min. Subsequently,
samples (2 µl) were spotted on a nylon membrane and blocked with 5%
skimmed milk for 1 h at room temperature. The membranes were
incubated with anti-5-hmC primary antibody (cat. no. 39769;
1:10,000; Active Motif, Inc.) overnight at 4°C. After washing, the
membrane was incubated with a horseradish peroxidase-conjugated
secondary antibody (anti-rabbit IgG; 1:2,000; cat. no. 7074; Cell
Signaling Technology, Inc.) for 1 h at room temperature. Detection
was performed using a chemiluminescence kit (cat. no. G2014; Wuhan
Servicebio Technology Co., Ltd.).
Cell Counting Kit-8 (CCK-8) assay
HaCaT cells (5×105/ml) were cultured in
96-well plates and transfected with plasmid or siRNA for 48 h at
37°C in a humidified atmosphere with 5% CO2. Cell
viability was assessed using the CCK-8 kit (cat. no. WC0037; Wuhan
Servicebio Technology Co., Ltd.), according to the manufacturer's
protocol. CCK-8 solution was added to each well and the plates were
incubated for 1–4 h at 37°C. The absorbance was measured at a
wavelength of 450 nm using a microplate reader. The assay was
performed in triplicate.
MTT assay
HaCaT cells (5×105/ml) were cultured in
96-well plates and transfected with plasmid DNA or siRNA for 48 h
at 37°C in a humidified atmosphere with 5% CO2. Cell
viability was measured using an MTT assay (cat. no. G4101; Wuhan
Servicebio Technology Co., Ltd.), according to the manufacturer's
protocol. MTT solution was added to each well and incubated for 4 h
at 37°C in a humidified atmosphere with 5% CO2.
Subsequently, the supernatant was removed and DMSO was used to
dissolve the purple formazan. The absorbance of each well was
measured at a wavelength of 570 nm using a microplate reader. Cell
viability was calculated as the ratio of absorbance at a wavelength
of 570 nm between each group and the control group.
Flow cytometric analysis
Following transfection, HaCaT cells
(5×105/ml) were cultured for 48 h at 37°C in a
humidified atmosphere with 5% CO2. Subsequently, cells
were trypsinized (0.25% trypsin at 37°C for 3–5 min), harvested and
washed twice with cold PBS. To measure apoptosis, the FITC Annexin
V Apoptosis Detection kit (cat. no. 556547; BD Biosciences) was
used, according to the manufacturer's protocol. After incubation
for 15 min at room temperature, the samples were analyzed by flow
cytometry (BD™ LSR-II; BD Biosciences). Data analysis was performed
using FlowJo software (v10.5.3; Becton, Dickinson and Company). The
apoptotic rate was the sum of the early and late apoptotic
rates.
For the analysis of the cell cycle,
cells were fixed in 70% pre-cooled ethanol at 4°C overnight
Subsequently, the cells were stained with propidium
iodide (PI)/RNase staining buffer (cat. no. 550825; BD Biosciences)
for 15 min at room temperature and analyzed using a FACScan flow
cytometer (Becton, Dickinson and Company), according to the
manufacturer's instructions. Data analysis was performed using
FlowJo software (v10.5.3; Becton, Dickinson and Company).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from HaCaT cells using
TRIzol® reagent (cat. no. 15596-026; Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
RNA (1 µg) was reverse transcribed into cDNA using the PrimeScript
RT Reagent kit (cat. no. RR036A; Takara Biotechnology Co., Ltd.),
according to the manufacturer's protocols. Subsequently, qPCR was
performed using a real-time PCR detection system and TB
Green® Premix Ex Taq™ (cat. no. RR420A; Takara
Biotechnology Co., Ltd.). The thermocycling conditions were:
Initial denaturation at 95°C for 10 min, followed by 40 cycles of
95°C for 15 sec, 60°C for 1 min and 72°C for 3 min. The primers
used for qPCR are presented in Table
SI. mRNA levels were quantified using the 2−ΔΔCq
method (18) and normalized to
GAPDH.
Western blotting
HaCaT cells were lysed using RIPA cell lysis buffer
(cat. no. P0013B; Beyotime Institute of Biotechnology), and the
supernatant was collected by centrifugation at 13,000 × g for 5 min
at 4°C. Total protein was quantified using a BCA protein assay kit
(cat. no. G2026; Wuhan Servicebio Technology Co., Ltd.). Equal
amounts of protein (30 µg) were separated by 10% SDS-PAGE and then
transferred to PVDF membranes. After blocking with 5% non-fat milk
for 1 h at room temperature, the membranes were incubated overnight
at 4°C with primary antibodies targeted against: GAPDH (cat. no.
118; 1:1,000; Cell Signaling Technology, Inc.), TET2 (cat. no.
ab94580; 1:1,000; Abcam), cyclin-dependent kinase inhibitor 2A
(P16INK4a; cat. no. ab108349; 1:2,000; Abcam),
interferon regulatory factor 7 (IRF7; cat. no. 22392-1-AP; 1:2,000;
ProteinTech Group, Inc.), S100 calcium binding protein A7 (S100A7;
cat. no. 13061-1-AP; 1:500; ProteinTech Group, Inc.), matrix
metallopeptidase 9 (MMP9; cat. no. 10375-2-AP; 1:1,000; ProteinTech
Group, Inc.), lipocalin 2 (LCN2; cat. no. 26991-1-AP; 1:1,000;
ProteinTech Group, Inc.), C-X-C motif chemokine ligand 1 (CXCL1;
cat. no. A5802; 1:1,000; ABclonal Biotech Co., Ltd.) and
interleukin-7 receptor (IL7R; cat. no. 17626-1-AP; 1:2,000;
ProteinTech Group, Inc.). Subsequently, the membranes were
incubated for 1 h at room temperature with corresponding
horseradish peroxidase-conjugated secondary antibodies (anti-rabbit
or anti-mouse IgG; cat. nos. 7074 and 7076; 1:2,000; Cell Signaling
Technology, Inc.). Protein bands were visualized using an ECL kit
(cat. no. G2014; Wuhan Servicebio Technology Co., Ltd.) and
GelCapture (v7.0; DNR Bio-Imaging Systems, Inc.), according to the
manufacturer's instructions. Protein expression was quantified
using ImageJ software (v1.8.0; National Institutes of Health) with
GAPDH as the loading control.
Statistical analysis
Data are presented as the mean ± SD. Statistical
analyses were performed using GraphPad Prism software (v6.0;
GraphPad Software, Inc.). Comparisons between data containing 2
groups were performed using an unpaired two-tailed Student's
t-test. Comparisons between data containing >2 groups were
performed using one-way ANOVA followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Effect of TET2 on the level of 5-hmC
in HaCaT cells
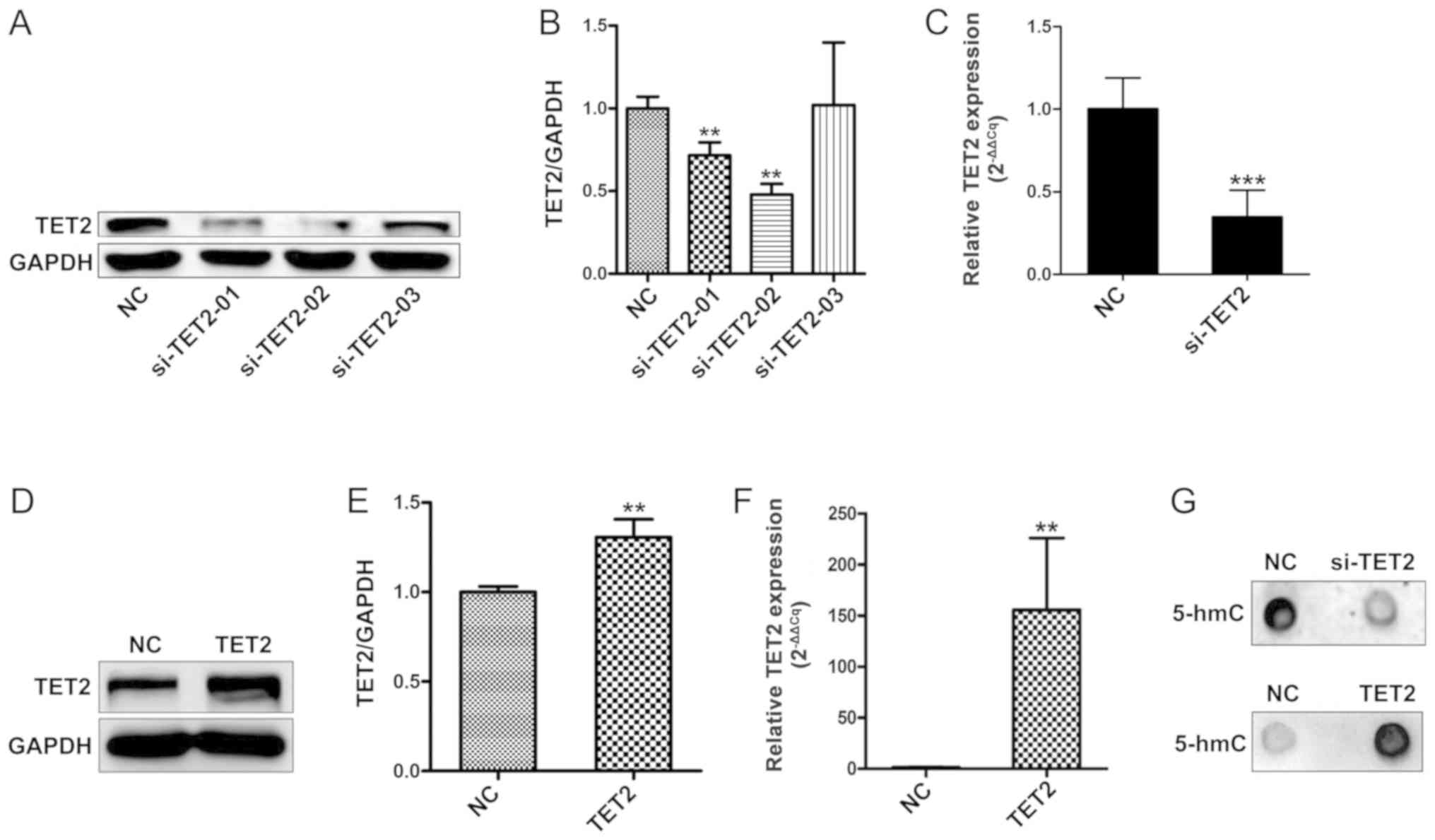
To investigate whether the levels of genomic 5-hmC
are associated with TET2 protein levels in HaCaT cells, three
siRNAs targeting TET2 were designed. TET2 knockdown efficiency was
evaluated by western blotting (Fig. 1A
and B). Compared with the control group, two siRNAs targeting
TET2 significantly decreased TET2 mRNA expression; siRNA-TET2-02
was the most efficient in reducing TET2 mRNA and protein expression
(Fig. 1A-C). Therefore,
siRNA-TET2-02 was selected for further experiments. Subsequently,
TET2 overexpression was performed, and transduction efficiency was
confirmed by western blotting and RT-qPCR (Fig. 1D-F). Previous studies have reported
that members of the TET family are able to reverse DNA methylation
patterns by converting 5-mC into 5-hmC (2,6,7).
Therefore, a dot blot analysis was performed to test the levels of
5-hmC following TET2 overexpression. The results suggested that
5-hmC expression was notably altered following TET2 knockdown or
overexpression, compared with the control group (Fig. 1G).
Effect of TET2 on HaCaT cell viability
and apoptosis
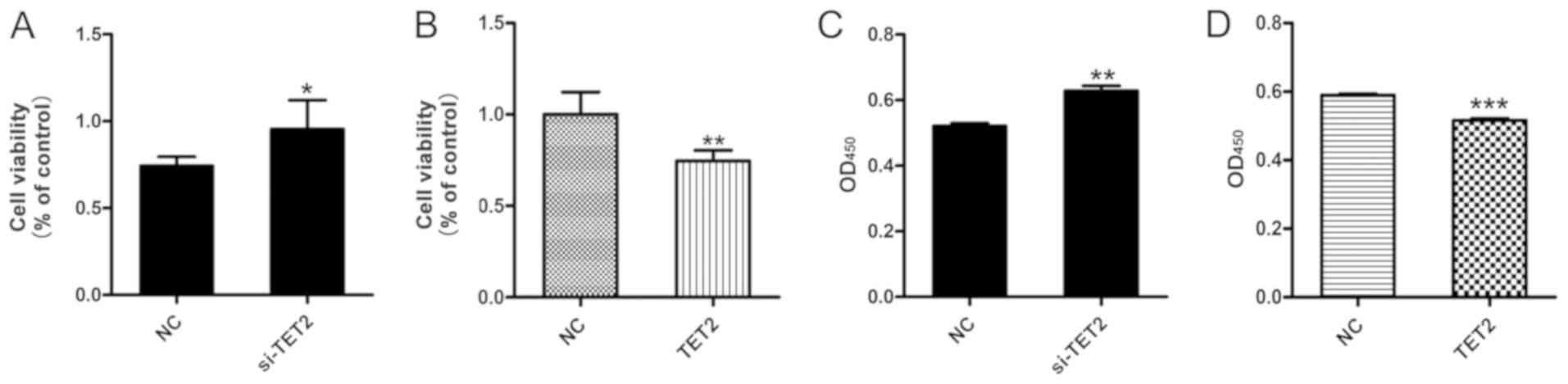
The viability of HaCaT cells following TET2
knockdown or overexpression was measured. The MTT assay results
suggested that si-TET2 significantly increased HaCaT cell viability
(Fig. 2A) and TET2 overexpression
significantly decreased HaCaT cell viability (Fig. 2B). In addition, the CCK-8 assay was
performed to further examine cell viability, and the results were
consistent with the MTT assay (Fig. 2C
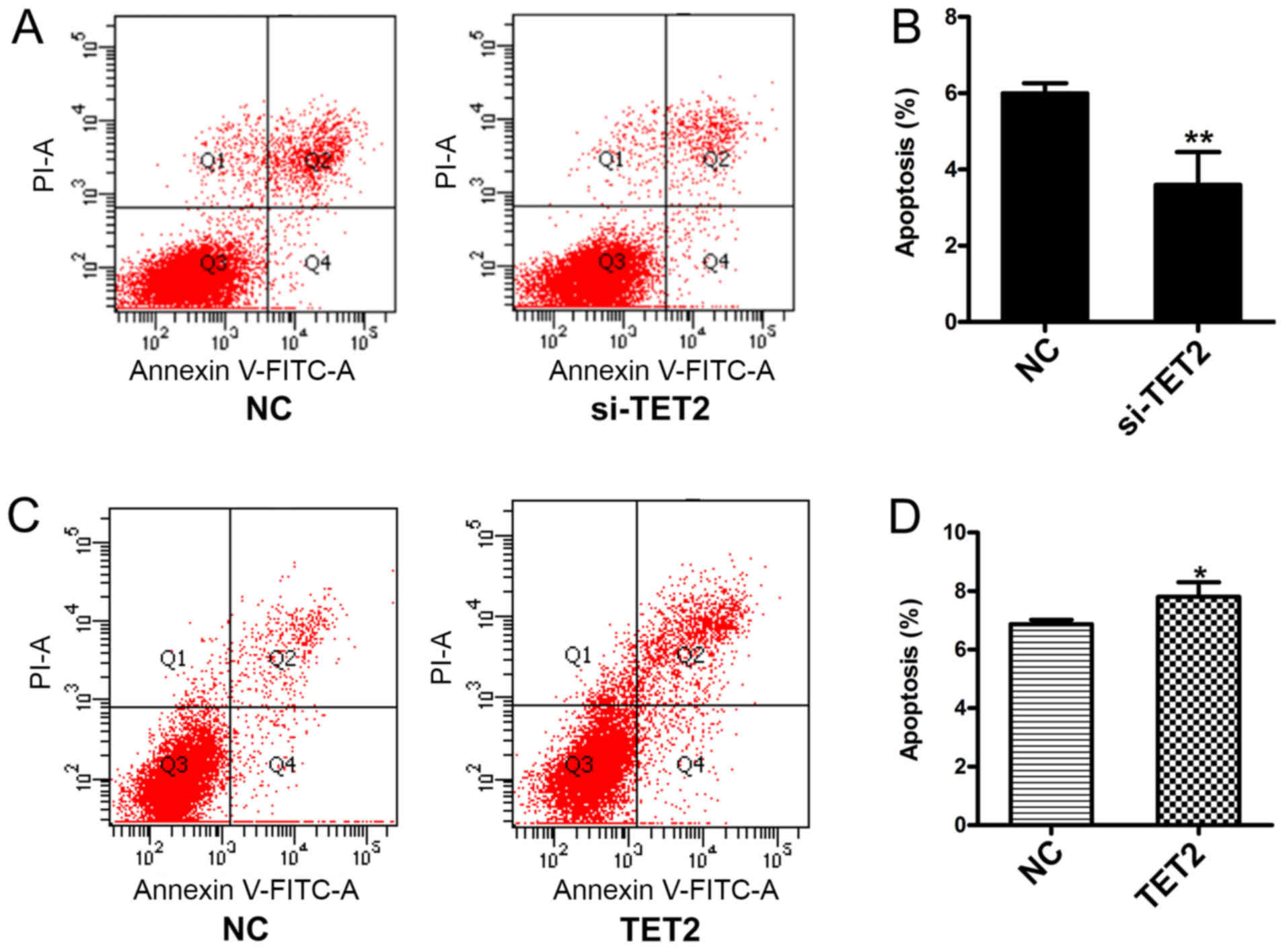
and D). To examine the effects of TET2 protein on HaCaT cell
apoptosis, a quantitative analysis of cell apoptosis was performed
by Annexin V-PI staining. The results indicated that the proportion
of apoptotic cells was significantly decreased in HaCaT cells
transfected with si-TET2 compared with the control group (Fig. 3A and B), whereas TET2
overexpression significantly increased the proportion of apoptotic
HaCaT cells (Fig. 3C and D). The
results suggested that TET2 may play an important role in
regulating HaCaT cell proliferation.
Effect of TET2 on the cell cycle in
HaCaT cells
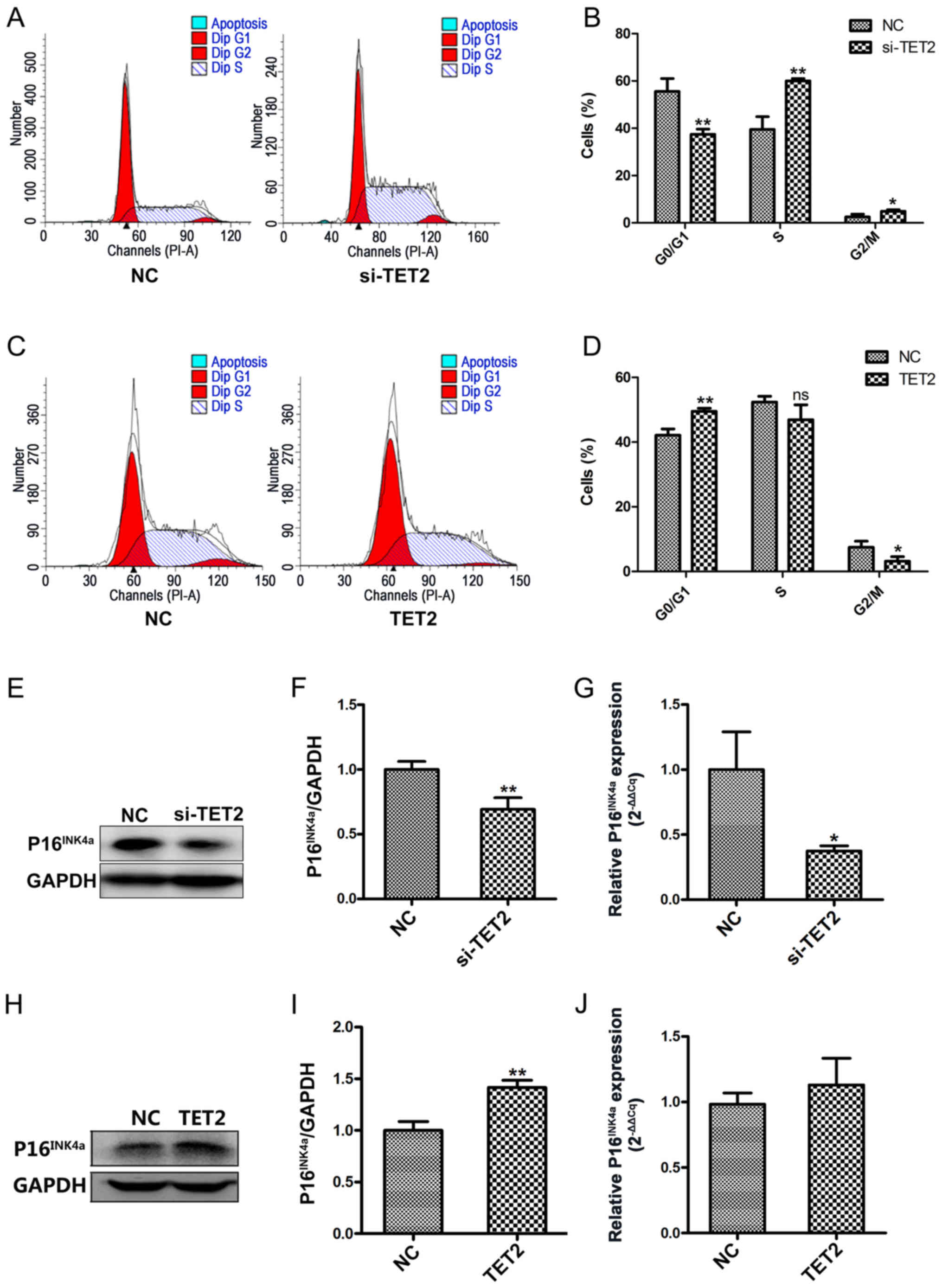
To further investigate the effects of TET2 protein
on HaCaT cell proliferation, flow cytometric analyses were
conducted to assess the cell cycle profile. Following TET2
knockdown, a significantly decreased proportion of cells was
observed in the G0/G1 phase, and a significantly increased
proportion of cells was observed in the S and G2/M phases, compared
with the control group (Fig. 4A and
B). By contrast, TET2 overexpression displayed the opposite
effect on the cell cycle in HaCaT cells (Fig. 4C and D). To further examine the
effects of TET2 on the cell cycle, P16INK4a protein and
mRNA expression levels were detected by western blotting and
RT-qPCR, respectively. Compared with the control group, TET2
knockdown significantly decreased the protein and mRNA expression
levels of P16INK4a (Fig.
4E-G); however, TET2 overexpression increased
P16INK4a protein expression compared with the control
group (Fig. 4H and I). mRNA
expression levels of P16INK4a were increased in HaCaT
cells following TET2 overexpression, but the difference was not
statistically significant (Fig.
4J). The results indicated that TET2 may inhibit HaCaT cell
proliferation by regulating cell cycle-related proteins, such as
P16INK4a.
Effect of TET2 on the expression
levels of proinflammatory cytokines in HaCaT cells
It has been reported that epigenetic modifications
may influence inflammatory mediators in chronic inflammatory
diseases, including psoriasis (19). Our previous integrated
bioinformatics analysis identified various hub genes that were
methylated in psoriatic tissues, including LCN2, S100A7, MMP9,
CXCL1, IRF7 and IL7R (20). To
further investigate the function of TET2 during the inflammatory
response in HaCaT cells, the expression levels of the
aforementioned proinflammatory cytokines were investigated by
RT-qPCR and western blotting (Fig.
5A-N). The expression of various inflammation-related cytokines
was significantly decreased in the si-TET2 group compared with
control group (Fig. 5A-F, M and
N). TET2 overexpression significantly increased the expression
of inflammation-related cytokines compared with the control group
(Fig. 5G-L). The results suggested
that TET2 acted as a transcriptional regulator of the
proinflammatory response in HaCaT cells.
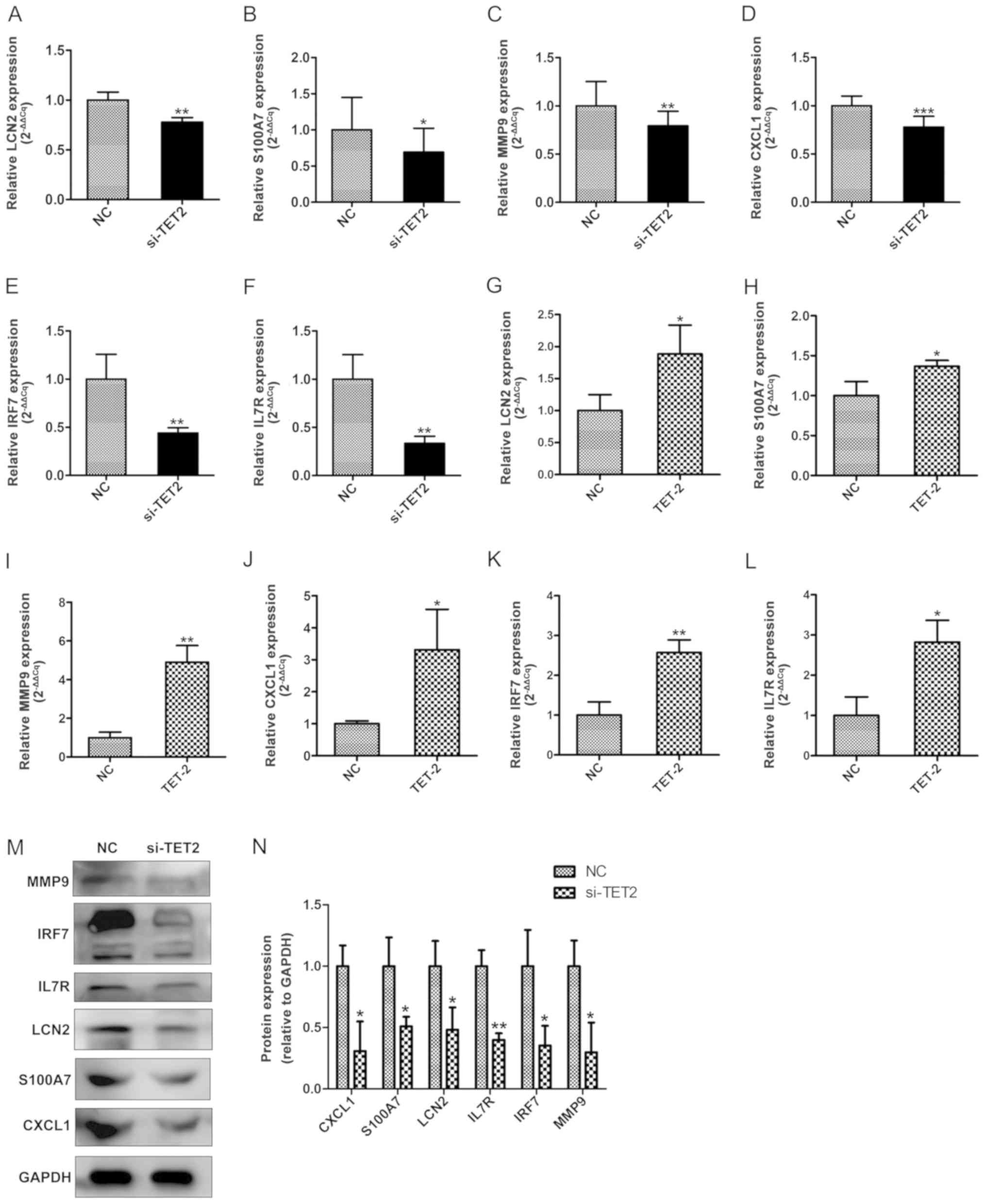 | Figure 5.TET2 regulates the expression levels
of proinflammatory cytokines at the mRNA and protein level in HaCaT
cells. RT-qPCR was performed to determine the mRNA expression
levels of (A) LCN2, (B) S100A7, (C) MMP9, (D) CXCL1, (E) IRF7 and
(F) IL7R following TET2 knockdown in HaCaT cells. RT-qPCR was
performed to determine the mRNA expression levels of (G) LCN2, (H)
S100A7, (I) MMP9, (J) CXCL1, (K) IRF7 and (L) IL7R following TET2
overexpression in HaCaT cells. Protein levels of proinflammatory
cytokines were (M) determined by western blotting and (N)
quantified following TET2 knockdown in HaCaT cells. Data are
presented as the mean ± SD of three independent experiments
performed in triplicate. *P<0.05, **P<0.01, ***P<0.001 vs.
NC group. TET2, ten-eleven transloation-2; RT-qPCR, reverse
transcription-quantitative PCR; LCN2, lipocalin 2; S100A7, S100
calcium binding protein A7; MMP9, matrix metallopeptidase 9; CXCL1,
C-X-C motif chemokine ligand 1; IRF7, interferon regulatory factor
7; IL7R, interleukin-7 receptor; NC, negative control; si, small
interfering RNA. |
Discussion
The epigenetic activity of TET2 in a number of
hematologic malignancies, and metabolic and autoimmune diseases has
been investigated (21–23). However, the role of TET2 in skin
disease remains unclear. Our previous study reported that 5-hmC and
TET2 levels were increased in epidermal keratinocytes in
psoriasiform dermatitis (24). To
further investigate the function of TET2 in inflammatory skin
diseases, the present study performed experiments using the
keratinocyte HaCaT cell line. Cell apoptosis, cell cycle and
proinflammatory cytokine expression were detected following TET2
knockdown or overexpression in HaCaT cells. The results suggested
that TET2 expression was positively associated with 5-hmC levels
and the proportion of apoptotic cells, but negatively associated
with cell viability and cell cycle progression. Furthermore, TET2
knockdown in HaCaT cells decreased the expression of several
proinflammatory cytokines, including LCN2, S100A7, MMP9, CXCL1,
IRF7 and IL7R. The data indicated that TET2 epigenetically
regulated the expression of inflammatory factors in
keratinocytes.
DNA methylation, the most common epigenetic
regulation, is partly controlled by DNA methyltransferases (DNMTs),
including DNMT1 and DNMT3A/3B (23,25).
The levels and patterns of DNA methylation are regulated by two
contrasting processes, methylation and demethylation, which have an
essential role in gene expression regulation (2). Since the crystal structure of TET2
and the 5mC-DNA complex were reported, knowledge of the mechanisms
underlying TET-mediated demethylation has gradually improved
(4,9). 5-hmC is an important intermediate
product of DNA demethylation, which is downregulated following TET2
knockdown (3,5,7,26).
Dot blot analysis indicated that TET2 knockdown or overexpression
decreased or increased 5-hmC expression levels, respectively. The
results of the present study were in line with previous studies
(5,7), thus suggesting that DNA
hydroxymethylation patterns in HaCaT cells are regulated by
TET2.
TET2 affects cellular proliferation and cell cycle
progression in various cell lines, such as CD4+T and
Hela cells, in other conditions, including autoimmune diseases and
development (5,27–30).
However, the precise mechanism underlying TET2 in HaCaT cells
remains unclear. The results suggested that HaCaT viability was
increased following TET2 knockdown and the proportion of apoptotic
cells decreased. Conversely, TET2 overexpression induced opposing
effects. The results suggested that TET2 may be involved in
regulating HaCaT cell proliferation. Furthermore, the flow
cytometric analysis indicated that TET2 regulated cell
proliferation by inducing cell cycle arrest at the G0/G1 phase.
Similar results have been reported in two TET2 inducible cell lines
(Ba/F3-EPOR and UT7) (30). A
limitation of the present study was that cell proliferation was not
analyzed using direct assays, for example 5-bromo-2-deoxyuridine
(BrdU) labeling. However, Prikrylova et al (27) reported that TET2-induced 5-hmC
expression was inversely proportional to the proportion of
apoptotic cells in HeLa cell lines using the BrdU assay, which is
in line with the results of the present study. Collectively, the
results suggested that TET2 may influence cell cycle arrest and
cell proliferation. To further investigate the underlying
mechanism, whether TET2-induced cell cycle arrest was mediated by
the cell cycle-associated protein P16INK4a, an inhibitor
of cyclin-dependent kinase 4 (31), was investigated. It has been
reported that P16INK4a affects cell proliferation and
differentiation by negatively regulating the cell cycle (32). Recently, several studies have
reported that TET2 regulates cell cycle-associated proteins in
neural and trophoblast stem cells (28,29).
In line with these previous studies, the results of the present
study suggested that the increase in P16INK4a expression
was mediated by TET2 overexpression, and si-TET2 decreased the
level of P16INK4a expression. The results indicated that
P16INK4a upregulation was decreased following TET2
knockdown.
Epigenetic factors are involved in various chronic
inflammatory diseases, including type 2 diabetes, Alzheimer's
disease and inflammatory bowel diseases (19,33).
DNA methylation and hydroxymethylation may be involved in
inflammation by altering the homeostasis of various inflammatory
mediators, such as interleukin (IL)-6, TNF receptor associated
factor 6 and IL-23 (19). Lagos
et al (34), reported that
DNA methylation and hydroxymethylation were associated with
glandular inflammation and dysfunction, which might be a novel
target for the treatment of Sjögren's syndrome. Wang et al
(35), revealed that TET2
knockdown by siRNAs not only affected the level of MyD88 innate
immune signal transduction adaptor hydroxymethylation, but also
inhibited lipopolysaccharide-induced inflammation in human dental
pulp cells. Our previous study suggested that TET2 modulates the
expression of proinflammatory cytokines in a mouse model of
psoriasiform dermatitis (24). In
the present study, the effects of DNA demethylation on inflammatory
factors were examined in vitro. The results of the present
study suggested that the expression levels of proinflammatory
mediators, including LCN2, S100A7, MMP9, CXCL1, IRF7 and IL7R, may
be positively associated with the level of DNA hydroxymethylation.
Therefore, the results suggested that epigenetic changes may affect
the progression of inflammatory skin diseases. The present study
investigated the mechanism underlying TET2 in HaCaT cells; however,
characterization of the hydroxymethylation pattern of specific
genes in keratinocytes requires further investigation.
The present study investigated the potential role
and mechanism underlying the effect of TET2 on cell proliferation
and expression levels of various inflammation mediators in HaCaT
cells. The results indicated that the regulation of the cell cycle
and expression of proinflammatory cytokines was mediated by DNA
hydroxymethylation. Further investigation is required to assess
whether TET2 protein plays an important role in the pathogenesis of
inflammatory skin diseases.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81673057 and
81502730).
Availability of data and materials
The datasets used and analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL, NL, YW and HC wrote the manuscript. XL, XW, NL,
KZ, SZ, XD, YH, HJ and ZJ performed the experiments and collected
the data. XL, XW, NL, KZ, YW and HC designed the study. HJ, YW and
HC edited the manuscript for English language. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shen J, Abu-Amer Y, O'Keefe RJ and
McAlinden A: Inflammation and epigenetic regulation in
osteoarthritis. Connect Tissue Res. 58:49–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu X and Zhang Y: TET-mediated active DNA
demethylation: Mechanism, function and beyond. Nat Rev Genet.
18:517–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li D, Guo B, Wu H, Tan L and Lu Q: TET
family of dioxygenases: Crucial roles and underlying mechanisms.
Cytogenet Genome Res. 146:171–180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M,
Shi YG, Zhu J, Wang P and Xu Y: Crystal structure of TET2-DNA
complex: Insight into TET-mediated 5mC oxidation. Cell.
155:1545–1555. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ichiyama K, Chen T, Wang X, Yan X, Kim BS,
Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, et al: The
methylcytosine dioxygenase Tet2 promotes DNA demethylation and
activation of cytokine gene expression in T cells. Immunity.
42:613–626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pastor WA, Aravind L and Rao A: TETonic
shift: Biological roles of TET proteins in DNA demethylation and
transcription. Nat Rev Mol Cell Biol. 14:341–356. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ito S, Shen L, Dai Q, Wu SC, Collins LB,
Swenberg JA, He C and Zhang Y: Tet proteins can convert
5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine.
Science. 333:1300–1303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rasmussen KD and Helin K: Role of TET
enzymes in DNA methylation, development, and cancer. Genes Dev.
30:733–750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q,
Ding J, Jia Y, Chen Z, Li L, et al: Tet-mediated formation of
5-carboxylcytosine and its excision by TDG in mammalian DNA.
Science. 333:1303–1307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weber AR, Krawczyk C, Robertson AB,
Kuśnierczyk A, Vågbø CB, Schuermann D, Klungland A and Schär P:
Biochemical reconstitution of TET1-TDG-BER-dependent active DNA
demethylation reveals a highly coordinated mechanism. Nat Commun.
7:108062016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li
X, Zhao D, Liu Y, Wang C, Zhang X, et al: Tet2 is required to
resolve inflammation by recruiting Hdac2 to specifically repress
IL-6. Nature. 525:389–393. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Yao B, Chen L, Kang Y, Li Y, Cheng
Y, Li L, Lin L, Wang Z, Wang M, et al: Ten-eleven translocation 2
interacts with forkhead box O3 and regulates adult neurogenesis.
Nat Commun. 8:159032017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun H, Miao Z, Wang H, Tao Y, Yang J, Cai
J, Wang J and Wang Y: DNA hydroxymethylation mediated traumatic
spinal injury by influencing cell death-related gene expression. J
Cell Biochem. 119:9295–9302. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Uchiyama R, Uhara H, Uchiyama A, Ogawa E,
Takazawa Y, Ashida A, Koga H, Hayashi K, Kiniwa Y and Okuyama R:
5-Hydroxymethylcytosine as a useful marker to differentiate between
malignant melanomas and benign melanocytic nevi. J Dermatol Sci.
73:161–163. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Benhadou F, Mintoff D and Del Marmol V:
Psoriasis: Keratinocytes or immune cells - Which is the trigger?
Dermatology. 235:91–100. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bitschar K, Wolz C, Krismer B, Peschel A
and Schittek B: Keratinocytes as sensors and central players in the
immune defense against Staphylococcus aureus in the skin. J
Dermatol Sci. 87:215–220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Asahina R and Maeda S: A review of the
roles of keratinocyte-derived cytokines and chemokines in the
pathogenesis of atopic dermatitis in humans and dogs. Vet Dermatol.
28:16–e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fogel O, Richard-Miceli C and Tost J:
Epigenetic changes in chronic inflammatory diseases. Adv Protein
Chem Struct Biol. 106:139–189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Liu X, Liu N and Chen H:
Prediction of crucial epigenetically-associated, differentially
expressed genes by integrated bioinformatics analysis and the
identification of S100A9 as a novel biomarker in psoriasis. Int J
Mol Med. 45:93–102. 2020.PubMed/NCBI
|
|
21
|
Poole CJ, Lodh A, Choi JH and van Riggelen
J: MYC deregulates TET1 and TET2 expression to control global DNA
(hydroxy)methylation and gene expression to maintain a neoplastic
phenotype in T-ALL. Epigenetics Chromatin. 12:412019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chiba S: Dysregulation of TET2 in
hematologic malignancies. Int J Hematol. 105:17–22. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Raghuraman S, Donkin I, Versteyhe S,
Barrès R and Simar D: The emerging role of epigenetics in
inflammation and immunometabolism. Trends Endocrinol Metab.
27:782–795. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang X, Liu X, Duan X, Zhu K, Zhang S, Gan
L, Liu N, Jaypaul H, Makamure JT, Ming Z, et al: Ten-eleven
translocation-2 regulates DNA hydroxymethylation status and
psoriasiform dermatitis progression in mice. Acta Derm Venereol.
98:585–593. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng X and Blumenthal RM: Mammalian DNA
methyltransferases: A structural perspective. Structure.
16:341–350. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ko M, An J, Bandukwala HS, Chavez L, Aijö
T, Pastor WA, Segal MF, Li H, Koh KP, Lähdesmäki H, et al:
Modulation of TET2 expression and 5-methylcytosine oxidation by the
CXXC domain protein IDAX. Nature. 497:122–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Prikrylova T, Robertson J, Ferrucci F,
Konorska D, Aanes H, Manaf A, Zhang B, Vågbø CB, Kuśnierczyk A,
Gilljam KM, et al: 5-hydroxymethylcytosine marks mammalian origins
acting as a barrier to replication. Sci Rep. 9:110652019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chrysanthou S, Senner CE, Woods L,
Fineberg E, Okkenhaug H, Burge S, Perez-Garcia V and Hemberger M: A
critical role of TET1/2 proteins in cell-cycle progression of
trophoblast stem cells. Stem Cell Reports. 10:1355–1368. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shimozaki K: Ten-eleven translocation 1
and 2 confer overlapping transcriptional programs for the
proliferation of cultured adult neural stem cells. Cell Mol
Neurobiol. 37:995–1008. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mahfoudhi E, Talhaoui I, Cabagnols X,
Della Valle V, Secardin L, Rameau P, Bernard OA, Ishchenko AA,
Abbes S, Vainchenker W, et al: TET2-mediated
5-hydroxymethylcytosine induces genetic instability and
mutagenesis. DNA Repair (Amst). 43:78–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
O'Neill CJ and McCluggage WG: p16
expression in the female genital tract and its value in diagnosis.
Adv Anat Pathol. 13:8–15. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aagaard L, Lukas J, Bartkova J, Kjerulff
AA, Strauss M and Bartek J: Aberrations of p16Ink4 and
retinoblastoma tumour-suppressor genes occur in distinct sub-sets
of human cancer cell lines. Int J Cancer. 61:115–120. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stylianou E: Epigenetics of chronic
inflammatory diseases. J Inflamm Res. 12:1–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lagos C, Carvajal P, Castro I, Jara D,
González S, Aguilera S, Barrera MJ, Quest AFG, Bahamondes V, Molina
C, et al: Association of high 5-hydroxymethylcytosine levels with
Ten Eleven Translocation 2 overexpression and inflammation in
Sjögren's syndrome patients. Clin Immunol. 196:85–96. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X, Feng Z, Li Q, Yi B and Xu Q: DNA
methylcytosine dioxygenase ten-eleven translocation 2 enhances
lipopolysaccharide-induced cytokine expression in human dental pulp
cells by regulating MyD88 hydroxymethylation. Cell Tissue Res.
373:477–485. 2018. View Article : Google Scholar : PubMed/NCBI
|