Introduction
Esophageal cancer (EC) is one of the most common
malignant tumors of the digestive system worldwide based on the
statistical analysis of 2008 (1).
Although its degree of malignancy is inferior to gastric and liver
cancer, according to statistics from the National Cancer Center in
2017, EC ranks as the fourth most common cause of malignant
tumor-associated mortality in China (1–3).
There are two main types of EC, esophageal squamous cell carcinoma
(ESCC), which accounts for >90% of cases, and esophageal
adenocarcinoma (4,5). At present, the primary treatment
strategy for EC involves surgery, radiotherapy and chemotherapy,
and the main causes of mortality are recurrence and metastasis
(6). Patients with EC typically
have a 5-year survival rate of 20–25%, even after radical
resection, which is caused by >90% of patients with EC being
diagnosed at the advanced stages, when the available treatment
strategies are no longer optimal (7,8).
Additionally, early diagnostic techniques used for EC, including
X-ray angiography, exfoliative cytology and endoscopy, have
complicated clinical operations and are often inaccurate;
therefore, the techniques cannot be used for routine screening
(6,9–11).
With the continuous development of molecular tumor markers, the
detection, simplicity, effectiveness and reliability of using
peripheral blood serum has aided in the early diagnosis and
treatment of patients with tumors (12,13).
However, the current markers for the early diagnosis and treatment
of EC are in the early stages of research and further investigation
is required to identify additional specific molecular
biomarkers.
The Wiskott-Aldrich syndrome protein (WASP) family
consists of novel actin-regulating proteins, which serve a
regulatory role in cell structure, cell membrane actin
polymerization regulation, cell migration and movement (14–16).
Previous studies have indicated that WASP deficiencies can result
in abnormal maintenance of morphological structure and cell
migration (17,18). As an important member of the WASP
family, Wiskott-Aldrich syndrome verprolin-homologous protein 3
(WAVE3) is a proline-rich protein, which influences cell migration,
the maintenance of cell structure and the regulation of actin
polymerization (19,20). A number of studies have
demonstrated the role of WAVE3 during tumor cell migration and
invasion (21,22). Davuluri et al (23,24)
reported that WAVE3 was associated with a variety of signaling
pathways, and cell migration regulation involved the NF-κB
signaling pathway. Further studies have indicated that WAVE3
affects proliferation, migration and invasion abilities in
pancreatic cancer cells via the AKT signaling pathway, suggesting
that the targeted inhibition of WAVE3 expression may serve as a
novel therapeutic strategy for pancreatic cancer (25,26).
Therefore, WAVE3, as a potential tumor marker, and may have broad
application prospects for the diagnosis, prognosis and treatment of
a number of different tumors (27). WAVE3 expression is upregulated in
pancreatic cancer (25,26) and breast cancer cells, particularly
in triple-negative breast cancer (27,28)
and human prostate cancer (29).
Numerous studies have investigated the roles of WAVE3, including
its effect on cell motility, migration and invasion, in a number of
different types of cancer, including hepatocellular carcinoma,
prostate cancer and colorectal cancer (22,29,30).
However, the expression of WAVE3 in EC and the associated
underlying mechanisms are yet to be fully elucidated.
MicroRNAs (miRNAs/miRs) are endogenous non-coding
RNAs which bind the 3′-untranslated regions (3′-UTR) of mRNAs to
regulate gene expression (31).
The miRNA-200 family includes three members: miR200a, miR200b and
miR200c. Sossey-Alaoui et al (32) demonstrated that miRNA200 binds to
the 3′-UTR of WAVE3 to inhibit WAVE3 protein expression and
influence the progression of tumors, with miRNA200b being
identified as the representative member. The present study aimed to
regulate the expression of WAVE3 using miRNA200b, and to further
explore the effect of WAVE3 on cell migration ability in ESCC.
Previous studies have indicated that WAVE3 is associated with
alterations to cell motility via the epithelial-mesenchymal
transition (EMT) process (21,33–35),
which is important during wound healing, embryonic development, and
cell migration and invasion (36).
A number of studies have also demonstrated that miRNA200 is
associated with the regulation of EMT, via the regulation of WAVE3
protein (30,32,37).
However, the mechanisms underlying the actions of WAVE3 in ESCC
require further investigation.
In the present study, the expression level of WAVE3
in ESCC tissues and serum, and its association with the progression
of ESCC was determined. miRNA200b mimics were transfected into ESCC
cell lines (EC109 and EC1), and the negative regulation of WAVE3
expression via miRNA200b was investigated. The present study
provided novel insight into the progression of ESCC and identified
a potential diagnostic biomarker and therapeutic target for
ESCC.
Materials and methods
Tissue samples and peripheral blood
collection
A total of 62 pairs of ESCC tissue samples and
corresponding adjacent normal tissues were collected between
September 2017 and December 2017 from the Pathology Laboratory of
the First Affiliated Hospital of Zhengzhou University (Zhengzhou,
China). A total of 80 patients with ESCC and 30 healthy volunteers
were enrolled between January 2018 and March 2018 from the Clinical
Laboratory of the First Affiliated Hospital of Zhengzhou University
(Zhengzhou, China). Serum samples were collected from patients and
healthy volunteers. The mean age of patients and volunteers was 60
years, ranging 37–75 years. The female-to-male ratio of tissue
samples was 0.7, and 0.5 in serum samples (Tables I and II). All samples were centrifuged at
1,370 × g for 5 min at 4°C to extract serum within 2 h of
collection, again at 16,000 × g for 5 min at 4°C to completely
remove cell debris and subsequently stored at −80°C until RNA
extraction. Tissue samples had not received anticancer treatment
prior to routine surgical resection, and all samples were confirmed
as ESCC via pathological analysis by two independent pathologists
blind to the clinical details of the patients. Serum samples were
obtained at the time of primary diagnosis, before treatment, by
collecting venous blood. The basic clinicopathological parameters
of the patients are presented in Tables I and II. The tumor staging was assessed
according to the TNM stage (38).
The present study was approved by the Institute Research Ethics
Committee of the First Affiliated Hospital of Zhengzhou University.
All patients provided written informed consent.
 | Table I.Association between clinicopathologic
parameters and WAVE3 expression. |
Table I.
Association between clinicopathologic
parameters and WAVE3 expression.
|
|
| WAVE3
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathologic
parameter | Patients (n) | High, n (%)
(n=13) | Low, n (%)
(n=49) | P-value |
|---|
| Sex |
|
|
| 0.775 |
|
Male | 36 | 8 (22.2) | 28 (77.8) |
|
|
Female | 26 | 5 (19.2) | 21 (80.8) |
|
| Age |
|
|
| 0.565 |
| <60
years | 29 | 7 (24.1) | 22 (75.9) |
|
| ≥60
years | 33 | 6 (18.2) | 27 (81.8) |
|
| TNM stage |
|
|
| 0.027a |
|
I/II | 40 | 5 (12.5) | 35 (87.5) |
|
|
III/IV | 22 | 8 (36.4) | 14 (63.6) |
|
| Infiltrate
depth |
|
|
| 0.027a |
|
Submucosal/superficial
layer | 15 | 0 (0)b | 15
(100.0) |
|
| Deep
muscle/outer layer | 47 | 13 (27.7) | 34 (72.3) |
|
| Lymphatic
invasion |
|
|
| 0.002a |
|
Positive | 21 | 9 (42.9) | 12 (57.1) |
|
|
Negative | 41 | 4 (9.8) | 37 (90.2) |
|
 | Table II.Association between WAVE3 mRNA
expression in serum and clinicopathologic parameters. |
Table II.
Association between WAVE3 mRNA
expression in serum and clinicopathologic parameters.
|
|
| WAVE3 mRNA
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathologic
parameter | Samples (n) | High | Low | P-value |
|---|
| Sex |
|
|
| 0.398 |
|
Male | 55 | 48 | 7 |
|
|
Female | 25 | 20 | 5 |
|
| Age, years |
|
|
| 0.803 |
|
<60 | 21 | 17 |
4b |
|
|
≥60 | 59 | 51 | 8 |
|
| TNM stage |
|
|
| 0.012a |
|
I/II | 40 | 21 | 19 |
|
|
III/IV | 28 | 23 | 5 |
|
| Lymphatic
invasion |
|
|
| 0.026a |
|
Yes | 22 | 19 |
3b |
|
| No | 45 | 25 | 20 |
|
Immunohistochemical (IHC)
staining
The patient tissue samples (paraffin sections of 4
µm fixed using 4% paraformaldehyde for 48 h at room temperature)
were used for IHC staining. For antigen retrieval, all paraffin
sections that had been deparaffinized in xylene for 15 min, were
rehydrated with a descending alcohol series (100, 95, 90, 75 and
70%) for 3 min each. stage, and treated with EDTA buffer in a
microwave for 10 min. Subsequently, the slides were treated with 3%
hydrogen peroxide to inhibit endogenous peroxidase activity and 10%
goat serum (Sangon Biotech Co., Ltd.) was used to block
non-specific binding sites for 30 min at room temperature. The
sections were incubated overnight at 4°C with the anti-WAVE3
primary antibody (1:400, cat. no. 2806S; Cell Signaling Technology,
Inc.), followed by incubation for 20 min at room temperature with a
HRP-conjugated goat anti-rabbit secondary antibody (1:100, cat. no.
A0208, Beyotime Institute of Biotechnology). Chromogen detection
was performed using 3,3′-diaminobenzidine. Following
counterstaining with hematoxylin for 3 min at room temperature,
dehydrated (descending alcohol series: 70, 75, 90, 95 and 100% for
1 min each, xylene for 15 min, all at room temperature), and
coverslipped with neutral gum,, sections were observed under a
light microscope (magnification, ×100) by two pathologists from the
Pathology Laboratory of the First Affiliated Hospital of Zhengzhou
University (Zhengzhou, China). The two had no knowledge of the
clinical data prior to evaluation were specialized in diagnosing of
ESCC. In the event of disagreement or difficulty, a third
pathologist was asked. Both intensity and extent were considered
using Image J (version 1.8.0; National Institutes of Health).
Intensity was scored from 0 to 3 and extent was scored from 0 to
100%. The score of intensity was defined: No positive staining=0,
light yellow=1, yellow-brown=2 and brown=3. For the area of
positive staining: No positive staining=0, 30% positive staining=1,
30–60% positive staining=2 and >60% positive staining=3. Final
quantitation of staining was calculated by multiplying the
intensity and extent scores. Sections with scores >1.5 were
considered as WAVE3-positive and sections were scores ≤1.5 were
considered as WAVE3-negative.
Cell culture and mimics
transfection
Human ESCC cell lines (EC109 and EC1) were obtained
from Basic Medical College of Zhengzhou University which had
purchased the cell lines from Shanghai Institute of Life Sciences
cell bank center (Shanghai, China). Cells were cultured in
RPMI-1640 medium (Sangon Biotech Co., Ltd.) supplemented with 10%
FBS (Sangon Biotech Co., Ltd.) and maintained at 37°C with 5%
CO2. Cells were passaged every 2 days to maintain
logarithmic growth until further analysis. The mature miRNA200b
sequence (access no. MIMAT0000318) and the putative miRNAs that
target WAVE3 was obtained from the miRBase database (version 22;
http://www.mirbase.org). miRNA200b mimic
(5′-UAAUACUGCCUGGUAAUGAUGA-3′) and mimic scrambled negative control
(access no. MIMAT0000295; 5′-UUUGUACUACACAAAAGUACUG-3′) were
synthesized by Guangzhou Ribobio Co., Ltd. Mimics and mimic
negative controls were diluted with riboFECT™ CPBuffer according to
the manufacturer's protocol (Guangzhou RiboBio Co., Ltd.) and
incubated for 15 min at room temperature. Subsequently, mimics and
mimic negative controls (final concentration 50 nM) were
transfected to the cells (4×105/well in a 6-well plate)
with riboFECT™ CPReagent according to the manufacturer's protocol
(Guangzhou Ribobio Co., Ltd.) at 37°C with 5% CO2 for 24
to 96 h. The control group received no treatment. After
transfection for 48 or 72 h, cells were collected for the later RNA
extraction and Transwell assay or western blotting.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from serum and transfected
cells with mimics and mimic negative controls using RNA isoBlood
reagent (Takara Biotechnology Co., Ltd.), according to the
manufacturer's protocol. Total RNA was reverse transcribed into
cDNA using the PrimeScript Reverse Transcription kit according to
the manufacturer's protocol (Takara Biotechnology Co., Ltd.).
Subsequently, qPCR was performed using the TB Green Premix Taq II
reagent kit according to the manufacturer's protocol (Takara
Biotechnology Co., Ltd.) to quantify WAVE3 mRNA in serum and
miRNA200b in serum and RNA extracted from transfected cells on a
LightCycler 480 II Real-Time PCR System (Roche Diagnostics). The
thermocycling conditions were: Pre-denaturation at 95°C 30 sec for
1 cycle, then 95°C for 5 sec, 60°C for 20 sec for 40 cycles and
followed by the melting curve and cooling stage at 65°C for 15 sec.
RNA levels were calculated using 2−ΔΔCq method (39). Patients with ESCC were classified
into two groups (high and low expression) according to the mean
value of serum WAVE3 expression in patients with ESCC. The
following primer pairs were used for qPCR: WAVE3 forward,
5′-GTGACGGTAGGAATGTGAGCA-3′ and reverse, 5′-CAAGCCCAACAGGTAGCCA-3′;
GAPDH forward, 5′-GAACGGGAAGCTCACTGG-3′ and reverse,
5′-GCCTGCTTCACCACCTTCT-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The primers of miRNA200b
were synthesized by Guangzhou Ribobio Co., Ltd. And the sequences
are proprietary. mRNA and miRNA expression levels were normalized
to the internal reference genes GAPDH and U6, respectively.
Western blot analysis
Total protein was extracted from cells transfected
with mimics and mimic negative controls using RIPA lysis buffer
(Sangon Biotech Co., Ltd.) and quantified using a bicinchoninic
acid protein assay kit (Sangon Biotech Co., Ltd.), according to the
manufacturer's protocol. Subsequently, proteins (25 µg/lane) were
separated via 12% SDS-PAGE and transferred onto PDVF membranes.
After blocking with 5% non-fat dry milk for 2 h at room
temperature, the membranes were incubated with the anti-WAVE3
primary antibody (1:1,000, cat. no. 2806S; Cell Signaling
Technology, Danvers, Inc.) and anti-GAPDH primary antibody
(1:6,000, cat. no. G8795; Sigma-Aldrich; Merck KGaA), overnight at
4°C. Subsequently, the membranes were washed with TBST and
incubated with the HRP-conjugated goat anti-rabbit secondary
antibody (1:1,000, A0208, Beyotime Institute of Biotechnology) for
2 h at room temperature. Protein band images were visualized with
an ECL kit (Sangon Biotech Co., Ltd.) and captured using a
chemiluminescence imaging system (EMD Millipore) and then analyzed
with ImageJ (version 1.8.0; National Institutes of Health). GAPDH
was used as the loading control.
Cell migration assay
Transwell plates (24-well; pore diameter, 0.8 µm)
were used to determine the cell migration ability. miRNA200b mimic
transfected cells (5×105 cells/ml) were plated into the
top chambers of the Transwell plates with RPMI-1640 medium.
RPMI-1640 medium containing 20% goat serum was plated into the
lower chambers as a chemoattractant. The Transwell plates were
incubated for 36 h at 37°C with 5% CO2. Subsequently,
non-migrated cells on the upper surface of the Transwell membrane
were removed using a cotton swab. After fixing with 4%
paraformaldehyde for 10 min at room temperature, migrating cells
were stained with gentian violet for 30 min at room temperature.
Stained cells were visualized under a light microscope
(magnification, ×100).
Statistical analysis
Statistical analyses were performed using SPSS
(version 21.0; IBM Corp.), GraphPad Prism (version 6; GraphPad
Software, Inc.) and Image J (version 1.8.0; National Institutes of
Health) software. Receiver operating characteristic curves were
constructed and the area under the curve (AUC) was analyzed to
evaluate the diagnostic value of serum WAVE3 expression levels in
ESCC. Normally distributed data are presented as the mean ±
standard deviation and non-normally distributed data are expressed
as the median and interquartile range. WAVE3 expression levels
between paired tumor and corresponding adjacent normal tissues was
compared using the paired Student's t-test. Data containing two
independent groups were compared using an unpaired Student's
t-test. Data containing >2 groups were compared using one-way
ANOVA followed by the LSD post hoc test. Pearson's χ2
test was used to analyze the association between WAVE3 expression
and clinicopathologic characteristics; however, when the number of
cases was <5, continuity correction and Fisher's Exact Test were
used. P<0.05 was considered to indicate a statistically
significant difference.
Results
IHC analysis of the expression of
WAVE3 protein in ESCC, and its association with clinical
parameters
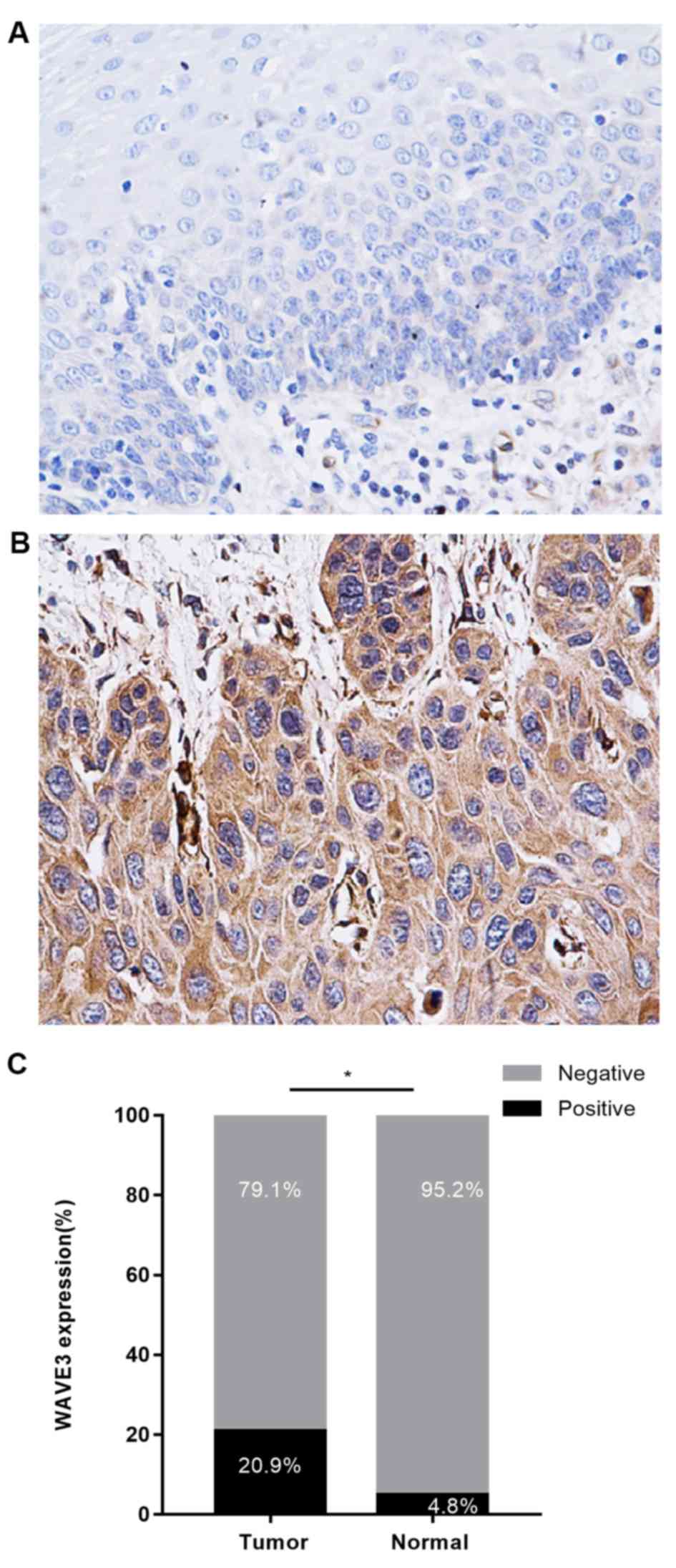
According to the immunohistochemical staining assay,
WAVE3 expression was primarily localized in the cytoplasm of ESCC
cells, and the expression of WAVE3 was increased in ESCC tissues
compared with normal tissues (Fig. 1A
and B). WAVE3-positive staining in ESCC tissues (20.9%) was
significantly higher compared with paired normal tissues (4.8%;
P<0.05; Fig. 1C). The present
study also assessed the association between WAVE3 protein
expression and clinicopathologic variables of patients with ESCC.
Increased WAVE3 expression was associated with TNM stage (P=0.027),
infiltrate depth (P=0.027) and lymphatic invasion (P=0.002)
(Table I). However, no significant
association was identified between WAVE3 expression and patient sex
or age (P>0.05; Table I).
WAVE3 mRNA expression in serum and its
association with clinical parameters
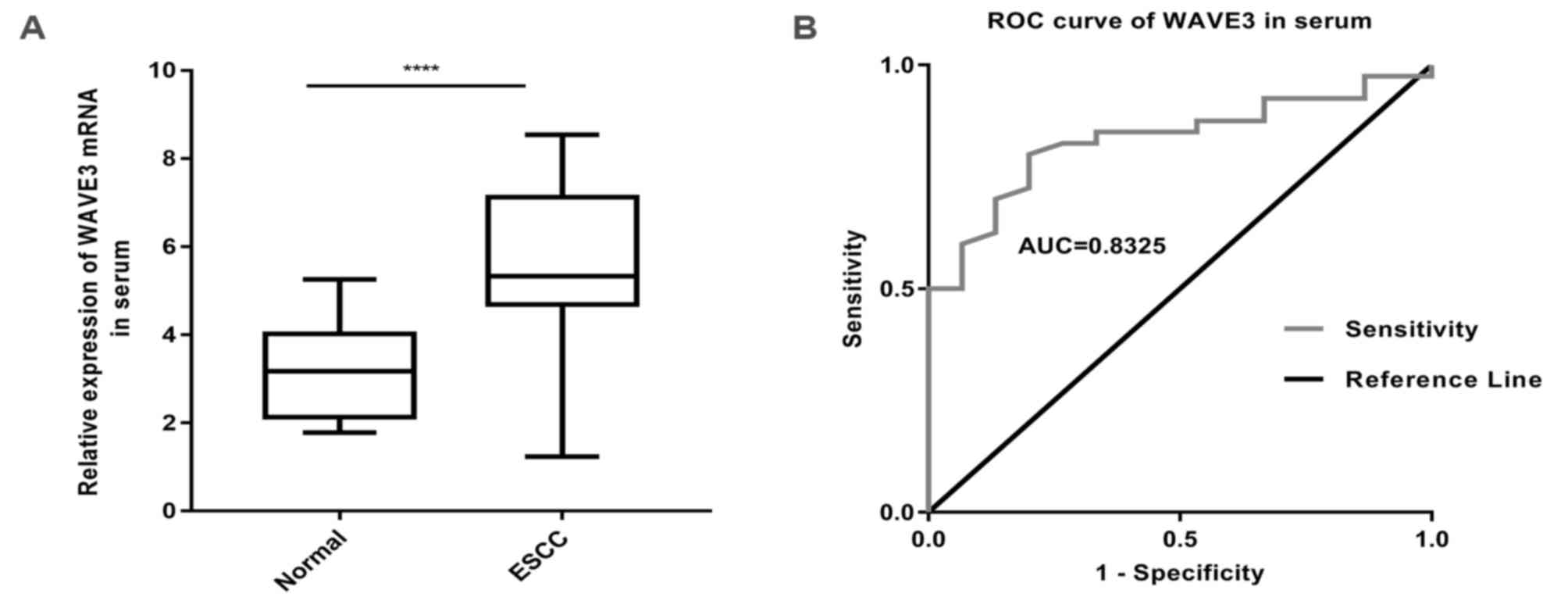
WAVE3 mRNA expression levels in the serum of
patients with ESCC were determined via RT-qPCR (Fig. 2A). The results indicated that WAVE3
expression was upregulated in serum of patients with ESCC compared
with healthy donors [5.3250; interquartile range (IQR),
4.6825–3.1225 vs. 3.1800; IQR, 2.1300–3.0200], which supported the
results of the IHC staining assay. As shown in Fig. 2B, the AUC of WAVE3 was 0.8325 [95%
confidence interval (CI); 0.7249–3.9401], which indicated that
WAVE3 exhibited a good diagnostic performance. Subsequently, the
association between the WAVE3 mRNA expression levels in serum and
clinicopathological features was assessed. High expression of WAVE3
mRNA was associated with TNM stage (P<0.05) and lymphatic
invasion (P<0.05) (Table II).
However, there was no significant association between serum WAVE3
expression and patient sex or age (P>0.05; Table II).
miRNA200b expression levels in serum
and its association with WAVE3
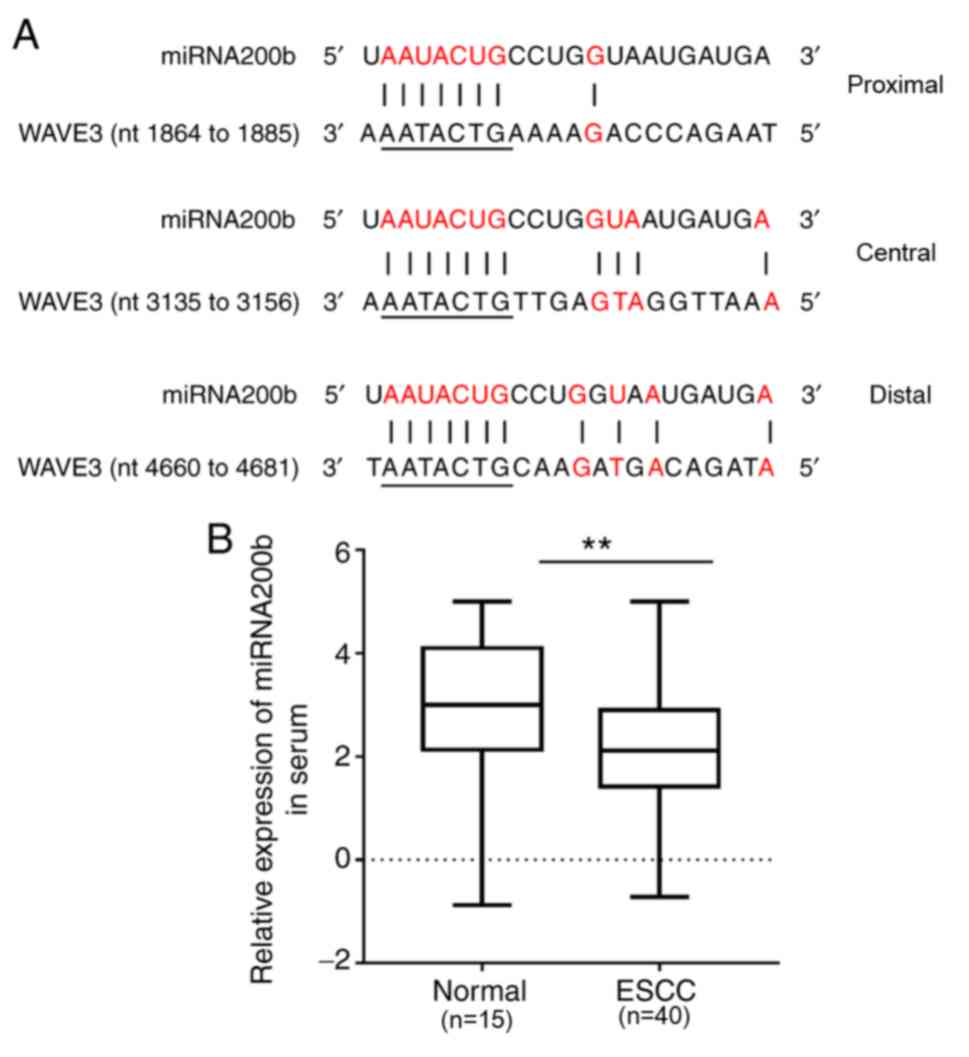
MiRBase was also used to search the putative miRNAs
that target WAVE3 (40,41), and it was indicated that miRNA200b
had three potential binding sites in the 3′-UTR of WAVE mRNA
(Fig. 3A). miRNA200b expression
levels were examined via RT-qPCR and the results suggested that the
expression of miRNA200b in ESCC serum was significantly decreased
compared with normal serum (P<0.0001; Fig. 3B). Together with the increased
level of WAVE3 in ESCC serum compared with the normal group, it was
hypothesized that miRNA200b expression is negatively associated
with WAVE3 expression.
WAVE3 expression is inhibited by
miRNA200b in ESCC cell lines
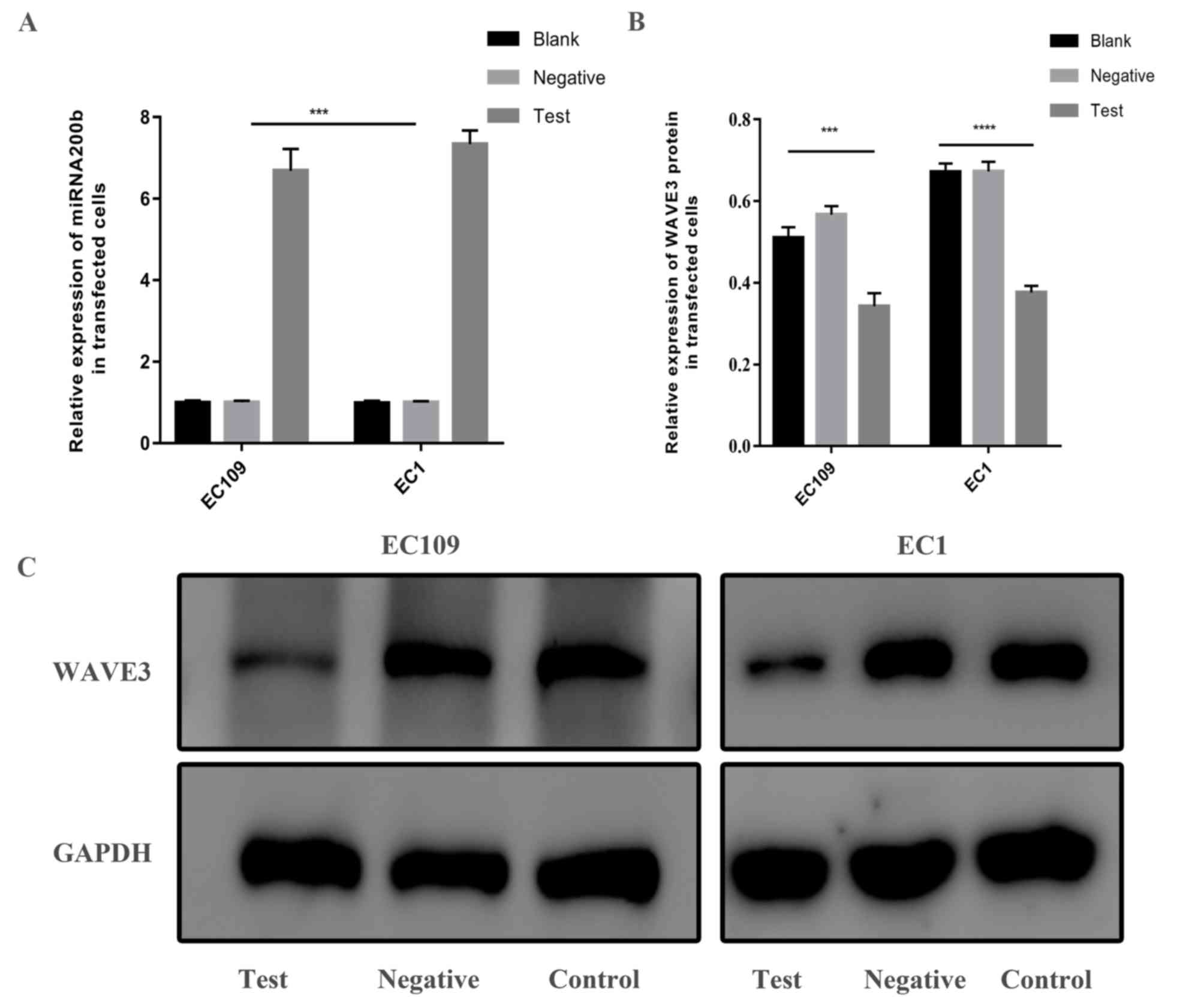
To investigate whether miRNA200b alters the
expression of WAVE3, EC109 and EC1 cell lines were transfected with
miRNA200b mimics. Subsequently, RT-qPCR was performed to analyze
the expression of miRNA200b and western blot analysis was used to
detect the expression of WAVE3 protein. Compared with the blank
control group and the mimic negative control group, the expression
of miRNA200b in the miRNA200b mimic group was increased (Fig. 4A) and the protein expression levels
of WAVE3 were decreased (Fig. 4B and
C; P<0.001). The results suggested that WAVE3 expression may
be inhibited by miRNA200b.
Effects of WAVE3 inhibition by
miRNA200b on cell migration
It has been previously reported that WAVE3 is
associated with cell migration, and displays a negative association
with the level of miRNA200b expression (31). Therefore, it was hypothesized that
high levels of miRNA200b expression may inhibit cell migration
(32). A Transwell assay was
performed to investigate this hypothesis. Compared with the blank
control group and the mimic negative control group, cell migration
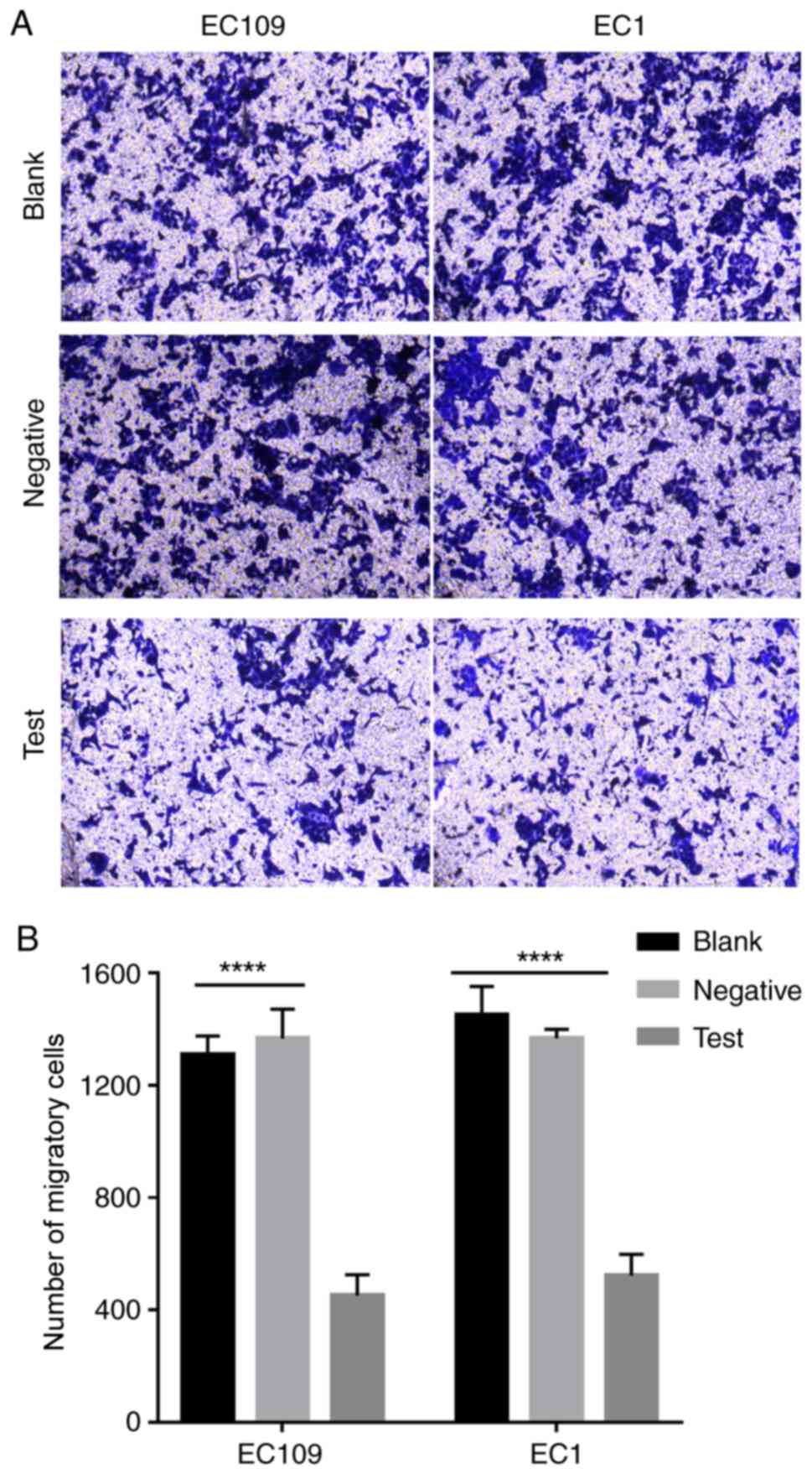
was decreased in the miRNA200b mimic group (Fig. 5A and B; P<0.01). Therefore, the
results suggest that miRNA200b inhibition of WAVE3 expression
decreases ESCC cell migration.
Discussion
A number of studies have indicated the significance
of WAVE3 in numerous types of cancer, including pancreatic, breast,
prostate, gastric, ovarian and bladder cancer (25,28,29,35,42,43).
WAVE3 has been reported to participate in a variety of signaling
pathways, including NF-κB signaling pathway, p38 pathway, PI3K
pathway and AKT pathway, displaying a regulatory role in cellular
processes, including proliferation and migration (20,21,24,28,35,43–45).
In the present study, increased WAVE3 expression was observed in
ESCC tissues and patient serum compared with the normal controls.
The results of the immunohistochemistry assay indicated that WAVE3
was primarily expressed in the cytoplasm, and expression in cancer
tissues was higher compared with non-cancerous tissues. The results
were consistent with previous studies, which also reported abnormal
expression of WAVE3 in tumors, such as breast, prostate, ovarian
and bladder cancer, indicating that WAVE3 expression may be
associated with the development of cancer (28,42,43,46).
A number of studies have indicated the role of WAVE3 in cellular
processes, including motility, migration, invasion and
proliferation, in a number of types of cancer such as
hepatocellular carcinoma, prostate cancer and colorectal cancer
(22,29,30).
The aforementioned studies indicate that the effect of WAVE3 on
cell migration occurs at the gene level and is not associated with
alterations to the number of cells.
Although the diagnosis and treatment of EC has
significantly improved in recent years, the 5-year survival rate
remains low, and infiltration and metastasis are still the primary
causes of EC-associated mortality (47). Sossey-Alaoui et al (20) indicated the upregulated WAVE3
expression was associated with poor prognosis in patients with
breast cancer. Further investigation indicated that the expression
of WAVE3 is closely associated with overall survival, survival rate
after recurrence, mortality of patients with breast cancer,
metastasis and progression (45).
WAVE3 upregulation is also associated with lymph node metastasis,
differentiation of pancreatic cancer cells and prognosis in
patients with pancreatic cancer (25). Furthermore, WAVE3 is highly
expressed in tissues of patients with colorectal cancer (30). However, in the analysis of
prognostic factors, patients without lymph node metastasis or
distant metastasis of other organs displayed increased WAVE3
expression levels compared with patients with poor prognosis, which
is contrary to the results reported in other studies investigating
breast, pancreatic and prostate cancer (25,28,46).
According to the analysis of ESCC clinical data conducted in the
present study, positive WAVE3 expression was higher in patients at
TNM stage III/IV (36.4%) compared with TNM stage I/II (12.5%). The
positive WAVE3 expression rate in deep muscle layer and outer layer
infiltration (27.7%) was also higher compared with the submucosa
and superficial layer (0%). Additionally, WAVE3 upregulation
displayed a positive association with lymph node metastasis. The
results of the present study indicated that the abnormal expression
of WAVE3 was associated with tumor stage, infiltration depth and
lymphatic invasion. Yue et al (35) reported high expression of WAVE3
mRNA in the serum of patients with gastric cancer, and revealed
that high WAVE3 expression was closely associated with lymph node
metastasis, depth of tumor invasion and TNM stage of gastric
cancer. Due to missing of patient clinical data on tumor invasion
depth of serum samples, the association between serum levels of
WAVE3 and depth of invasion were not analyzed in the present study.
However, the expression of WAVE3 in serum was also positively
associated with TNM stage and lymphatic invasion. The results of
the present study are consistent with the previous studies that
investigated breast, prostate, bladder and ovarian cancer (37,42,43).
According to the aforementioned data, WAVE3 may display an
important role in the aggressive progression of ESCC, and may be
associated with poor prognosis.
Previous studies have demonstrated that the
expression of the miRNA200 family is downregulated in a number of
different types of cancer, including breast, gastric and bladder
cancer (28,32,35,43).
The present study indicated that miRNA200b was downregulated in the
serum of patients with ESCC compared with healthy subjects.
Considering the higher serum level of WAVE3 in patients with ESCC
compared with healthy controls, it was hypothesized that miRNA200
expression was negatively associated with WAVE3 expression in
patients with ESCC. The results are consistent with a previous
study that indicated that WAVE3 is a direct target of miRNA200b,
using a luciferase reporter assay (32). The ESCC cell lines EC109 and EC1
were transfected with a miRNA200b mimic, and the expression of
WAVE3 protein and miRNA200b were determined. The results suggested
that higher levels of miRNA200b and lower levels of WAVE3 protein
were observed in the miRNA200b mimic group compared with the
negative control group, which indicated that exogenous miRNA200b
inhibits the expression of WAVE3 protein in ESCC cells. Therefore,
it was hypothesized that miRNA200b downregulation may contribute to
the upregulation of WAVE3 in ESCC cells; however, the underlying
mechanisms require further investigation.
Previous studies have demonstrated that the
expression of WAVE3 is positively correlated with tumor metastasis,
such as breast, pancreatic, prostate and colon cancer, and that
miRNA200b may inhibit the expression of WAVE3 (20,25,32,46).
In the present study, cells transfected with miRNA200b mimic were
used to analyze cell migration. According to the Transwell assay,
the number of migratory cells was significantly reduced at 48 h in
the miRNA200b mimic group compared with the negative control group,
which indicated that cell migration decreased when WAVE3 expression
was downregulated. WAVE3 may serve as a novel diagnostic biomarker
and therapeutic target for ESCC; however, the underlying mechanisms
require further investigation. By investigating WAVE3, its altered
expression in digestive system tumors and its association with
microRNAs, knowledge of the role of WAVE3 in tumor cell migration
may be improved.
Although the diagnostic and therapeutic strategies
for malignant tumors have improved in recent years, the association
between tumors and clinicopathological parameters has not yet been
determined (6,7,9). At
present, the migration mechanism mediated by WAVE3 has not been
reported. An increasing number of studies have demonstrated that
WAVE3 protein expression is associated with the development and
progression of tumors (27,45,48).
To the best of our knowledge, the present study is the first to
investigate the role of WAVE3 in ESCC. Further research is required
to support the use of WAVE3 as a biological indicator of tumor
diagnosis and prognosis. Moreover, future studies assessing WAVE3
expression in ESCC may support the use of WAVE3 as a potential
therapeutic target to reduce metastasis, improve the survival rate
of patients with cancer and decrease the rate of cancer
recurrence.
In conclusion, the present study indicated that
WAVE3 expression was upregulated in ESCC tissues and serum, and
increased levels of WAVE3 were associated with poor prognosis.
Furthermore, the expression of WAVE3 was negatively associated with
miRNA200b expression. miRNA200b overexpression decreased the
expression of WAVE3 protein, thereby inhibiting the migration of
ESCC. Therefore, the present study provided a novel insight into
ESCC progression, and identified a potential diagnostic biomarker
and therapeutic target for ESCC.
Acknowledgements
The authors would like to thank Professor Chunfeng
Ren of the First Affiliated Hospital of Zhengzhou University, for
providing laboratory support.
Funding
The present study was supported by the Henan
Provincial Department of Education Key Science and Technology
Project (grant no. 18A320007), the Ministry of Health and Welfare
Committee (grant no. SBGJ2018013) and the Key Medical Science and
Technology Project of Health Department (grant no. 201503008).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL and JG performed the experiments. XL wrote the
manuscript, and analyzed and interpreted the data. ZR and CX
participated in performing the experiments and analysis of the
data. YL designed the study and participated the revision of the
manuscript. As the corresponding author, HL participated in the
design of the research, and was responsible for the accuracy of all
aspects of the work and the final decision to submit the article
for publication. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institute
Research Ethics Committee of the First Affiliated Hospital of
Zhengzhou University (permit no. SS-2019-060). All patients
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liang H, Fan JH and Qiao YL: Epidemiology,
etiology, and prevention of esophageal squamous cell carcinoma in
China. Cancer Biol Med. 14:33–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Malhotra GK, Yanala U, Ravipati A, Follet
M, Vijayakumar M and Are C: Global trends in esophageal cancer. J
Surg Oncol. 115:564–579. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arnold M, Soerjomataram I, Ferlay J and
Forman D: Global incidence of oesophageal cancer by histological
subtype in 2012. Gut. 64:381–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen J, Kwong DL, Cao T, Hu Q, Zhang L,
Ming X, Chen J, Fu L and Guan X: Esophageal squamous cell carcinoma
(ESCC): Advance in genomics and molecular genetics. Dis Esophagus.
28:84–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsukada Y, Higashi T, Shimada H, Kikuchi Y
and Terahara A: The use of neoadjuvant therapy for resectable
locally advanced thoracic esophageal squamous cell carcinoma in an
analysis of 5016 patients from 305 designated cancer care hospitals
in Japan. Int J Clin Oncol. 23:81–91. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang L, Ma J, Han Y, Liu J, Zhou W, Hong
L and Fan D: Targeted therapy in esophageal cancer. Expert Rev
Gastroenterol Hepatol. 10:595–604. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Katada C, Muto M, Momma K, Arima M, Tajiri
H, Kanamaru C, Ooyanagi H, Endo H, Michida T, Hasuike N, et al:
Clinical outcome after endoscopic mucosal resection for esophageal
squamous cell carcinoma invading the muscularis mucosae-a
multicenter retrospective cohort study. Endoscopy. 39:779–783.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin D, Ma L, Ye T, Pan Y, Shao L, Song Z,
Jiang S, Chen H and Xiang J: Results of neoadjuvant therapy
followed by esophagectomy for patients with locally advanced
thoracic esophageal squamous cell carcinoma. J Thorac Dis.
9:318–326. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fink U, Stein HJ, Bochtler H, Roder JD,
Wilke HJ and Siewert JR: Neoadjuvant therapy for squamous cell
esophageal carcinoma. Ann Oncol. 5 (Suppl 3):S17–S26. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Heise JW, Heep H, Frieling T, Sarbia M,
Hartmann KA and Röher HD: Expense and benefit of neoadjuvant
treatment in squamous cell carcinoma of the esophagus. BMC Cancer.
1:202001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu SY, Liu Z, Ma WJ, Sheyhidin I, Zheng ST
and Lu XM: New potential biomarkers in the diagnosis of esophageal
squamous cell carcinoma. Biomarkers. 14:340–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lo YM, Tein MS, Lau TK, Haines CJ, Leung
TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM and Hjelm NM:
Quantitative analysis of fetal DNA in maternal plasma and serum:
Implications for noninvasive prenatal diagnosis. Am J Hum Genet.
62:768–775. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thrasher AJ and Burns SO: WASP: A key
immunological multitasker. Nat Rev Immunol. 10:182–192. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takenawa T and Suetsugu S: The WASP-WAVE
protein network: Connecting the membrane to the cytoskeleton. Nat
Rev Mol Cell Biol. 8:37–48. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Machesky LM and Insall RH: Scar1 and the
related Wiskott-Aldrich syndrome protein, WASP, regulate the actin
cytoskeleton through the Arp2/3 complex. Curr Biol. 8:1347–1356.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Imai K, Morio T, Zhu Y, Jin Y, Itoh S,
Kajiwara M, Yata J, Mizutani S, Ochs HD and Nonoyama S: Clinical
course of patients with WASP gene mutations. Blood. 103:456–464.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Miki H, Suetsugu S and Takenawa T: WAVE, a
novel WASP-family protein involved in actin reorganization induced
by Rac. EMBO J. 17:6932–6941. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sossey-Alaoui K, Ranalli TA, Li X, Bakin
AV and Cowell JK: WAVE3 promotes cell motility and invasion through
the regulation of MMP-1, MMP-3, and MMP-9 expression. Exp Cell Res.
308:135–145. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sossey-Alaoui K, Li X, Ranalli TA and
Cowell JK: WAVE3-mediated cell migration and lamellipodia formation
are regulated downstream of phosphatidylinositol 3-kinase. J Biol
Chem. 280:21748–21755. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu Z, Chen W, Yin X, Lai J, Wang Q, Liang
L, Wang W, Wang A and Zheng C: WAVE3 induces EMT and promotes
migration and invasion in intrahepatic cholangiocarcinoma. Dig Dis
Sci. 61:1950–1960. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ji Y, Li B, Zhu Z, Guo X, He W, Fan Z and
Zhang W: Overexpression of WAVE3 promotes tumor invasiveness and
confers an unfavorable prognosis in human hepatocellular carcinoma.
Biomed Pharmacother. 69:409–415. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Davuluri G, Sossey-Alaoui K and Plow E:
Abstract 2702: WAVE3 regulates NF B signaling and sensitizes cancer
cells to apoptosis and cell death driven by TNF. Cancer Res. 73
(Suppl 8):27022013.
|
|
24
|
Davuluri G, Augoff K, Schiemann WP, Plow
EF and Sossey-Alaoui K: WAVE3-NFκB interplay is essential for the
survival and invasion of cancer cells. PLoS One. 9:e1106272014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang S, Huang C, Chen W, Liu Y, Yin X,
Lai J, Liang L, Wang Q, Wang A and Zheng C: WAVE3 promotes
proliferation, migration and invasion via the AKT pathway in
pancreatic cancer. Int J Oncol. 53:672–684. 2018.PubMed/NCBI
|
|
26
|
Jaganathan S, Yue P, Paladino DC,
Bogdanovic J, Huo Q and Turkson J: A functional nuclear epidermal
growth factor receptor, SRC and Stat3 heteromeric complex in
pancreatic cancer cells. PLoS One. 6:e196052011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kulkarni S, Augoff K, Rivera L, McCue B,
Khoury T, Groman A, Zhang L, Tian L and Sossey-Alaoui K: Increased
expression levels of WAVE3 are associated with the progression and
metastasis of triple negative breast cancer. PLoS One.
7:e428952012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sossey-Alaoui K, Safina A, Li X, Vaughan
MM, Hicks DG, Bakin AV and Cowell JK: Down-regulation of WAVE3, a
metastasis promoter gene, inhibits invasion and metastasis of
breast cancer cells. Am J Pathol. 170:2112–2121. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moazzam M, Ye L, Sun PH, Kynaston H and
Jiang WG: Knockdown of WAVE3 impairs HGF induced migration and
invasion of prostate cancer cells. Cancer Cell Int. 15:512015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Paterson EL, Kazenwadel J, Bert AG,
Khew-Goodall Y, Ruszkiewicz A and Goodall GJ: Down-regulation of
the miRNA-200 family at the invasive front of colorectal cancers
with degraded basement membrane indicates EMT is involved in cancer
progression. Neoplasia. 15:180–191. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sossey-Alaoui K, Bialkowska K and Plow EF:
The miR200 family of microRNAs regulates WAVE3-dependent cancer
cell invasion. J Biol Chem. 284:33019–33029. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Honor H, Ackland ML, Blick T, Lawrence MG,
Clements JA, Williams ED and Thompson EW: Epithelial-mesenchymal
and mesenchymal-epithelial transitions in carcinoma progression. J
Cell Physiol. 213:374–383. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Taylor MA, Davuluri G, Parvani JG,
Schiemann BJ, Wendt MK, Plow EF, Schiemann WP and Sossey-Alaoui K:
Upregulated WAVE3 expression is essential for TGF-β-mediated EMT
and metastasis of triple-negative breast cancer cells. Breast
Cancer Res Treat. 142:341–353. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yue Z, Feng W, Xiangke L, Liuxing W,
Qingxia F and Jianbo G: WAVE3 promotes epithelial-mesenchymal
transition of gastric cancer through upregulation of Snail. Cancer
Gene Ther. 21:499–506. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Savagner P: Leaving the neighborhood:
Molecular mechanisms involved during epithelial-mesenchymal
transition. Bioessays. 23:912–923. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen Y, Sun Y, Chen L, Xu X, Zhang X, Wang
B, Min L and Liu W: miRNA-200c increases the sensitivity of breast
cancer cells to doxorubicin through the suppression of
E-cadherin-mediated PTEN/Akt signaling. Mol Med Rep. 7:1579–1584.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cowherd SM: Tumor staging and grading: A
primer. Methods Mol Biol. 823:1–18. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lu J, Wang SL, Wang YC, Wu YN, Yu X, Zhao
WZ and Wang JH: High WAVE3 expression correlates with
proliferation, migration and invasion in human ovarian cancer.
Oncotarget. 8:41189–41201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jin H, Xie Q, Guo X, Xu J, Wang A, Li J,
Zhu J, Wu XR, Huang H and Huang C: p63α protein up-regulates heat
shock protein 70 expression via E2F1 transcription factor 1,
promoting Wasf3/Wave3/MMP9 signaling and bladder cancer invasion. J
Biol Chem. 292:15952–15962. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sossey-Alaoui K, Li X and Cowell JK:
c-Abl-mediated phosphorylation of WAVE3 is required for
lamellipodia formation and cell migration. J Biol Chem.
282:26257–26265. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sossey-Alaoui K: Surfing the big WAVE:
Insights into the role of WAVE3 as a driving force in cancer
progression and metastasis. Semin Cell Dev Biol. 24:287–297.
2013.PubMed/NCBI
|
|
46
|
Fernando HS, Sanders AJ, Kynaston HG and
Jiang WG: WAVE3 is associated with invasiveness in prostate cancer
cells. Urol Oncol. 28:320–327. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Condeelis J and Segall JE: Intravital
imaging of cell movement in tumours. Nat Rev Cancer. 3:921–930.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Takenawa T and Miki H: WASP and WAVE
family proteins: Key molecules for rapid rearrangement of cortical
actin filaments and cell movement. J Cell Sci. 114:1801–1809.
2001.PubMed/NCBI
|