Introduction
Esophageal cancer (EC) is one of the most common
types of malignant tumor worldwide and it is associated with a high
incidence (5.9 per 100,000) and patient mortality (5 per 100,000)
(1). China exhibits one of the
highest incidence rates of EC (12.5 per 100,000) and a high patient
mortality (~9.1 per 100,000), with a total of ~50% new EC cases
identified in 2012 (2,3). Esophageal squamous cell carcinoma
(ESCC) is the major histological type of EC, accounting for >
90% of new diagnoses (4,5). However, clinical practices, including
the diagnosis and treatment of EC remain challenging worldwide,
thus there is an urgent requirement to identify novel biomarkers
that can be used to improve the diagnosis and treatment of EC.
MicroRNAs (miRNAs) are widely reported to
participate in the pathogenesis of tumor development, and are often
used as biomarkers to monitor disease progression (4). miRNAs are small, single-stranded
non-coding RNA molecules of ~22 nucleotides in length that can
inhibit the expression of target mRNAs through binding to
complementarity sites in the 3′-untranslated region (UTR) to induce
RNA silencing and post-transcriptional regulation (5,6).
Each miRNA can target multiple genes, thus functioning in complex
regulatory networks involved in a variety of biological processes,
including cell proliferation, migration and invasion, especially
within tumors (7–9). In EC, it was reported that miRNA
(miR)-100 significantly inhibited EC cell proliferation, migration
and invasion through targeting the C-X-C chemokine receptor type 7
(10). miR-373 is highly expressed
in EC tissues and was demonstrated to increase cell proliferation,
migration and invasion through directly targeting metalloproteinase
inhibitor 3, with opposite results occurring following miR-373
silencing (11). In a cohort of
102 patients, miR-451a, miR-144-3p and miR-144-5p expression levels
in EC tissues were reported to be significantly lower compared with
normal tissues (12). Furthermore,
low expression levels of miR-144-3p and miR-144-5p is an
independent risk factor for the occurrence of EC (12). Multiple other miRNAs, including
miR-133a, miR-138, miR-375 and miR-593 serve as tumor suppressors,
whereas miR-16, miR-21, miR-31, miR-34b, miR-208, miR-223, miR-373
and miR-423 exhibit oncogenic abilities (13–18).
It has been reported that miR-106b, miR-204, miR-371-3p,
miR-574-3p, miR-886-3p, miR-1203, miR-1303 and miR-1909 were
differentially expressed between patients with and without tumor
relapse following surgery (13).
miR-21 and miR-375 have previously been used as diagnostic and
prognostic biomarkers of EC (14)
and miR-100 has been suggested as a promising treatment owing to
its tumor suppressor role in EC (10). These studies provide reasoning to
identify additional miRNAs that may contribute to EC, alongside
determining their mechanism of action in EC pathogenesis. The
present study used bioinformatics analysis to reanalyze the miRNA
expression microarray dataset of EC tissues (GSE113776), with the
aim of identifying novel miRNAs to use as biomarkers for this
disease. Since miRNAs demonstrated potential to be used for
treatment and as biomarkers of EC according to the research cited
above, the current study aimed to further investigate their
functions in vitro through identifying key miRNAs that
contribute to the development of EC, and to identify possible miRNA
biomarkers for use in EC diagnosis, prognosis and therapy.
Materials and methods
Bioinformatics analysis
The miRNA profile dataset GSE113776 was downloaded
from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Considering
the identical tissue variation and potential mutual effects or
interactions in ESCC and neuroendocrine carcinoma (NEC), the
GSE113776 dataset contained profiled miRNAs expressed in paired NEC
and normal tissues based on an Agilent-041686 Unrestricted Human
miRNA Microarray platform was used. Since only one sample was
collected for sequencing in each group, no related research was
cited in the dataset summary. R version 3.6 software (RStudio,
Inc.) was used to analyze and visualize the dataset. To select
miRNAs, the cut-off value of absolute fold change (|FC|) was set to
2. In addition, differentially expressed miRNAs (DEMs) that are
associated with EC development were extracted from The Cancer
Genome Atlas (TCGA; http://portal.gdc.cancer.gov) to intersect with the
selected miRNAs. Verification of DEM expression was based on the
miRNA profile dataset GSE112840 of ESCC (19).
miRNA target gene extraction and
associated pathway enrichment
Using the miRNA databases TargetScanHuman
(http://www.targetscan.org), miRTarBase
(http://mirtarbase.mbc.nctu.edu.tw)
and mirRDB (http://mirdb.org), intersections were
filtered as the ready-for-test genes. Representative
immunohistochemistry images of these genes in normal esophageal
tissues were identified using the Human Protein Atlas (http://proteinatlas.org) to determine their protein
expression. The Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.ncifcrf.gov) was used for pathway
enrichment of the filtered genes and ClueGO and Search Tool for
Recurring Instances of Neighbouring Genes (STRING; http://string-db.org) were used for visualization,
which was performed using Cytoscape version 3.7.0 (https://cytoscape.org/) software.
Cell culture and transfection
The human ESCC cell line KYSE150 was purchased from
The Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. KYSE150 cells were incubated in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.), supplemented with 10% FBS
(HyClone; GE Healthcare Life Sciences) and 1%
penicillin-streptomycin (Thermo Fisher Scientific, Inc.), and
maintained in a humidified incubator at 37°C with 5%
CO2. A total of 5×104 KYSE150 cells/well were
seeded in a 6-well plate and were transfected with 50 nM miR-200a
mimic (5′-UAACACUGUCUGGUAACGAUGU-3′), miR-200a inhibitor
(5′-UAACCUCAUGGUGUACGAAUGU-3′) or scramble control sequences
(5′-UUGUACUACACAAAAGUACUG-3′) (Shanghai GenePharma Co., Ltd.)
separately for 24 h at 37°C using 10 nM Lipofectamine®
3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturers' protocol. Cell proliferation and
Transwell assays were subsequently performed and transfected cells
were harvested at 48 h for reverse transcription-quantitative PCR
(RT-qPCR) or western blot analysis.
Cell proliferation assay
Following transfection, a total of 1×104
KYSE150 cells/well were seeded into 96-well plates to assess cell
viability. Cells were cultured for 2 days post-transfection in
RPMI-1640 medium prior to the addition of 10 µl Cell Counting kit-8
(CCK-8) solution (WST-8, Dojindo Molecular Technologies, Inc.) to
each well. Following continuous incubation for 2 h at 37°C, cell
viability was determined by measuring the absorbance at 450 nm
using an ELISA reader (Tecan Group, Ltd.) at 0, 12, 24 and 48
h.
Migration and invasion assays
Migratory and invasive abilities of transfected
KYSE150 cells were examined using Transwell permeable supports
(Corning, Inc.). Following cell transfection for 24 h at 37°C,
cells were subsequently cultured in 200 µl serum-free RPMI-1640
medium before being transferred into 24-well plates separated into
upper and lower chambers. A total of 1×105 KYSE150
cells/well were plated in the upper chambers of Transwell plates
with serum-free RPMI-1640 medium, of which membranes were or were
not precoated with Matrigel (BD Biosciences) for the invasion and
migration assay, respectively. A total of 800 µl RPMI-1640 medium
supplemented with 10% FBS was plated in the lower chambers. After
24 h incubation in wells without Matrigel or 48 h with Matrigel for
the migration and invasion assay, respectively, cells in the lower
chambers were fixed for 20 min with absolute methanol and
subsequently stained with 0.1% crystal violet solution
(Sigma-Aldrich; Merck KGaA) for 10 min at room temperature. Stained
cells were counted in three randomly selected fields using an
inverted microscope (magnification, ×200). ImageJ version 1.49
software (National Institutes of Health) was used for image
analysis and quantification. Experiments were performed in
triplicate.
RT-qPCR
Total RNA was extracted from KYSE150 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Total RNA was
reverse transcribed into cDNA using PrimeScript RT Reagent kit
purchased from Takara Biotechnology Co., Ltd., and the
concentration of RNA was determined as described in a previous
study (20). qPCR was subsequently
performed in 96-well reaction plates using the ABI
StepOnePlus™ Real-Time PCR system (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Briefly, in each well, a total of 1 µl cDNA template, 0.2 µl primer
(0.1 µl forward and 0.1 µl reverse primer), 3.6 µl diethyl
pyrocarbonate-H2O and 5 µl SYBR® Green dye
were mixed. The primers (Sangon Biotech Co., Ltd.) used for the
qPCR are presented in Table SI.
The following thermocycling conditions were used for the qPCR:
Initial denaturation at 95°C for 30 sec; followed by 40 cycles of
denaturation at 95°C for 5 sec, annealing at 60°C for 10 sec and
extension at 72°C for 30 sec. Expression levels were quantified
using the 2−ΔΔCq method and normalized to the internal
reference gene, U6 for miR-200a expression or GAPDH for miR-200a
target genes (21).
Western blotting
Total protein was extracted from KYSE150 cells using
RIPA lysis buffer (Beyotime Institute of Biotechnology) and
protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). Total
protein was quantified using a bicinchoninic acid assay kit (Thermo
Fisher Scientific, Inc.) and western blot analysis was performed
according to previous protocols (22). A total of 30 µg protein/lane was
separated by 6 or 12% SDS-PAGE. The separated proteins were
subsequently transferred onto a PVDF membrane (Merck KGaA) and
blocked for 1 h at room temperature using 5% non-fat milk. The
membranes were incubated overnight at 4°C with the following
primary antibodies diluted in 5% milk: Anti-catenin β1 (CTNNB1;
1:1,000; cat. no. sc-59737; Santa Cruz Biotechnology, Inc.),
anti-cadherin-1 (CDH1; 1:500; cat. no. sc-71009; Santa Cruz
Biotechnology, Inc.), anti-PTEN (1:100; cat. no. sc-73420; Santa
Cruz Biotechnology, Inc.), anti-adenomatous polyposis coli (APC;
1:100; cat. no. sc-393704; Santa Cruz Biotechnology, Inc.),
anti-catenin α1 (CTNNA1; 1:500; cat. no. sc-47753; Santa Cruz
Biotechnology, Inc.), anti-superoxide dismutase 2 (SOD2; 1:200;
cat. no. sc-130345; Santa Cruz Biotechnology, Inc.) and anti-GAPDH
(1:3,000; cat. no. sc-47724; Santa Cruz Biotechnology, Inc.).
Following the primary antibody incubation, membranes were incubated
with horseradish peroxidase-conjugated goat anti-mouse secondary
antibodies (1:5,000; cat. no. A0216; Beyotime Institute of
Biotechnology) for 45 min at room temperature. Protein bands were
visualized using the ECL luminol reagent (PerkinElmer Inc.) and a
ChemiDoc image analyzer (Bio-Rad Laboratories, Inc.). Protein
expression was quantified using ImageJ software (version 1.49v;
National Institutes of Health) and normalized to the internal
reference gene GAPDH.
Dual-luciferase reporter assay
The luciferase reporter assay was performed as
previously described (22). The
3′-UTR sequence of miR-200a target genes (CTNNB1, CDH1, PTEN, APC,
CTNNA1 and SOD2) was separately amplified and inserted into the
luciferase reporter vector pGL3-enhancer (Promega Corporation). The
primers of the 3′-UTR of miR-200a target genes were designed and
are presented in Table SII. A
total of 1×104 KYSE150 cells/well were seeded into
24-well plates and incubated for 24 h at 37°C. Wild-type or mutant
miR-200a target gene 3′-UTR vectors, combined with the miR-200a
mimic, were subsequently co-transfected into KYSE150 cells using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
construction of the miR-200a target genes' 3′-UTR wild-type or
mutant reporter genes were performed using the miRNA databases
(Fig. S1). Following incubation
for 48 h at 37°C, KYSE150 cells were lysed and firefly and
Renilla luciferase activity was detected using a Luciferase
assay system (Promega Corporation), according to the manufacturer's
protocol. Firefly luciferase activity was normalized to
Renilla luciferase activity.
Statistical analysis
Data were analyzed using GraphPad Prism version 6.0
software (GraphPad Software, Inc.). Statistical differences between
two groups were compared using a Student's t-test, whereas three
groups were compared using a one-way ANOVA with Bonferroni
correction post hoc test. Data were presented as the mean ± SEM
from 3 independent experiments. P<0.05 was considered to
indicate a statistically significant difference.
Results
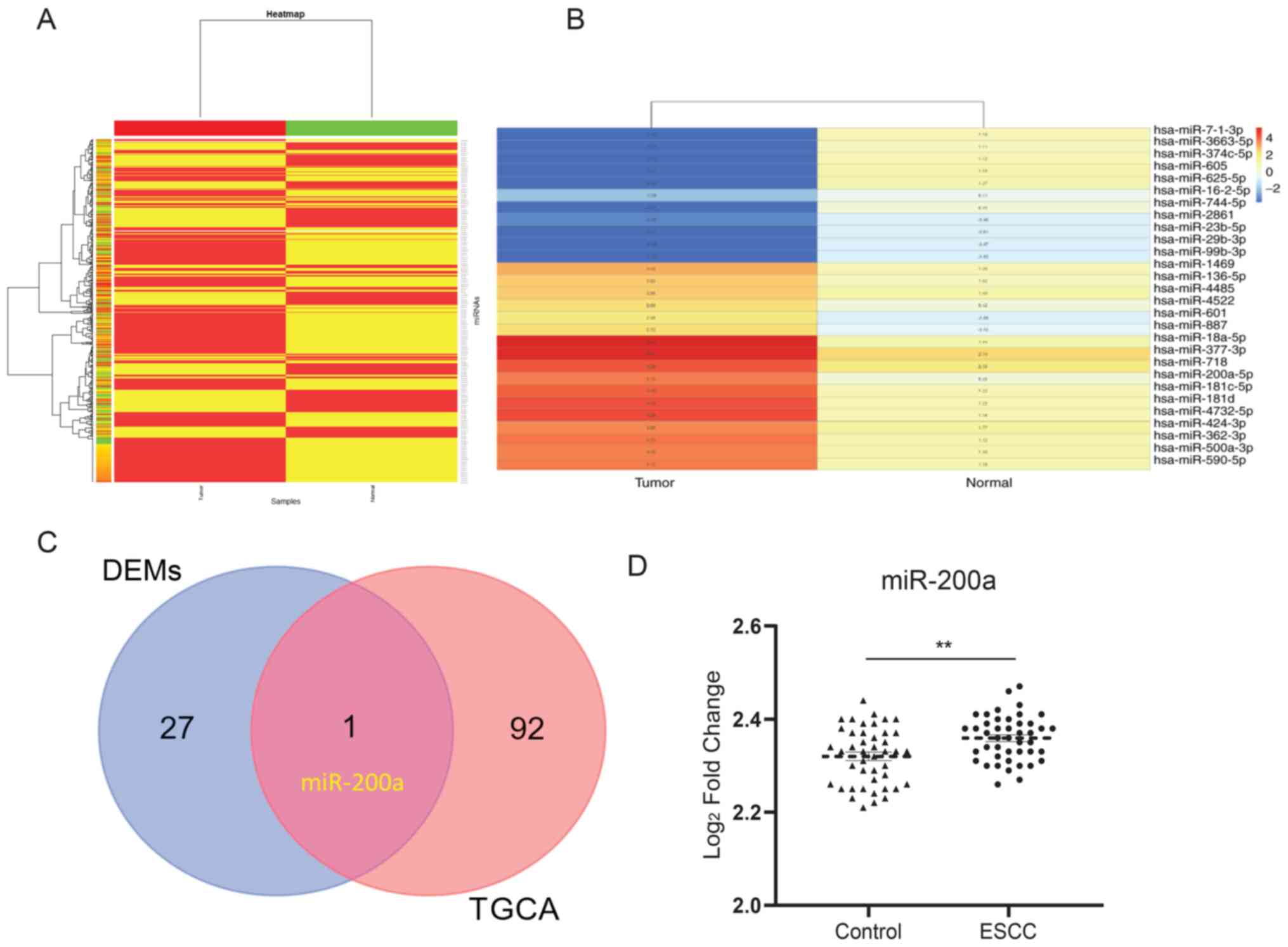
Heat map of miRNA expression and
identification of DEMs in a paired NEC sample
ESCC and neuroendocrine carcinoma (NEC) are from the
identical sites in esophageal tissues, indicating a potentially
similar mechanism. A heat map was constructed to identify the
expression of miRNAs in esophageal tissues using a Heat map package
in R. For the miRNA y dataset GSE113776, 494 miRNAs were detected
(Fig. 1A). A total of 28 miRNAs
were selected, of which the absolute expression value threshold was
|FC|>2 (Fig. 1B). In addition,
93 DEMs that were associated with the development of EC were
identified from TCGA (Table I).
One miRNA was observed to overlap between the selected miRNAs and
DEMs from TCGA, which was miR-200a (Fig. 1C). In addition, under the
cross-verification with the GSE112840 dataset, miR-200a was
confirmed to be upregulated in ESCC compared with the control
(Fig. 1D), which suggests the
critical role of miR-200a in ESCC.
 | Table I.miRNAs identified from the tissue
microarray and TCGA. |
Table I.
miRNAs identified from the tissue
microarray and TCGA.
| Origin | Count | Elements |
|---|
| miRNAs
profiling | 28 | hsa-miR-136
hsa-miR-1469 hsa-miR-16 hsa-miR-181c hsa-miR-181d hsa-miR-18a
hsa-miR-200a hsa-miR-23b hsa-miR-2861 hsa-miR-29b hsa-miR-362
hsa-miR-3663 hsa-miR-374c hsa-miR-377 hsa-miR-424 hsa-miR-4486
hsa-miR-4522 hsa-miR-4732 hsa-miR-500a hsa-miR-590 hsa-miR-601
hsa-miR-605 hsa-miR-625 hsa-miR-7-1 hsa-miR-718 hsa-miR-744
hsa-miR-887 hsa-miR-99b |
| TCGA | 93 | microRNA 663b
microRNA 770 microRNA 520b microRNA 485 microRNA 1270 AC116165.1
microRNA 1302-3 microRNA 498 AC005631.1 microRNA 3156-3 AC100757.1
microRNA 516a-1 microRNA 920 AC233702.1 microRNA 519d microRNA 5787
microRNA 515-2 microRNA 6511a-3 microRNA 181b-1 microRNA 4697
AL391261.1 microRNA 6511a-1 AC005071.4 microRNA 1208 AC106782.1
AF254983.1 AJ271736.1 AC010203.1 microRNA 600 microRNA 6511a-2
AL354833.1 microRNA 4300 microRNA 1468 AL078621.1 microRNA 513c
microRNA 3142 AC011453.4 microRNA 412 microRNA let-7f-1 microRNA
221 AC245033.1 microRNA 6859-3 AL354820.1 microRNA 300 microRNA
519b microRNA 182 AC106788.1 microRNA 514a-2 microRNA 622 microRNA
507 AC126544.1 microRNA 205 microRNA 15a Z83819.1 microRNA 409
microRNA 6859-4 AC090825.1 microRNA 2682 microRNA 410 microRNA 154
microRNA 411 microRNA 548× microRNA 519a-1 microRNA 6859-1 microRNA
548i-4 microRNA 1197 microRNA 495 AL161651.1 microRNA 892a microRNA
3680-2 AC133555.1 AC092375.1 microRNA 509-2 AC105339.2 AC023310.1
AC024937.1 microRNA 570 microRNA 517b AC092017 microRNA 6080
microRNA 509-3 microRNA 3147 AC215219.2 microRNA 466 microRNA 129-2
microRNA 548i-1 AC011467.1 microRNA 1-1 microRNA 520d AC011453.3
microRNA 518f microRNA 758 |
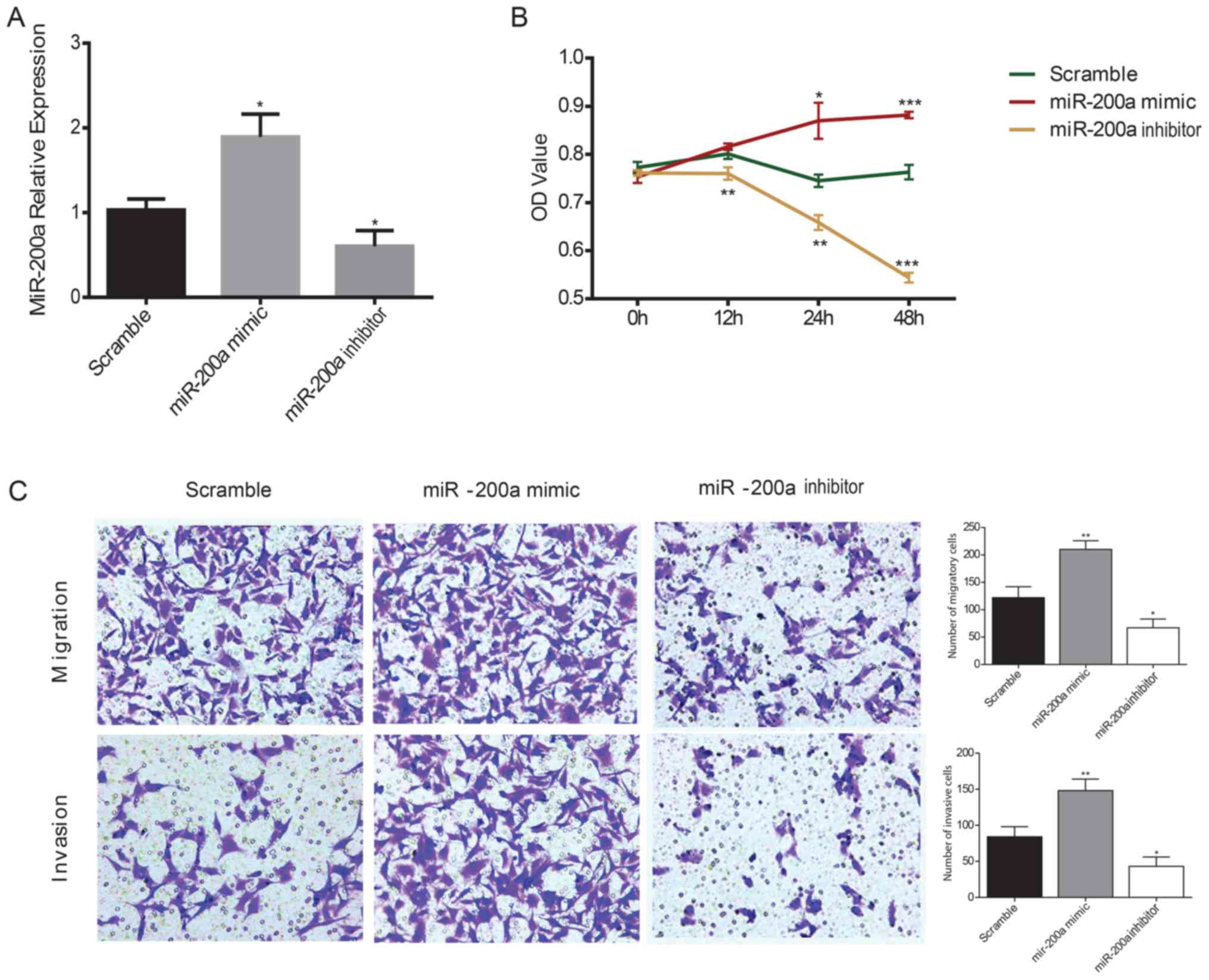
Effects of miR-200a on the
proliferation, migration and invasion of ESCC cells
KYSE150 ESCC cells were transiently transfected with
miR-200a mimics or inhibitors and the effects of miR-200a on ESCC
cell proliferation were assessed using a CCK-8 assay. RT-qPCR was
used to confirm the transfection efficiency of the mimic and
inhibitor in KYSE150 ESCC cells; the miR-200a mimic significantly
increased miR-200a expression levels compared with the scramble
control, whereas the miR-200a inhibitor significantly decreased
miR-200a expression levels compared with the scramble controls in
ESCC cells (Fig. 2A). The
viability of ESCC cells was significantly increased in miR-200a
mimic-transfected cells compared with the scramble control at 24
and 48 h, whereas the inhibition of miR-200a expression
significantly decreased ESCC cell viability compared with the
scramble control at all of the time points (Fig. 2B). To evaluate the effects of
miR-200a on cell migration and invasion, a Transwell assay was
performed in ESCC cells. miR-200a mimics transfection significantly
increased the migratory and invasive ability of ESCC cells compared
with the scramble control, whereas the miR-200a
inhibitor-transfected cells exhibited significantly reduced
migratory and invasive capacity compared with the scramble control
(Fig. 2C).
Identification of target genes and
pathway enrichment analysis
As miR-200a was the only miRNA selected from the two
groups, the current study aimed to identify target genes that
miR-200a may regulate in ESCC. The miRNA databases TargetScanHuman,
miRTarBase and mirRDB were searched and the intersections were
filtered using 54 genes. DAVID was used for pathway enrichment and
ClueGO and STRING were used for visualization. The results of the
GO analysis indicated that the main biological processes affected
by miR-200a were embryonic heart tube development and
transcriptional regulation, and the main cellular components
regulated were transcription factor complexes and cell-cell
adherens junctions (Table II).
The most significant molecular functions associated with miR-200a
were DNA binding, transcription factor binding and cell-cell
adhesion. The results of the KEGG pathway analysis reported that
basal cell carcinoma pathways in cancer and carbon metabolism were
the most significantly enriched pathways (P<0.05), whereas the
Wnt and the Hippo signaling pathways were classed as critical
pathways despite not being significant (P=0.05 and P=0.06,
respectively; Table II).
Subsequently, miR-200a target genes were assigned to ClueGO to
assess the association between miR-200a target genes and GO
biological process, molecular function and cellular component
terms, which yielded the terms ‘regulation of transcription’,
‘positive regulation of core promoter region’ and others (Fig. 3A) and GO terms distribution
associated with the number of genes and percentage (Fig. 3B). According to the relationships,
the protein-protein interaction map of target genes was created
using STRING, and the network visualization was performed using
Cytoscape (Fig. 3C). In the
network, CTNNB1, CDH1, TP53, PTEN, CDK1, APC, CTNNA1, FYN and SOD2
were the most prominent, as these genes exhibited multiple
associations with other genes.
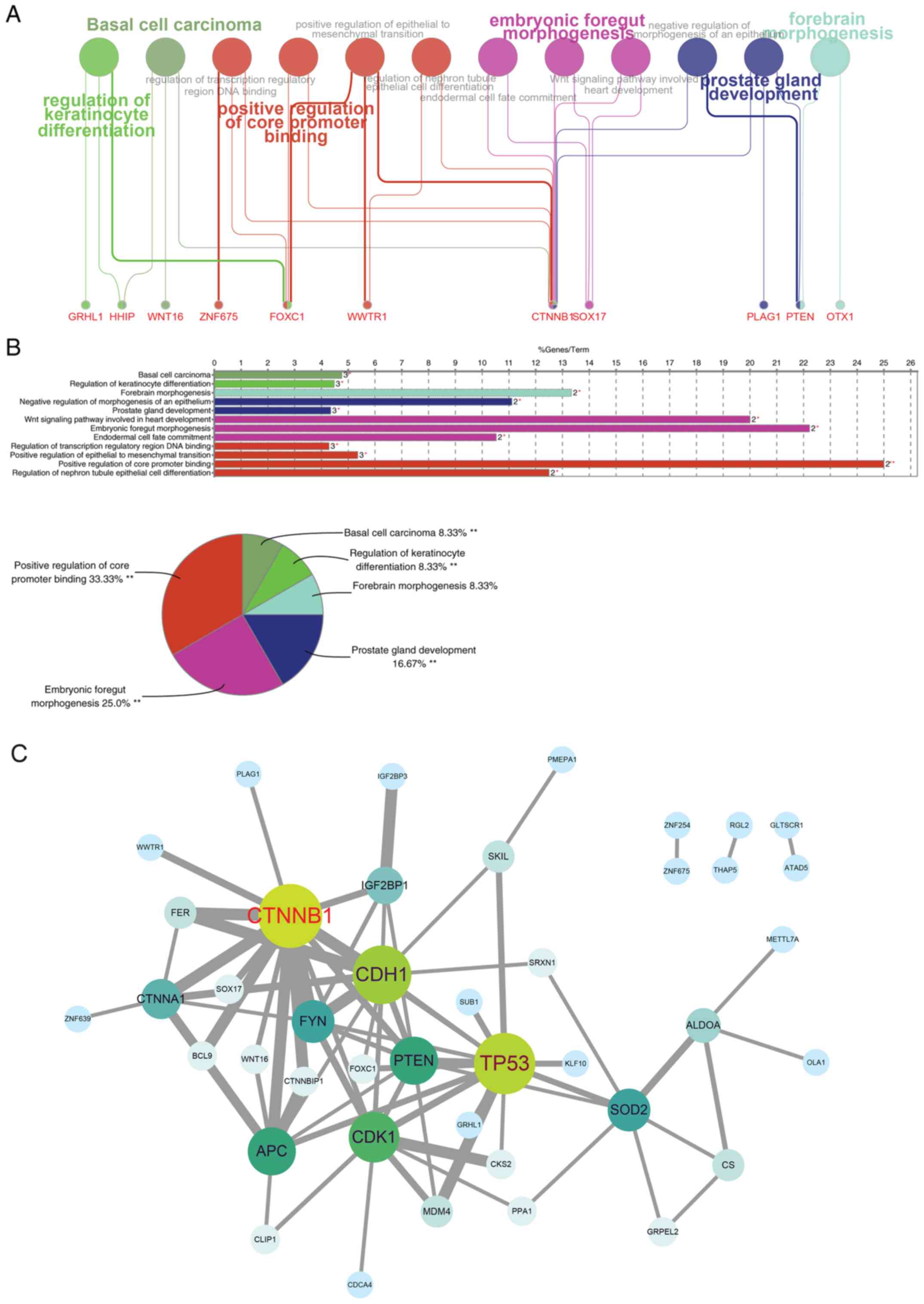 | Figure 3.Pathway enrichment analysis and
identification of key target genes of miR-200a. miR-200a target
genes from the intersection of miRNAs databases TargetScanHuman,
miRTarBase and mirRDB were input into ClueGO (version 2.54) for (A)
pathway enrichment and (B) target genes related GO term
distribution. (C) A protein-protein interaction network of target
genes was evaluated using STRING according to the gene-interaction
degree (in grey line), and the networks visualization was performed
using Cytoscape software with the size of circle (from green to
blue) representing the interaction count. *P<0.05, **P<0.01
for enrichment P-value from χ2 test. GO, gene ontology;
miR, microRNA; STRING, Search Tool for the Retrieval of Interacting
Genes. GRHL1, grainyhead like transcription factor 1; HHIP,
hedgehog interacting protein; WNT16, Wnt family member 16; ZNF675,
zinc finger protein 675; FOXC1, forkhead box C1; WWTR1, WW domain
containing transcription regulator 1; CTNNB1, catenin β1; SOX17,
SRY-box transcription factor 17; PLAG1, PLAG1 zinc finger; PTEN,
phosphatase and tensin homolog; OTX1, orthodenticle homeobox 1. |
 | Table II.GO analysis and KEGG pathway
enrichment analyses of miR-200a target genes. |
Table II.
GO analysis and KEGG pathway
enrichment analyses of miR-200a target genes.
| Category | Term | Count | P-value |
|---|
| GO
term-biological | Embryonic heart
tube development | 3 | <0.001 |
| processes | Negative regulation
of transcription from RNA polymerase II promoter | 9 | 0.001 |
|
| Positive regulation
of transcription from RNA polymerase II promoter | 10 | 0.002 |
|
| Transcription from
RNA polymerase II promoter | 7 | 0.004 |
|
| Response to zinc
ion | 3 | 0.005 |
|
| Regulation of
transcription, DNA-templated | 11 | 0.01 |
|
| Response to
drug | 5 | 0.01 |
|
| Cell
proliferation | 5 | 0.02 |
|
| Prostate gland
growth | 2 | 0.02 |
|
| Canonical Wnt
signaling pathway | 3 | 0.02 |
|
| Embryonic foregut
morphogenesis | 2 | 0.02 |
|
| Positive regulation
of skeletal muscle tissue development | 2 | 0.03 |
|
| Negative regulation
of cell proliferation | 5 | 0.03 |
|
| Positive regulation
of sequence-specific DNA binding transcription factor activity | 3 | 0.04 |
|
| Epithelial tube
branching involved in lung morphogenesis | 2 | 0.04 |
| GO term-cellular
components | Nucleus | 29 | <0.001 |
|
| Transcription
factor complex | 5 | 0.001 |
|
| Nuclear
transcription factor complex | 2 | 0.03 |
|
| Cell-cell adherens
junction | 4 | 0.05 |
|
| Mitochondrial
matrix | 4 | 0.05 |
| GO term-molecular
function | RNA polymerase II
core promoter proximal region sequence-specific DNA binding | 10 | <0.0001 |
|
| Nucleic acid
binding | 12 | 0.0001 |
|
| Transcription
factor activity, sequence-specific DNA binding | 10 | 0.001 |
|
| Transcription
regulatory region DNA binding | 5 | 0.003 |
|
| Transcriptional
activator activity, RNA polymerase II core promoter proximal region
sequence-specific binding | 5 | 0.005 |
|
| Metal ion
binding | 13 | 0.01 |
|
| Transcription
coactivator activity | 4 | 0.04 |
|
| Transcription
factor binding | 4 | 0.05 |
|
| DNA binding | 10 | 0.05 |
|
| Cadherin binding
involved in cell-cell adhesion | 4 | 0.05 |
| KEGG_Pathway | Signaling pathways
regulating pluripotency of stem cells | 4 | 0.006 |
|
| Basal cell
carcinoma | 3 | 0.009 |
|
| Biosynthesis of
amino acids | 3 | 0.01 |
|
| Pathways in
cancer | 5 | 0.02 |
|
| Carbon
metabolism | 3 | 0.04 |
|
| Wnt signaling
pathway | 3 | 0.05a |
|
| Hippo signaling
pathway | 3 | 0.06a |
Interactions of miR-200a and its
target genes
As previously described, CTNNB1, CDH1, TP53, PTEN,
CDK1, APC, CTNNA1, FYN and SOD2 were identified as the most
significant genes targeted by miR-200a in the network.
Representative immunohistochemistry images of these genes were
obtained from the Human Protein Atlas and were used to determine
the protein expression of each gene in normal esophageal tissues.
These proteins were highly expressed in normal tissues according to
the Human Protein Atlas (Fig.
S2). Accordingly, the highly expressed genes (CTNNB1, CDH1,
PTEN, APC, CTNNA1 and SOD2) were selected for subsequent analysis.
To detect the effects of miR-200a on the target genes, CTNNB1,
CDH1, PTEN, APC, CTNNA1 and SOD2 mRNA and protein expression levels
were examined in KYSE150 ESCC cells transfected with a miR-200a
mimic or inhibitor. RT-qPCR and western blot analysis demonstrated
that target genes and their protein levels were significantly
decreased in cells transfected with miR-200a mimic compared with
the scramble-transfected cells, whereas expression levels were
significantly increased in cells transfected with the miR-200a
inhibitor compared with the control (Fig. 4A). A dual-luciferase reporter assay
was performed to evaluate the direct interaction between miR-200a
and the 3′-UTR of the target genes. The results indicated that
KYSE150 ESCC cells transfected with the miR-200a mimic
significantly suppressed the luciferase activity of the wild-type
reporters containing the 3′-UTR of all target genes (CTNNB1, CDH1,
APC, PTEN, CTNNA1 and SOD2) compared with the scramble control;
this inhibition disappeared when the miR-200a target site was
mutated (Fig. 4B).
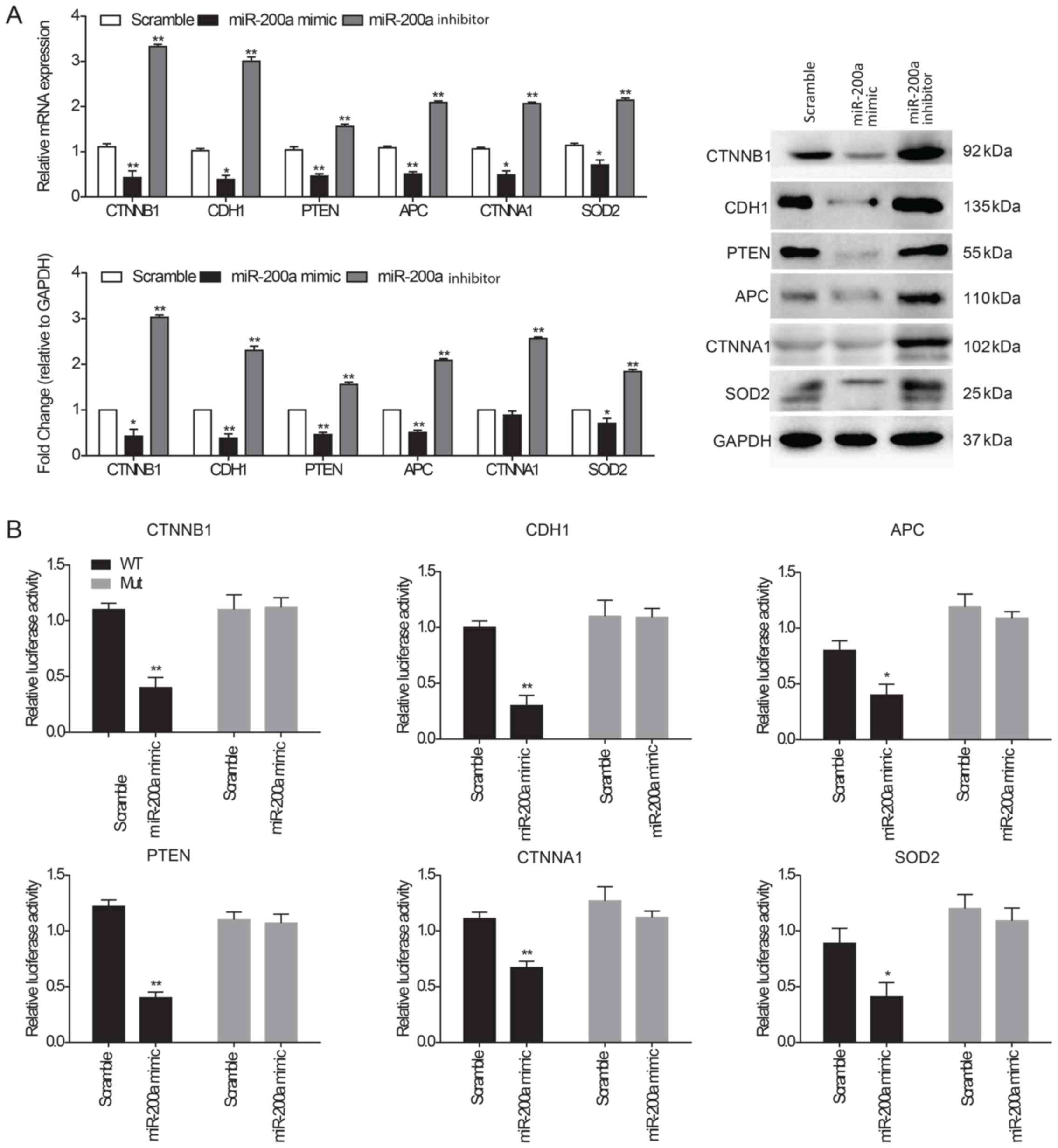 | Figure 4.Interaction of miR-200a and its
target genes. (A) CTNNB1, CDH1, PTEN, APC, CTNNA1 and SOD2 mRNA and
protein expression levels were determined in KYSE150 ESCC cells
following transfection with scramble control miRNA, miR-200a mimic
or inhibitor using reverse transcription-quantitative PCR and
western blot analysis. GAPDH was used as an internal loading
control for both analyses. (B) Dual-luciferase assay using
wild-type or mutated 3′-UTR sequences of target genes
co-transfected with miR-200a mimics into ESCC cells. The cells were
evaluated for luciferase activity following 48 h incubation. Data
are presented as the mean ± SEM of three independent experiments.
*P<0.05 and **P<0.01 vs. Scramble. APC, adenomatous polyposis
coli protein; CDH1, cadherin-1; CTNNA1, catenin α1; CTNNB1, catenin
β1; ESCC, esophageal squamous cell carcinoma; SOD2, superoxide
dismutase; UTR, untranslated region. |
Discussion
ESCC is the main histological type of EC, accounting
for >90% of new cases (23);
the majority of new ESCC cases occur in Asia, especially in China
(2). Clinical treatment for ESCC
remains challenging, and novel diagnosis and treatment strategies
are urgently required. miRNAs have been widely reported to be
associated with tumor development, and are often regarded as
disease biomarkers; it has been demonstrated that miRNAs affect
ESCC cell behaviors, including cell proliferation, migration and
invasion (7–9). However, miRNAs function in a
complicated regulatory network, which involves a number of
different biological processes (24). The present study analyzed the miRNA
expression microarray of NEC (with its incidence being rare but of
similar effects with ESCC) tissues from dataset GSE113776 of the
GEO database. A total of 28 miRNAs were selected according to the
inclusion criteria. miR-200a was selected following the
identification of an intersection between the 28 selected miRNAs
and 93 DEMs using the TCGA. Subsequent in vitro experiments
demonstrated that miR-200a may be involved in promoting ESCC cell
proliferation, migration and invasion. Furthermore, the results
provided evidences that miR-200a interacts with its target genes,
CTNNB1, CDH1, PTEN, APC, CTNNA1 and SOD2, which may contribute to
the abnormal cell behaviors in EC.
A number of previous studies have performed
high-throughput sequencing, while a total of 140 DEMs have been
extracted, with 113 upregulated and 27 downregulated miRNAs being
identified in ESCC tissue (25).
In a previous study, only five miRNAs (miR-103-1, miR-18a, miR-324,
miR-369 and miR-320b-2), which have previously been associated with
survival rates (25), were
studied. In another study, out of a total of 136 DEMs identified in
EC, the top five DEMs were revealed to be miRNA-21, miRNA-93,
miRNA-196a-1, miRNA-196a-2 and miRNA-4746 (26). In addition, within a cohort of 102
patients with EC, miR-451a, miR-144-3p and miR-144-5p were
identified; other miRNAs including miR-133a miR-138, miR-375 and
miR-593 were observed to serve as tumor suppressors, whereas
miR-16, miR-21, miR-31, miR-34b, miR-208, miR-223, miR-373 and
miR-423 exhibited oncogenic properties (13–18).
Meanwhile, researchers identified miR-28-5p, miR-34a-5p and
miR-186-5p as the significant biomarkers of ESCC (27). Furthermore, previous studies
explored the underlying miRNAs that can play key roles in ESCC
development. For instance, miRNA-10b can promote ESCC cell
proliferation, migration, invasion and colony formation, as well as
metastasis; miRNA-548 and miRNA-576 enhance the migration and
invasion of ESCC cells; miRNA-1 can suppress the proliferation,
migration and invasion of ESCC cells (28–30).
Based on these studies, a number of miRNAs have been reported to
serve a role in EC; however, to the best of our knowledge, no
previous studies have predicted the potential role of miR-200a in
EC pathogenesis using bioinformatics. In the present study, a total
of 28 miRNAs were identified in the NEC microarray dataset
GSE113776, including miR-200a. Additionally, a number of important
miRNAs, including miR-181c-5p, hsa-miR-500a-3p, hsa-miR-601 and
hsa-miR-605 were matched with identified DEMs in previous research
(25,26).
To the best of our knowledge, no previous
bioinformatics study has identified miR-200a as a crucial miRNA,
and this may be due to the fact that a single sample of NEC tissue
was used. The GSE113776 dataset may not have been investigated
owing to the difficulty of using statistical analysis to assess it.
However, this disadvantage permitted the identification of novel
miRNAs because the high expression of miRNAs in the tissue may have
excluded the significance of the less expressed, but critical
miRNAs. In addition, previous studies may not have focused on the
most important miRNAs, but instead on determining DEMs. Owing to
the single sample comparison in the present study, miR-200a was
identified and its was demonstrated to be upregulated in ESCC
according to the miRNA profile GSE112840 (19). The importance of miR-200a was
confirmed by matching with TGCA records and RT-qPCR verification.
It is hypothesized that different types of cells exhibit mutual
effects or interactions with each other (31–34);
NEC, squamous cell carcinoma and sarcoma exhibit metastatic and
site-transfer-direction (32,33),
and definitive chemoradiotherapy promoted the conversion of NEC to
squamous cell carcinoma (34). In
addition, NEC, squamous cell carcinoma and adenocarcinoma can
coexist (31,35,36).
Although miR-220a was singled out from NEC tissues, the present
study demonstrated the effects of miR-200a in ESCC cells; however,
the direct association between ESCC and NEC and adenocarcinoma was
not assessed due to the lack of tissue.
The alterations of ESCC cell behaviors following
miR-200a overexpression are not well determined. The role of miRNAs
depends on the type of miRNAs. It is reported that miR-100 inhibits
EC cell proliferation, migration and invasion, (10), while miR-373 enhances cell
proliferation, migration and invasion (11). The present study demonstrated that
the upregulation of miR-200a could promote ESCC cell proliferation,
migration and invasion. This was a similar role to the effects of
miR-373 in decreasing the proliferation, migration and invasion of
ESCC cells. In addition to the effects of miR-200a in ESCC,
miR-200a has been studied in a number of tumor types, including
breast, ovarian, gastric, colorectal and pancreatic cancers
(37–41); miR-200a is also associated with
cell proliferation, migration and invasion in these tumor types
(37–41).
The present study investigated the putative target
genes of miR-200a, as miRNAs function in the development of ESCC
via binding to the UTR region of target genes: miRNA-10b targeting
FOXO3, micRNA-548 and miRNA-576 downregulating NRIP1 and miRNA-1
binding to Notch2 (28–30). Bioinformatics analysis revealed
that the main biological processes that miR-200a was involved in
included embryonic development and transcription regulation, the
main cellular components were transcription factor complexes and
cell-cell adherence junctions and the main molecular functions were
related to DNA binding, transcription factor binding and cell-cell
adhesion, which showed the similar results with the findings of
previous studies enriched from other miRNAs (25,26,42).
The results of KEGG pathway enrichment, except for the suggestions
of basic pathological pathways, also indicated that miR-200a may
serve a role in the Wnt signaling pathway and the Hippo signaling
pathway, which was consistent with previous findings (43–47).
According to target gene prediction and their expression in normal
tissues from the Human Protein Atlas, miR-200a was identified to
target CTNNB1, CDH1, PTEN, APC, CTNNA1 and SOD2. CTNNB1 is a
crucial downstream component of the Wnt signaling pathway, forming
a complex to promote phosphorylation on N-terminal serine and
threonine residues in the absence of Wnt, while acting as a
coactivator for Wnt responsive genes in the presence of the Wnt
ligand (48,49). Cadherins are calcium-dependent cell
adhesion proteins that interact in a homophilic manner during cell
communications and CDH1 is associated with cell-cell adhesions
regulation, mobility and proliferation (50). The major function of CTNNB1 is to
regulate cell adhesion by participating in the E-cadherin/catenin
adhesion complex (51). CTNNA1,
which is an isotype of the catenin protein, is associated with a
number of cadherins, including E- and N-cadherins, and stabilizes
E-cadherin/catenin adhesion complexes (52). APC, which is correlated with its
phosphorylation state, participates in the Wnt signaling pathway as
a negative regulator and promotes the rapid degradation of CTNNB1
(53,54). PTEN is a dual-specificity protein
phosphatase, which mediates the phosphorylation of N-terminal
tyrosine, serine and threonine residues (55). The dephosphorylation of
tyrosine-phosphorylated focal adhesion kinase inhibits focal
adhesion formation, cell migration and integrin-mediated cell
spreading (56). SOD2 has been
reported to delay tumor cell growth in a number of tumor types
(57,58); its activity affects different
stages of the cell cycle and its downregulation may stimulate cell
cycle progression (59–61). In addition, it has been
demonstrated through immunohistochemical staining that the protein
expression levels of CTNNB1, CDH1, TP53 and PTEN are decreased in
ESCC tissues compared with normal tissues (62–64),
while the results of the present study showed the high expression
of CTNNB1, CDH1, TP53 and PTEN in normal tissue with IHC staining
(Fig. S2). Previous studies
reported that miR-200a regulated CTNNB1, CDH1 and PTEN, but not
APC, CTNNA1 and SOD2 in tumors. miR-200a was demonstrated to
downregulate CTNNB1 and inhibit nasopharyngeal carcinoma cell
growth, migration and invasion (65). miR-200a suppressed CDH1 and
resulted in the induction of EMT, which has critical functions in
tumor cell migration and invasion (66). miR-200a inhibited the expression of
PTEN and promoted the invasion and migration of ovarian cancer
cells (67). Thus, determining the
regulation of miR-200a on APC, CTNNA1 and SOD2 during EC
oncogenesis may be useful in the future. Since these target genes
have been reported to be associated with the development of a
number of types of cancer by influencing the cell cycle and cell
adhesion, it is suggested that miR-200a may alter the cell
behaviors of ESCC due to the role that the downstream genes have in
cell proliferation, migration and invasion pathways.
In conclusion, the present study demonstrated that
miR-200a participated in promoting ESCC cell proliferation,
migration and invasion, and provided novel evidence for the direct
interaction between miR-200a and CTNNB1, CDH1, PTEN, APC, CTNNA1
and SOD2, which may be responsible for this observed cell
behavior.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
BY performed the experiments and wrote the
manuscript. YL and LL performed the bioinformatics analysis. HD
contributed to the interpretation of the data and helped write and
revise the manuscript. LX conceived and designed the experiments.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Karamanou M, Markatos K, Papaioannou TG,
Zografos G and Androutsos G: Hallmarks in history of esophageal
carcinoma. J BUON. 22:1088–1091. 2017.PubMed/NCBI
|
|
2
|
Klingelhöfer D, Zhu Y, Braun M, Brüggmann
D, Schöffel N and Groneberg DA: A world map of esophagus cancer
research: A critical accounting. J Transl Med. 17:1502019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu F, Wu K, Wu W, Chen Y, Wu H, Wang H
and Zhang W: miR203 contributes to preeclampsia via inhibition of
VEGFA expression. Mol Med Rep. 17:5627–5634. 2018.PubMed/NCBI
|
|
5
|
Liu F, Wu W, Wu K, Chen Y, Wu H, Wang H
and Zhang W: MiR-203 participates in human placental angiogenesis
by inhibiting VEGFA and VEGFR2 Expression. Reprod Sci. 25:358–365.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jopling CL, Schütz S and Sarnow P:
Position-dependent function for a tandem microRNA miR-122-binding
site located in the hepatitis C virus RNA genome. Cell Host
Microbe. 4:77–85. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
8
|
Tutar L, Özgür A and Tutar Y: Involvement
of miRNAs and pseudogenes in cancer. Methods Mol Biol. 1699:45–66.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou SM, Zhang F, Chen XB, Jun CM, Jing X,
Wei DX, Xia Y, Zhou YB, Xiao XQ, Jia RQ, et al: miR-100 suppresses
the proliferation and tumor growth of esophageal squamous cancer
cells via targeting CXCR7. Oncol Rep. 35:3453–3459. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu DD, Zhou PJ, Wang Y, Zhang L, Fu WY,
Ruan BB, Xu HP, Hu CZ, Tian L, Qin JH, et al: Reciprocal activation
between STAT3 and miR-181b regulates the proliferation of
esophageal cancer stem-like cells via the CYLD pathway. Mol Cancer.
15:402016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao Z, Liu R, Liao J, Yang M, Pan E, Yin L
and Pu Y: Possible tumor suppressive role of the miR-144/451
cluster in esophageal carcinoma as determined by principal
component regression analysis. Mol Med Rep. 14:3805–3813. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Okumura T, Kojima H, Miwa T, Sekine S,
Hashimoto I, Hojo S, Nagata T and Shimada Y: The expression of
microRNA 574-3p as a predictor of postoperative outcome in patients
with esophageal squamous cell carcinoma. World J Surg Oncol.
14:2282016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lv H, He Z, Wang H, Du T and Pang Z:
Differential expression of miR-21 and miR-75 in esophageal
carcinoma patients and its clinical implication. Am J Transl Res.
8:3288–3298. 2016.PubMed/NCBI
|
|
15
|
Sun J, Song K, Feng X and Gao S:
MicroRNA-367 is a potential diagnostic biomarker for patients with
esophageal squamous cell carcinoma. Biochem Biophys Res Commun.
473:363–369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun L, Dong S, Dong C, Sun K, Meng W, Lv
P, Yin H, Ming L and He F: Predictive value of plasma miRNA-718 for
esophageal squamous cell carcinoma. Cancer Biomark. 16:265–273.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Matsuzaki J and Suzuki H: Role of
MicroRNAs-221/222 in digestive systems. J Clin Med. 4:1566–1577.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Phatak P, Byrnes KA, Mansour D, Liu L, Cao
S, Li R, Rao JN, Turner DJ, Wang JY and Donahue JM: Overexpression
of miR-214-3p in esophageal squamous cancer cells enhances
sensitivity to cisplatin by targeting survivin directly and
indirectly through CUG-BP1. Oncogene. 35:2087–2097. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng D, Ding Y, Ma Q, Zhao L, Guo X, Shen
Y, He Y, Wei W and Liu F: Identification of serum MicroRNAs as
novel biomarkers in esophageal squamous cell carcinoma using
feature selection algorithms. Front Oncol. 8:6742018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Di Stefano V, Wang B, Parobchak N, Roche N
and Rosen T: RelB/p52-mediated NF-κB signaling alters histone
acetylation to increase the abundance of corticotropin-releasing
hormone in human placenta. Sci Signal. 8:ra852015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu X, Er K, Mao C, Yan Q, Xu H, Zhang Y,
Zhu J, Cui F, Zhao W and Shi H: miR-203 suppresses tumor growth and
angiogenesis by targeting VEGFA in cervical cancer. Cell Physiol
Biochem. 32:64–73. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lepage C, Rachet B, Jooste V, Faivre J and
Coleman MP: Continuing rapid increase in esophageal adenocarcinoma
in England and Wales. Am J Gastroenterol. 103:2694–2699. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chekouo T, Stingo FC, Doecke JD and Do KA:
miRNA-target gene regulatory networks: A Bayesian integrative
approach to biomarker selection with application to kidney cancer.
Biometrics. 71:428–438. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao JY, Wang F, Li Y, Zhang XB, Yang L,
Wang W, Xu H, Liu DZ and Zhang LY: Five miRNAs considered as
molecular targets for predicting esophageal cancer. Med Sci Monit.
21:3222–3230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zeng JH, Xiong DD, Pang YY, Zhang Y, Tang
RX, Luo DZ and Chen G: Identification of molecular targets for
esophageal carcinoma diagnosis using miRNA-seq and RNA-seq data
from The Cancer Genome Atlas: A study of 187 cases. Oncotarget.
8:35681–35699. 2017.PubMed/NCBI
|
|
27
|
Chen L, Jin Y, Wang L, Sun F, Yang X, Shi
M, Zhan C, Shi Y and Wang Q: Identification of reference genes and
miRNAs for qRT-PCR in human esophageal squamous cell carcinoma. Med
Oncol. 34:22017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu YF, Yu JR, Yang Z, Zhu GX, Gao P, Wang
H, Chen SY, Zhang J, Liu MY, Niu Y, et al: Correction to: Promoter
hypomethylation mediated upregulation of MicroRNA-10b-3p targets
FOXO3 to promote the progression of esophageal squamous cell
carcinoma (ESCC). J Exp Clin Cancer Res. 39:192020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ni XF, Zhao LH, Li G, Hou M, Su M, Zou CL
and Deng X: MicroRNA-548-3p and MicroRNA-576-5p enhance the
migration and invasion of esophageal squamous cell carcinoma cells
via NRIP1 down-regulation. Neoplasma. 65:881–887. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu W, Li M, Chen X, Zhu S, Shi H, Zhang
D, Cheng C and Li B: MicroRNA-1 suppresses proliferation, migration
and invasion by targeting Notch2 in esophageal squamous cell
carcinoma. Sci Rep. 8:51832018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kaneko Y, Saito S, Takahashi K, Kanamaru
R, Hosoya Y, Yamaguchi H, Kitayama J, Niki T, Lefor AK and Sata N:
Neuroendocrine carcinoma of the esophagus with an adenocarcinoma
component. Clin J Gastroenterol. 12:534–538. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsuchihashi K, Arita S, Fujiwara M,
Iwasaki K, Hirano A, Yoshihiro T, Nio K, Koga Y, Esaki M, Ariyama
H, et al: Metastatic esophageal carcinosarcoma comprising
neuroendocrine carcinoma, squamous cell carcinoma, and sarcoma: A
case report. Medicine (Baltimore). 97:e127962018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fujihara S, Kobayashi M, Nishi M, Yachida
T, Yoshitake A, Deguchi A, Muraoka A, Kobara H and Masaki T:
Composite neuroendocrine carcinoma and squamous cell carcinoma with
regional lymph node metastasis: A case report. J Med Case Rep.
12:2272018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Morita M, Saeki H, Nakaji YU, Zaitsu Y,
Hirahashi M, Ohguri T, Oki E, Toh Y, Oda Y and Maehara Y:
Conversion to neuroendocrine carcinoma from squamous cell carcinoma
of the esophagus after definitive chemoradiotherapy. Anticancer
Res. 36:4045–4049. 2016.PubMed/NCBI
|
|
35
|
Yazıcı O, Aksoy S, Özhamam EU and Zengin
N: Squamous cell and neuroendocrine carcinoma of esophagus:
Collision versus composite tumor: A case report and review of
literature. Indian J Cancer. 52:603–604. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang L, Sun X, Zou Y and Meng X: Small
cell type neuroendocrine carcinoma colliding with squamous cell
carcinoma at esophagus. Int J Clin Exp Pathol. 7:1792–1795.
2014.PubMed/NCBI
|
|
37
|
Yu SJ, Yang L, Hong Q, Kuang XY, Di GH and
Shao ZM: MicroRNA-200a confers chemoresistance by antagonizing
TP53INP1 and YAP1 in human breast cancer. BMC Cancer. 18:742018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun Q, Zou X, Zhang T, Shen J, Yin Y and
Xiang J: The role of miR-200a in vasculogenic mimicry and its
clinical significance in ovarian cancer. Gynecol Oncol.
132:730–738. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu X, Du P, Han L, Zhang A, Jiang K and
Zhang Q: Effects of miR-200a and FH535 combined with taxol on
proliferation and invasion of gastric cancer. Pathol Res Pract.
214:442–449. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang W, Ning N and Jin X: The lncRNA H19
promotes cell proliferation by competitively binding to miR-200a
and Derepressing β-catenin expression in colorectal cancer. Biomed
Res Int. 2017:27674842017.PubMed/NCBI
|
|
41
|
Hu B, Qiu-Lan H, Lei RE, Shi C, Jiang HX
and Qin SY: Interleukin-9 promotes pancreatic cancer cells
proliferation and migration via the miR-200a/Beta-catenin axis.
Biomed Res Int. 2017:28310562017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen F, Zhou H, Wu C and Yan H:
Identification of miRNA profiling in prediction of tumor recurrence
and progress and bioinformatics analysis for patients with primary
esophageal cancer: Study based on TCGA database. Pathol Res Pract.
214:2081–2086. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang L, Zhang Z, Yu X, Huang X, Liu Z,
Chai Y, Yang L, Wang Q, Li M, Zhao J, et al: Unbalanced YAP-SOX9
circuit drives stemness and malignant progression in esophageal
squamous cell carcinoma. Oncogene. 38:2042–2055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qi B, Wang Y, Chen ZJ, Li XN, Qi Y, Yang
Y, Cui GH, Guo HZ, Li WH and Zhao S: Down-regulation of
miR-30a-3p/5p promotes esophageal squamous cell carcinoma cell
proliferation by activating the Wnt signaling pathway. World J
Gastroenterol. 23:7965–7977. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gao Y, Yi J, Zhang K, Bai F, Feng B, Wang
R, Chu X, Chen L and Song H: Downregulation of MiR-31 stimulates
expression of LATS2 via the hippo pathway and promotes
epithelial-mesenchymal transition in esophageal squamous cell
carcinoma. J Exp Clin Cancer Res. 36:1612017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zang B, Huang G, Wang X and Zheng S:
HPV-16 E6 promotes cell growth of esophageal cancer via
downregulation of miR-125b and activation of Wnt/β-catenin
signaling pathway. Int J Clin Exp Pathol. 8:13687–13694.
2015.PubMed/NCBI
|
|
47
|
Ge C, Wu S, Wang W, Liu Z, Zhang J, Wang
Z, Li R, Zhang Z, Li Z, Dong S, et al: miR-942 promotes cancer stem
cell-like traits in esophageal squamous cell carcinoma through
activation of Wnt/β-catenin signalling pathway. Oncotarget.
6:10964–10977. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Weiske J, Albring KF and Huber O: The
tumor suppressor Fhit acts as a repressor of beta-catenin
transcriptional activity. Proc Natl Acad Sci USA. 104:20344–20349.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lillehoj EP, Lu W, Kiser T, Goldblum SE
and Kim KC: MUC1 inhibits cell proliferation by a
beta-catenin-dependent mechanism. Biochim Biophys Acta.
1773:1028–1038. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Meigs TE, Fedor-Chaiken M, Kaplan DD,
Brackenbury R and Casey PJ: Galpha12 and Galpha13 negatively
regulate the adhesive functions of cadherin. J Biol Chem.
277:24594–24600. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu Y, Wu J, Wang Y, Zhao T, Ma B, Liu Y,
Fang W, Zhu WG and Zhang H: Kindlin 2 forms a transcriptional
complex with β-catenin and TCF4 to enhance Wnt signalling. EMBO
Rep. 13:750–758. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Escobar DJ, Desai R, Ishiyama N, Folmsbee
SS, Novak MN, Flozak AS, Daugherty RL, Mo R, Nanavati D, Sarpal R,
et al: α-Catenin phosphorylation promotes intercellular adhesion
through a dual-kinase mechanism. J Cell Sci. 128:1150–1165. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zaoui K, Benseddik K, Daou P, Salaün D and
Badache A: ErbB2 receptor controls microtubule capture by
recruiting ACF7 to the plasma membrane of migrating cells. Proc
Natl Acad Sci USA. 107:18517–18522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sagara M, Kawasaki Y, Iemura SI, Natsume
T, Takai Y and Akiyama T: Asef2 and Neurabin2 cooperatively
regulate actin cytoskeletal organization and are involved in
HGF-induced cell migration. Oncogene. 28:1357–1365. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vazquez F, Ramaswamy S, Nakamura N and
Sellers WR: Phosphorylation of the PTEN tail regulates protein
stability and function. Mol Cell Biol. 20:5010–5018. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Costa HA, Leitner MG, Sos ML, Mavrantoni
A, Rychkova A, Johnson JR, Newton BW, Yee MC, De La Vega FM, Ford
JM, et al: Discovery and functional characterization of a
neomorphic PTEN mutation. Proc Natl Acad Sci USA. 112:13976–13981.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Weydert C, Roling B, Liu J, Hinkhouse MM,
Ritchie JM, Oberley LW and Cullen JJ: Suppression of the malignant
phenotype in human pancreatic cancer cells by the overexpression of
manganese superoxide dismutase. Mol Cancer Ther. 2:361–369.
2003.PubMed/NCBI
|
|
58
|
Zhong W, Oberley LW, Oberley TD and St
Clair DK: Suppression of the malignant phenotype of human glioma
cells by overexpression of manganese superoxide dismutase.
Oncogene. 14:481–490. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sarsour EH, Kalen AL and Goswami PC:
Manganese superoxide dismutase regulates a redox cycle within the
cell cycle. Antioxid Redox Signal. 20:1618–1627. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Oberley LW, Oberley TD and Buettner GR:
Cell division in normal and transformed cells: The possible role of
superoxide and hydrogen peroxide. Med Hypotheses. 7:21–42. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Oberley LW, Oberley TD and Buettner GR:
Cell differentiation, aging and cancer: The possible roles of
superoxide and superoxide dismutases. Med Hypotheses. 6:249–268.
1980. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Taghavi N, Biramijamal F, Sotoudeh M,
Khademi H, Malekzadeh R, Moaven O, Memar B, A'rabi A and
Abbaszadegan MR: p16INK4a hypermethylation and p53, p16 and MDM2
protein expression in esophageal squamous cell carcinoma. BMC
Cancer. 10:1382010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lu J, Pan Y, Xia X, Gu Y and Lei Y:
Prognostic significance of mTOR and PTEN in patients with
esophageal squamous cell carcinoma. Biomed Res Int.
2015:4172102015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ishiguro H, Wakasugi T, Terashita Y,
Sakamoto N, Tanaka T, Mizoguchi K, Sagawa H, Okubo T and Takeyama
H: Decreased expression of CDH1 or CTNNB1 affects poor prognosis of
patients with esophageal cancer. World J Surg Oncol. 14:2402016.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Xia H, Ng SS, Jiang S, Cheung WK, Sze J,
Bian XW, Kung HF and Lin MC: miR-200a-mediated downregulation of
ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma
cell growth, migration and invasion. Biochem Biophys Res Commun.
391:535–541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Asakura T, Yamaguchi N, Ohkawa K and
Yoshida K: Proteasome inhibitor-resistant cells cause EMT-induction
via suppression of E-cadherin by miR-200 and ZEB1. Int J Oncol.
46:2251–2260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Suo HB, Zhang KC and Zhao J: MiR-200a
promotes cell invasion and migration of ovarian carcinoma by
targeting PTEN. Eur Rev Med Pharmacol Sci. 22:4080–4089.
2018.PubMed/NCBI
|